Abstract
The molecular basis of different outcomes in pediatric acute lymphoblastic leukemia (ALL) remains poorly understood. We addressed the clinical significance and mechanisms behind in vitro cellular responses to ionizing radiation (IR)–induced DNA double-strand breaks in 74 pediatric patients with ALL. We found an apoptosis-resistant response in 36% of patients characterized by failure to cleave caspase-3, -7, -9, and PARP1 by 24 hours after IR and an apoptosis-sensitive response with the cleavage of the same substrates in the remaining 64% of leukemias. Resistance to IR in vitro was associated with poor early blast clearance at day 7 or 15 and persistent minimal residual disease (MRD) at day 28 of induction treatment. Global gene expression profiling revealed abnormal up-regulation of multiple prosurvival pathways in response to IR in apoptosis-resistant leukemias and differential posttranscriptional activation of the PI3-Akt pathway was observed in representative resistant cases. Importantly, pharmacologic inhibition of selected prosurvival pathways sensitized apoptosis-resistant ALL cells to IR in vitro. We suggest that abnormal prosurvival responses to DNA damage provide one of the mechanisms of primary resistance in ALL, and that they should be considered as therapeutic targets in children with aggressive disease.
Introduction
Despite considerable progress in cure rates in the past 2 decades, 20% of children with B-precursor acute lymphoblastic leukemia (ALL) still experience disease progression with current treatment, and ALL remains the leading cause of cancer death in children between 1 and 15 years of age.1
Current treatment relies on the stratification of ALL based on known prognostic markers, which include both clinical and genetic features.2 Reduced rates of early blast clearance during induction therapy represent an important indicator of high-risk ALL.3-5 Furthermore, monitoring of minimal residual disease (MRD) has refined the identification of high-risk disease and provided a 100-fold increase in sensitivity of blast detection compared with morphologic assessment.6 Despite these dramatic improvements in the ability to detect and predict aggressive ALL, there remains a lack of understanding of the molecular defects behind reduced treatment-induced blast killing and disease progression.
In the past decade, gene expression array studies have facilitated the creation of molecular taxonomies and increased understanding of treatment responses and drug resistance in ALL. Specific gene signatures have been assigned to subsets of ALL with resistance to individual or combined cytotoxic treatments.7-12 In addition, gene signatures associated with persistence of MRD at different points throughout the treatment have been identified.13,14 These studies provide valuable information regarding prediction of treatment outcome, but their role in defining new therapeutic targets remains limited. The mechanisms by which most currently used individual or combined chemotherapeutics exert their cytotoxic effect are complex, and it is difficult to use responses to these compounds in vitro and in vivo to evaluate the integrity of specific cellular pathways in leukemic cells.
Induction of DNA double-strand breaks (DSBs) is a common end effect of many chemotherapeutic agents used in ALL treatment, and contributes to their mechanism of cytotoxicity.15 The principal form of DNA damage caused by ionizing radiation (IR) is also the induction of DNA DSBs, thus providing a reproducible model system with which to address the integrity of defined and critical cellular responses. We have previously shown that in leukemic cells, DNA DSBs initiate a cascade of cellular responses that activate both proapoptotic and prosurvival signals.16 The ability of a tumor cell to undergo apoptosis depends on the level of DNA DSBs and integrity of DSB response pathways. Thus, if the function of damage response pathways in ALL cells is intact, a high level of IR-induced DSBs would be expected to facilitate p53-driven proapoptotic responses and override prosurvival signals.17 Consequently, the fidelity of DSB response pathways is an important factor preventing both occurrence and progression of lymphoid tumors, and it is conceivable that compromised integrity of these responses might be an important determinant of ALL chemosensitivity.
We have previously reported that a subset of childhood ALL exhibits defective induction of apoptosis following IR, despite normal up-regulation of p53.18 We have also shown that defects in the DSB response pathway in chronic lymphocytic leukemia (CLL) cause both chemoresistance and shorter patient survival.19,20 It is conceivable that altered response to DSBs in ALL could also influence clinical responses.
In this study, we have considered the clinical relevance and molecular mechanisms behind defective responses to IR-induced DSBs in ALL. We explored the relationship between responses to IR in vitro and early blast clearance and the level of MRD following induction therapy in patients with ALL. We show that DSB response profiles in vitro reflect the early response to treatment as measured at a morphologic and molecular level. To further advance our understanding of resistance to DSB-induced apoptosis, we compared the IR-induced global gene expression profiles in apoptosis-sensitive and apoptosis-resistant ALL tumors and observed an association between the apoptosis-resistant ALL phenotype and differential induction of defined prosurvival pathways in response to IR. Finally, we show that prosurvival pathways are also differentially up-regulated at the posttranscriptional level, and that pharmacologic inhibition of these pathways can sensitize resistant ALL cells to IR-induced DSBs.
Methods
B-precursor ALL tumor samples
Diagnosis of B-precursor ALL was based on standard morphologic and immunophenotypic evaluation. Diagnostic bone marrow samples were collected from 74 children with ALL who were enrolled in UK protocols ALL97 and ALL2003 (www.ctsu.ox.ac.uk/projects/leuk/ukall2003/ and http://www.icnet.uk/trials/children/mrcall97_mar00.pdf). In both of these protocols, patients aged 1 to 18 years were initially stratified and treated according to 3 risk categories: high risk (hypodiploidy less than 45 chromosomes, and presence of either BCR-ABL or MLL rearrangement), intermediate risk (age 10 years and older and/or white cell count [WCC] greater than 50 × 109/L), and standard risk. Induction treatment included vincristine, dexamethasone, asparaginase, intrathecal methotrexate, and cytarabine, without daunorubicin (standard-risk patients) or with daunorubicin (intermediate- and high-risk patients). Patients were included in the study on the “intention to treat” basis. Standard- and intermediate-risk patients with poor blast clearance at days 15 and 8, respectively, were then reclassified as high risk. ALL2003 patients were also assessed for MRD by fluorescence-activated cell sorting (FACS) and molecular analysis at day 28,21,22 and consolidation/maintenance treatment was adjusted accordingly. Patients with bone marrow leukemic blasts equal to or greater than 25% at day 8 or 15 were classified as “poor early responders” (PERs), while those whose blasts were reduced below this threshold were classed as “good early responders” (GERs). Patients with 0.01% or more leukemic blasts identified by either FACS or molecular analysis at day 28 of induction treatment were classified as MRD+.
Ethical approval was obtained from South Birmingham Research Ethics Committee (REF 2003/043), and written consent was obtained from all patients before material was collected in accordance with the principles of the Declaration of Helsinki.
Cell culture and γ irradiation
Mononuclear cells were isolated from bone marrow aspirate using Ficoll-Paque (Amersham Biosciences, Piscataway, NJ), and cells were cultured in RPMI 1640. Aliquots of 2 × 106 cells per time point were irradiated with 5 Gy IR (cobalt Co60) and incubated at 37°C. Only samples with more than 90% blast cells were used for analysis.
Western blot analysis
Western blotting was performed as previously described23 using sheep polyclonal p53 antibody (donated by Prof David Lane, University of Dundee, Dundee, United Kingdom), rabbit polyclonal antibody against p21, goat polyclonal against PARP1, mouse monoclonal against PTEN and Bad (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit polyclonal antibodies against procaspase-7 and -9, cleaved caspase-3, phospho–serine 473 Akt, phospho–serine 112/136 Bad, rabbit monoclonal against phospho–serine 235/236 S6; mouse monoclonal antibody against S6 (Cell Signaling Technology, Beverly, MA); and rabbit polyclonal antibody against phospho–serine 2056 DNA PKCS (Abcam, Cambridge, United Kingdom). Mouse monoclonal antibody against β-actin (Sigma-Aldrich, St Louis, MO) was used as a loading control. Protein of interest–actin expression ratios were determined by densitometry for each condition and for each patient. IR-induced change in protein levels in relation to actin, of plus or minus 50%, was considered a significant response.
Phosphorylation of RTKs
Parallel determination of the relative levels of phosphorylation of 42 receptor tyrosine kinases (RTKs) was performed using the human Phospho RTK array system according to the manufacturer's instructions (R&D Systems, Abingdon, United Kingdom). The phosphorylation signals were compared by densitometry and expressed as the pixel density (OD/mm2). The pixel density of the background was subtracted from the pixel density of each spot, and the average of duplicate spots was determined. Fold change in density signal was calculated by dividing the pixel density for each spot for each ALL tumor after IR by the pixel density of the same spot in the same tumor before IR.
Immunofluorescence
Repair of IR-induced DSBs was assessed by quantification of γH2AX and MDC1 foci as previously described24 using mouse monoclonal antibody against γH2AX (Millipore, Watford, United Kingdom) and rabbit polyclonal antibody against MDC1.24 Primary ALLs were incubated for 1 hour at 37°C and then for a further hour with or without 30 μM LY294002. Subsequently, ALL cells were exposed to 2 Gy ionizing radiation and γH2AX and MDC1 foci assessed at 0, 0.5, and 4 hours after irradiation.
Annexin V/FACS analysis of IR-induced apoptosis following pharmacologic inhibition of survival pathways
Primary ALL cells were irradiated with 5 Gy and treated with 20 μM MEK inhibitor UO126 (Promega, Madison, WI), 500 nM mTOR inhibitor rapamycin (Calbiochem, San Diego, CA), 30 μM PI3K inhibitor LY294002 (Calbiochem), or 30 μM IGF-R1 tyrosine kinase and IGF1 inhibitor AG1024 (Calbiochem) at 37°C for 24 hours. Apoptosis was assayed using an Annexin V Apoptosis Kit (BD Pharmingen, San Diego, CA) according to the manufacturer's instructions and analyzed using a Coulter Epics XL-MCL flow cytometer (Beckman Coulter, Fullerton, CA). At least 30 000 events were scored for each condition. Experiments were performed in triplicate.
Gene expression profiling
A total of 22 B-precursor ALL tumors (11 responsive to IR-induced DNA damage and 11 resistant) were analyzed before and 8 hours after exposure to 5 Gy IR. RNA extraction, first- and second-strand cDNA synthesis, in vitro transcription, and hybridization to Affymetrix chips HG U133A (Affymetrix, Santa Clara, CA) were performed according to the standard Affymetrix protocol.16 Expression values were obtained for all 44 hybridizations using Affymetrix Microarray Suite (MAS) 5.0 software. Raw data were exported from MAS 5.0, and values were normalized to the median signal value for each array or for gene set enrichment analysis (GSEA) to the mean level of gene expression in the unirradiated partner sample. Differentially expressed probe sets were identified using significance analysis of microarrays (SAM).25-28 All microarray data has been deposited with Gene Expression Omnibus (GEO)29 under accession number GSE13280.
GSEA
GSEA is designed to test whether differentially expressed genes belong to specific functionally related gene sets (e.g., genes belonging to common pathways or responding to specific stimuli). GSEA is based on an enrichment score that reflects the level at which a functional set is overrepresented in a gene list that has been ranked by the degree of differential gene expression.30 We have applied this methodology in the software implementation available at the Molecular Signatures Database (MsigDB).31
To decrease the complexity of the data, normalized expression values for 22 215 probe sets were reduced to 17 249 probe sets by excluding genes with expression values less than 50 in all analyzed samples. Subsequently, multiple probe sets for a given gene were replaced by the probe set with the maximum expression value. This further lowered the dataset to 11 125 genes. GSEA analysis was then carried out to determine the enrichment score for a total of 276 pathways (with a minimum of 15 genes in each) in the MsigDB_vs.1 genesets database.31 Significance values (P values) were calculated by comparing the observed enrichment score with the set of scores computed using phenotype labels randomly assigned to samples over 1000 permutations. Adjustment for multiple hypothesis testing produced a false discovery rate (FDR) score as an indicator of the probability that an observed result represents a false positive finding. Significance was determined using a cutoff point of FDR less than 0.20.
Identification of differentially expressed pathway modules
To identify functional modules differentially regulated between apoptosis-sensitive and apoptosis-resistant tumors, initially the genes belonging to each given Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway32 were clustered according to their expression profile. A correlation threshold was then used to define distinct clusters within each functional pathway. The correlation threshold was defined as correspondent to a 5% FDR estimated from a randomized data set. The expression profile of a supergene was then constructed by averaging the profiles of the genes in each cluster. Only functional clusters containing at least 3 different genes were used to derive supergenes. Thus, the profile of the supergene represented the average behavior of a pathway module and significantly reduced variance and the complexity of the dataset.
Subsequently, the established expression profiles of supergenes were subjected to SAM analysis using a permutation strategy to estimate the FDR. We implemented this procedure in the statistical environment R.33
Statistical analysis
We tested for a relationship between in vitro sensitivity to IR and patients' per-protocol risk classification, age, sex, blast clearance, and MRD using the Fisher exact test,34 and with WCC using the Mann-Whitney U test.34 Differences in event-free survival (EFS) were analyzed by log-rank test and Kaplan-Meier survival curves.35 An event was defined as relapse or disease-related death without relapse. The Student t test was used to compare the percentage of cells with more than 5 γH2AX/MDC1 foci induced by IR in the sensitive and resistant tumors.34
Results
IR-induced cellular responses in vitro correlate with blast clearance in vivo
Using a previously devised classification based on the activation status of the intrinsic cascade of apoptotic responses,18 we first stratified the in vitro DNA damage responses of 74 primary ALL tumors into apoptosis-sensitive or -resistant phenotypes. Apoptosis-sensitive tumors revealed cleavage of procaspase-3, -7, -9, and PARP1 between 8 and 24 hours after IR, whereas apoptosis-resistant cases failed to do so. All tumors exhibited comparable increases in p53 and p21 levels after IR (Figure 1). We found that 47 (64%) of 74 of our cohort of pediatric ALL samples were apoptosis sensitive, whereas 27 (36%) of 74 were apoptosis resistant.
Profiling of in vitro IR response in ALL by Western blotting identifies apoptosis-sensitive and apoptosis-resistant tumors. Left panel shows a representative IR-sensitive ALL exhibiting gradual cleavage of PARP1 and disappearance of full-size PARP1 protein by 24 hours after 5 Gy IR. This is accompanied by up-regulation of p53 protein and its target p21 between 4 and 8 hours after IR, cleavage and disappearance of full-length proteins procaspase-9 and -7, and occurrence of the cleaved caspase-3 by 24 hours after IR. Right panel shows a representative IR-resistant ALL, exhibiting absence of cleavage of PARP1 and procaspase-9, -7, and -3 by 24 hours after 5 Gy IR despite abundant up-regulation of proteins p53 and p21 between 4 and 8 hours after IR. Expression of actin shows equal protein loading. The same Western blot for either representative IR-sensitive or IR-resistant ALL was reprobed with different antibodies multiple times. For each of the tumors, each horizontal group of lanes represents probing with a single antibody, and these individual probings are separated by black boxes.
Profiling of in vitro IR response in ALL by Western blotting identifies apoptosis-sensitive and apoptosis-resistant tumors. Left panel shows a representative IR-sensitive ALL exhibiting gradual cleavage of PARP1 and disappearance of full-size PARP1 protein by 24 hours after 5 Gy IR. This is accompanied by up-regulation of p53 protein and its target p21 between 4 and 8 hours after IR, cleavage and disappearance of full-length proteins procaspase-9 and -7, and occurrence of the cleaved caspase-3 by 24 hours after IR. Right panel shows a representative IR-resistant ALL, exhibiting absence of cleavage of PARP1 and procaspase-9, -7, and -3 by 24 hours after 5 Gy IR despite abundant up-regulation of proteins p53 and p21 between 4 and 8 hours after IR. Expression of actin shows equal protein loading. The same Western blot for either representative IR-sensitive or IR-resistant ALL was reprobed with different antibodies multiple times. For each of the tumors, each horizontal group of lanes represents probing with a single antibody, and these individual probings are separated by black boxes.
We observed a significant association with older age (P < .001), but no association between WCC or sex and apoptosis resistance. However, when individual prognostic features were combined according to the trial protocol risk stratification, a strong association with apoptotic sensitivity was noted both before and after early blast clearance assessment (P = .02 and .002, respectively).
We next assessed whether there was a correlation between the DNA DSB response in vitro and blast clearance from patients' bone marrow at days 8 and 15 of treatment induction. Notably, we observed that among 44 apoptosis-sensitive patients, 3 were classified as PERs (7%) with 25% or more blast cells present, whereas 8 (38%) of 21 patients with in vitro apoptosis-resistant ALL phenotype were classified as PERs. This difference was statistically significant (P = .005 by Fisher exact test; Table 1; Figure 2). The same trend was observed when blast clearance was analyzed separately on day 8 for intermediate-risk patients and on day 15 for low-risk patients (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Clinical features of pediatric B-precursor ALLs with 2 types of in vitro response to DNA DSBs
| Parameter . | Apoptosis-sensitive ALL (n = 47) . | Apoptosis-resistant ALL (n = 27) . | Difference between the 2 subgroups, P . |
|---|---|---|---|
| Clinical trial protocol, % (no.) | |||
| ALL97/99 | 38 (18/47) | 48 (13/27) | .468 |
| UKALL2003 | 62 (29/47) | 52 (14/27) | |
| Age at diagnosis ≥ 10 y | 9 (4/47) | 48 (13/27) | < .001 |
| WCC × 109, median (iqr) | 10.8 (6-49) | 10.6 (6-51) | .955 |
| Sex, % male (no.) | 55 (26/47) | 63 (17/27) | .627 |
| Initial risk classification, % (no.) | |||
| Low risk | 70 (33/47) | 44 (12/27) | .022 |
| Intermediate risk | 30 (14/47) | 52 (14/27) | |
| High risk | 0 (0/47) | 4 (1/27) | |
| Risk classification after early blast clearance, % (no.) | |||
| Low risk | 69 (31/45) | 41 (9/22) | .002 |
| Intermediate risk | 24 (11/45) | 18 (4/22) | |
| High risk | 7 (3/45) | 41 (9/227) | |
| Blast clearance at day 8/15, % (no.)* | 93 (41/44) GER 7 (3/44) PER | 62 (13/21) GER 38 (8/21) PER | .005 |
| Positive molecular MRD at day 28, % (no.)† | 19 (5/26) | 69 (9/13) | .004 |
| Positive FACS MRD at day 28, % (no.)† | 21 (8/38) | 59 (10/17) | .014 |
| Median follow-up, mo (range) | 40.0 (29.9-55.2) | 37.9 (22.8-53.1) | .475 |
| 3-y event-free survival, %‡ | 93.4 | 87.5 | .176 |
| Parameter . | Apoptosis-sensitive ALL (n = 47) . | Apoptosis-resistant ALL (n = 27) . | Difference between the 2 subgroups, P . |
|---|---|---|---|
| Clinical trial protocol, % (no.) | |||
| ALL97/99 | 38 (18/47) | 48 (13/27) | .468 |
| UKALL2003 | 62 (29/47) | 52 (14/27) | |
| Age at diagnosis ≥ 10 y | 9 (4/47) | 48 (13/27) | < .001 |
| WCC × 109, median (iqr) | 10.8 (6-49) | 10.6 (6-51) | .955 |
| Sex, % male (no.) | 55 (26/47) | 63 (17/27) | .627 |
| Initial risk classification, % (no.) | |||
| Low risk | 70 (33/47) | 44 (12/27) | .022 |
| Intermediate risk | 30 (14/47) | 52 (14/27) | |
| High risk | 0 (0/47) | 4 (1/27) | |
| Risk classification after early blast clearance, % (no.) | |||
| Low risk | 69 (31/45) | 41 (9/22) | .002 |
| Intermediate risk | 24 (11/45) | 18 (4/22) | |
| High risk | 7 (3/45) | 41 (9/227) | |
| Blast clearance at day 8/15, % (no.)* | 93 (41/44) GER 7 (3/44) PER | 62 (13/21) GER 38 (8/21) PER | .005 |
| Positive molecular MRD at day 28, % (no.)† | 19 (5/26) | 69 (9/13) | .004 |
| Positive FACS MRD at day 28, % (no.)† | 21 (8/38) | 59 (10/17) | .014 |
| Median follow-up, mo (range) | 40.0 (29.9-55.2) | 37.9 (22.8-53.1) | .475 |
| 3-y event-free survival, %‡ | 93.4 | 87.5 | .176 |
iqr indicates interquartile range; GER, good early response (ie, < 25% blasts at days 8/15 after start of induction therapy); and PER, poor early response (ie, ≥ 25% blasts at days 8/15). Significant P values are given in bold.
Assessment day depends on protocol—day 15 for initial low-risk patients, day 8 for intermediate-/high-risk patients.
Minimal residual disease as classified by both FACS and molecular analysis at day 28 after the start of induction therapy (positive, ≥ 0.01% leukemic blasts; negative, < 0.01% blasts).
Event indicates relapse or treatment-related death. A total of 3 relapses and 1 treatment-related death occurred in the apoptosis-sensitive group and 5 relapses in the apoptosis-resistant group.
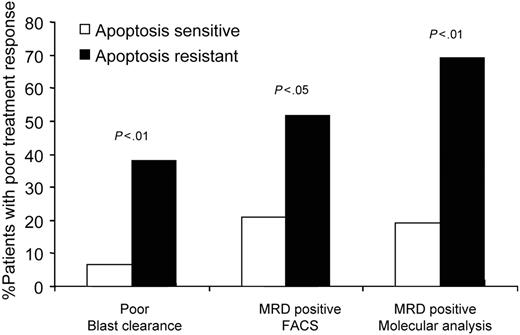
In vitro IR-induced response in primary ALL reflects clinical response. A total of 7% of patients with apoptosis-sensitive ALL are PERs exhibiting more than 25% of leukemic blasts in bone marrow on the days 8/15 of induction treatment. In contrast, 38% of apoptosis-resistant patients are PERs. A total of 21% of apoptosis-sensitive samples are MRD+ (0.01% or more blasts in bone marrow on day 28 of induction treatment) by FACS analysis. In comparison, 59% of patients with apoptosis-resistant ALL are MRD+. A total of 19% of apoptosis-sensitive patients and 69% of apoptosis-resistant patients exhibit MRD positivity as measured by molecular MRD analysis.
In vitro IR-induced response in primary ALL reflects clinical response. A total of 7% of patients with apoptosis-sensitive ALL are PERs exhibiting more than 25% of leukemic blasts in bone marrow on the days 8/15 of induction treatment. In contrast, 38% of apoptosis-resistant patients are PERs. A total of 21% of apoptosis-sensitive samples are MRD+ (0.01% or more blasts in bone marrow on day 28 of induction treatment) by FACS analysis. In comparison, 59% of patients with apoptosis-resistant ALL are MRD+. A total of 19% of apoptosis-sensitive patients and 69% of apoptosis-resistant patients exhibit MRD positivity as measured by molecular MRD analysis.
Subsequently, we compared the in vitro DNA DSB response and MRD at day 28 of induction treatment, as measured either by flow cytometry or by molecular detection of leukemia-specific immunoglobulin gene rearrangements. Flow cytometric and molecular analysis identified altogether 18 of 55 and 14 of 39 patients, respectively, as MRD+, revealing blast cells 0.001% or greater in their bone marrow. Concordance between the 2 MRD techniques was 79%. Within the apoptosis-sensitive group, 8 (21%) of 38 were classified as MRD+ by flow cytometry and 5 (19%) of 26 by molecular analysis. In contrast, 10 (59%) of 17 patients with apoptosis-resistant ALLs were assessed as MRD+ by flow cytometry and 9 (69%) of 13 by molecular MRD assessment. There was a highly significant difference between patients with apoptosis-sensitive and -resistant ALL with respect to MRD (Fisher exact test for flow MRD, P = .014; for molecular MRD, P = .004; Table 1).
Thus, we observed a significant correlation between in vitro DNA DSB response in pediatric ALL and risk of relapse as determined by in vivo early blast clearance and persistence of MRD.
ALL resistant to IR-induced apoptosis in vitro display differential transcriptional up-regulation of multiple pathways with prosurvival function
We next addressed the nature of the transcriptional responses associated with IR-induced apoptosis in 11 apoptosis-resistant and 11 apoptosis-sensitive cases in which viable leukemic blasts were available.
Initially, we compared the transcription profiles of untreated cells between the 2 groups and observed no significant differences. We concluded that differential baseline transcription does not indicate the mechanism for ALL resistance to IR-induced apoptosis in vitro.
We subsequently compared the IR-induced transcription profiles of the 2 subgroups of ALL. Apoptosis-sensitive cases exhibited an abundant transcription response to IR, with 528 genes significantly changing expression following IR. Interestingly, the predominant response was gene down-regulation (376 genes were down-regulated and 152 were up-regulated). In contrast, apoptosis-resistant cells revealed a less abundant transcription response, with only 196 genes significantly changing expression (115 genes were up-regulated and 81 genes were down-regulated).
Notably, in both the apoptosis-resistant and -sensitive groups, there was a comparable up-regulation of a number of p53-transcriptional targets in response to IR. These targets included proapoptotic genes BAX, FDXR, FAS, and TNFRSF10B; cell-cycle and DNA repair genes GADD45A, PCNA, and DDB2; and IGFBP4 as well as the p53 repressor MDM2 (Figure 3). This observation was consistent with our in vitro DNA DSB response profiling by Western blotting, which revealed normal IR-induced up-regulation of p53 protein and its downstream target p21 in the apoptosis-resistant leukemias.
Apoptosis-sensitive and apoptosis-resistant ALL tumors exhibit comparable p53-dependent transcriptional responses to IR. Microarray analysis reveals comparable up-regulation IR of a set of p53-dependent genes (PCNA, IFFBP4, DDB2, BCL2, GADD45A, FAS MDM2, FDXR, BAX, TNFRSF10B, and BBC3) 8 hours following 5 Gy IR in 11 representative apoptosis-sensitive and 11 apoptosis-resistant ALLs. The error bars represent two times the standard deviation of the values.
Apoptosis-sensitive and apoptosis-resistant ALL tumors exhibit comparable p53-dependent transcriptional responses to IR. Microarray analysis reveals comparable up-regulation IR of a set of p53-dependent genes (PCNA, IFFBP4, DDB2, BCL2, GADD45A, FAS MDM2, FDXR, BAX, TNFRSF10B, and BBC3) 8 hours following 5 Gy IR in 11 representative apoptosis-sensitive and 11 apoptosis-resistant ALLs. The error bars represent two times the standard deviation of the values.
Interestingly, no single gene response to IR was found to be significantly discriminative between apoptosis-resistant and -sensitive ALLs. We therefore investigated potential differences in coordinated responses of functionally related genes using 2 complementary approaches. First, we applied GSEA to test whether functional pathways might be differentially enriched in response to IR depending on the propensity to undergo apoptosis. The second approach directly tested the differential expression of functional modules defined as an average expression profile of genes regulated in a coordinated manner within a given pathway.
Using GSEA analysis, we identified 13 gene sets with prosurvival function that were enriched in resistant tumors in response to IR (Table 2). We noted an overrepresentation of sets of genes involved in epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) signaling, both supporting cellular proliferation and survival.36,37 Similarly, 3 of the enriched gene sets included genes involved in insulin growth factor (IGF) signaling, which has a role in protection from apoptosis.38 Furthermore, gene sets involved in sphingolipid metabolism and regulation of survival and apoptosis through the ceramide pathway,39 sets involved in B cell–receptor signaling crucial for transducing survival and proliferative signals in B cells,40 and sets of genes involved in oxidative phosphorylation that can provide a survival advantage to cancer cells41 were all differentially up-regulated in the apoptosis-resistant group. Finally, the role of PI3K-related signaling in apoptosis resistance was inferred not only from overrepresentation of PTEN and inositol phosphate metabolism-associated gene sets, but also from the presence of PI3K in most (9 of 13) of the significantly enriched gene sets in the resistant group (Table 2). The same was true for MAPK genes, which were found in 8 of 13 gene sets enriched in response to IR in apoptosis-resistant blasts.
Differential regulation of cellular pathways in response to IR between apoptosis-sensitive and -resistant ALL tumors by GSEA analysis
| Pathway name . | No. genes in the pathway . | NOM P . | FDR q . |
|---|---|---|---|
| PDGF | 20 | 0 | .005 |
| PTEN | 16 | 0 | .007 |
| Insulin | 18 | 0 | .007 |
| EGF | 20 | 0 | .006 |
| B-cell receptor complexes | 16 | .002 | .02 |
| IGF1 | 17 | 0 | .027 |
| Oxidative phosphorylation | 29 | .01 | .052 |
| CREB | 19 | .002 | .064 |
| mRNA processing | 18 | 0 | .092 |
| Inositol phosphate metabolism | 15 | .008 | .095 |
| S1P signaling | 16 | .004 | .088 |
| Insulin receptor pathway in cardiac myocytes | 43 | .008 | .134 |
| mRNA splicing | 20 | .017 | .162 |
| TPO pathway | 17 | .031 | .215 |
| Insulin signaling | 19 | .014 | .201 |
| IL6 pathway | 18 | .051 | .238 |
| G13 signaling pathway | 17 | .027 | .226 |
| IL2 | 17 | .017 | .243 |
| TRKA receptor | 15 | .036 | .247 |
| Pathway name . | No. genes in the pathway . | NOM P . | FDR q . |
|---|---|---|---|
| PDGF | 20 | 0 | .005 |
| PTEN | 16 | 0 | .007 |
| Insulin | 18 | 0 | .007 |
| EGF | 20 | 0 | .006 |
| B-cell receptor complexes | 16 | .002 | .02 |
| IGF1 | 17 | 0 | .027 |
| Oxidative phosphorylation | 29 | .01 | .052 |
| CREB | 19 | .002 | .064 |
| mRNA processing | 18 | 0 | .092 |
| Inositol phosphate metabolism | 15 | .008 | .095 |
| S1P signaling | 16 | .004 | .088 |
| Insulin receptor pathway in cardiac myocytes | 43 | .008 | .134 |
| mRNA splicing | 20 | .017 | .162 |
| TPO pathway | 17 | .031 | .215 |
| Insulin signaling | 19 | .014 | .201 |
| IL6 pathway | 18 | .051 | .238 |
| G13 signaling pathway | 17 | .027 | .226 |
| IL2 | 17 | .017 | .243 |
| TRKA receptor | 15 | .036 | .247 |
NOM P indicates nominal probability.
The results of the functional module analysis of gene expression following IR were remarkably similar to that observed with GSEA. Of 15 pathways found to be overexpressed in apoptosis-resistant ALL by functional module analysis, 5 were also identified by GSEA. The 5 overlapping pathways included genes involved in phosphatidyl inositol, IGF receptor, and MAPK and PI3K signaling (Figure 4), all of which are crucial prosurvival pathways. In addition, overexpression of genes from prosurvival signaling pathways such as calcium and Wnt signaling were also identified in the apoptosis-resistant ALL by functional module analysis.
Prosurvival pathway modules are differentially up-regulated in resistant ALL 8 hours after IR. At 8 hours after 5 Gy IR, 11 representative samples of apoptosis-resistant ALL show differential coordinate up-regulation of genes involved in prosurvival pathways compared with the apoptosis-sensitive tumors. Each column represents a different patient sample, and each row represents a pathway consisting of a number of genes given in parentheses. Color changes within a row indicate expression levels relative to the average of the same population. Red indicates up-regulation; green, down-regulation.
Prosurvival pathway modules are differentially up-regulated in resistant ALL 8 hours after IR. At 8 hours after 5 Gy IR, 11 representative samples of apoptosis-resistant ALL show differential coordinate up-regulation of genes involved in prosurvival pathways compared with the apoptosis-sensitive tumors. Each column represents a different patient sample, and each row represents a pathway consisting of a number of genes given in parentheses. Color changes within a row indicate expression levels relative to the average of the same population. Red indicates up-regulation; green, down-regulation.
Thus, our expression profiling suggests that the transcriptional response of several critical prosurvival pathways after induction of DNA damage contributes to the differences in the DNA DSB response between apoptosis-sensitive and -resistant tumors.
Postranscriptional activation of prosurvival pathways in apoptosis-resistant ALLs
We next addressed the possibility that prosurvival pathways might also be differentially activated at the posttranscriptional level in resistant ALL tumors during the IR response. PI-3/Akt activation in patients with apoptosis-resistant leukemias was strongly inferred from the transcription analysis; therefore, we addressed phosphorylation events in this pathway that are known to provide antiapo-ptotic stimuli.42,43 Compared with a representative apoptosis-sensitive leukemia (ALL 114), during the response to ionizing radiation, all 3 representative resistant tumors (ALLs 102, 106, and 111) displayed a moderately increased phosphorylation of Akt and ribosomal protein S6, a substrate for the Akt target p70S6 kinase (Figure 5A). Interestingly, phosphorylation of the Bcl-2 family member Bad, an Akt target with an antiapoptotic activity, was comparable between apoptosis-sensitive and -resistant ALLs and therefore could not provide an explanation for the apoptosis-resistant phenotype.
Differential phosphorylation of PI3-kinase/Akt pathway during IR response in apoptosis-resistant ALLs. (A) Increased phosphorylation at 0 hours, 30 minutes, and 4 hours after IR of DNA-PK at serine 2056, Akt at serine 473, and ribosomal protein S6 at serine 235/6 in apoptosis-resistant ALL tumors (102, 106, and 111) compared with the representative apoptosis-sensitive tumor ALL 114. (B) Comparable phosphorylation of Bad at residues serine 112 and 136 and expression of PTEN protein at 0 hours, 4 hours, and 8 hours after IR in apoptosis-resistant ALL tumors (102, 106, and 111) compared with the representative apoptosis-sensitive tumor ALL 114. Numerical values represent expression ratios of the phosphorylated-total protein for DNA-PKCS, Akt, S6, and Bad, and protein/actin for PTEN. R indicates apoptosis-resistant ALL; S, apoptosis-sensitive ALL.
Differential phosphorylation of PI3-kinase/Akt pathway during IR response in apoptosis-resistant ALLs. (A) Increased phosphorylation at 0 hours, 30 minutes, and 4 hours after IR of DNA-PK at serine 2056, Akt at serine 473, and ribosomal protein S6 at serine 235/6 in apoptosis-resistant ALL tumors (102, 106, and 111) compared with the representative apoptosis-sensitive tumor ALL 114. (B) Comparable phosphorylation of Bad at residues serine 112 and 136 and expression of PTEN protein at 0 hours, 4 hours, and 8 hours after IR in apoptosis-resistant ALL tumors (102, 106, and 111) compared with the representative apoptosis-sensitive tumor ALL 114. Numerical values represent expression ratios of the phosphorylated-total protein for DNA-PKCS, Akt, S6, and Bad, and protein/actin for PTEN. R indicates apoptosis-resistant ALL; S, apoptosis-sensitive ALL.
It has recently emerged that PI3/Akt activation could confer radio resistance through another mechanism (ie, activation of DNA DSB repair).44,45 Consistent with these observations, we noted that differential Akt activation in apoptosis-resistant tumors (ALLs 102, 106, and 111) was associated with increased DNA-PK phosphorylation and more efficient repair of IR-induced DSBs. Unrepaired DNA DSBs are associated with the retention of DNA repair proteins at the sites of DNA damage. Consequently, quantification of cells with IR-induced intranuclear foci of γH2AX and MDC1, used as markers of DNA DSBs, can provide an insight into kinetics of DSB repair.46 We observed that 4 hours after IR, the percentage of cells with more than 5 γH2AX or MDC1 intranuclear foci was significantly reduced in apoptosis-resistant ALL tumors compared with the sensitive ALL 114 (Figure 6A). Furthermore, addition of the PI3-kinase inhibitor LY294002 notably delayed disappearance of IR-induced foci in a resistant tumor but failed to do so in apoptosis-sensitive ALL (Figure 6B). Thus, we concluded that differential up-regulation of the PI3/Akt pathway might render primary ALL tumors resistant to IR by an increased efficiency of DNA DSB repair.
Kinetics of IR-induced γH2AX suggests more efficient DNA DSB repair in apoptosis-resistant ALLs. (A) (i) Following treatment with 2 Gy of IR and induction of a comparable number of DNA DSBs measured by γH2AX/MDC1 foci at 30 minutes, by 4 hours the resistant ALL 102 shows more efficient DNA repair and resolution of these foci, whereas sensitive ALL 114 retains foci at the same time point. Primary antibody binding was detected using Alexa Fluor 488 and Alexa Fluor 594–labeled secondary antibodies. Images were captured with a Nikon Eclipse E600 microscope (100×/1.40 NA oil objective) with a Hamamatsu digital camera C4742-95 and Volocity 4.2 software. (ii) Percentage of cells with more than 5 γH2AX/MDC1 foci 4 hours after IR is significantly higher in sensitive ALL 114 compared with resistant ALLs 102 and 106 (P = .04 and .02, respectively). The number of foci per cell was counted in 150 to 200 cells in randomly selected fields. The data are expressed as a percentage of cells with greater than 5 foci and the error bars represent the standard deviation from 3 independent counts. (B) Addition of PI3-kinase inhibitor LY294002 significantly delays disappearance of IR-induced foci at 4 hours after IR with 2 Gy in resistant tumor 102 (bottom panel), but shows no effect on resolution of foci in apoptosis-sensitive ALL 114 at the same time point (top panel).
Kinetics of IR-induced γH2AX suggests more efficient DNA DSB repair in apoptosis-resistant ALLs. (A) (i) Following treatment with 2 Gy of IR and induction of a comparable number of DNA DSBs measured by γH2AX/MDC1 foci at 30 minutes, by 4 hours the resistant ALL 102 shows more efficient DNA repair and resolution of these foci, whereas sensitive ALL 114 retains foci at the same time point. Primary antibody binding was detected using Alexa Fluor 488 and Alexa Fluor 594–labeled secondary antibodies. Images were captured with a Nikon Eclipse E600 microscope (100×/1.40 NA oil objective) with a Hamamatsu digital camera C4742-95 and Volocity 4.2 software. (ii) Percentage of cells with more than 5 γH2AX/MDC1 foci 4 hours after IR is significantly higher in sensitive ALL 114 compared with resistant ALLs 102 and 106 (P = .04 and .02, respectively). The number of foci per cell was counted in 150 to 200 cells in randomly selected fields. The data are expressed as a percentage of cells with greater than 5 foci and the error bars represent the standard deviation from 3 independent counts. (B) Addition of PI3-kinase inhibitor LY294002 significantly delays disappearance of IR-induced foci at 4 hours after IR with 2 Gy in resistant tumor 102 (bottom panel), but shows no effect on resolution of foci in apoptosis-sensitive ALL 114 at the same time point (top panel).
We next addressed the upstream mechanisms of PI-3/Akt up-regulation in resistant ALLs. We first analyzed the genetic status of upstream regulators of the PI3/Akt pathway RAS and PTEN. We observed no evidence of PTEN mutation in any of 15 representative apoptosis-resistant ALL tumors and an equal but low frequency of H-RAS mutations (14%) in codons 12/13 in both apoptosis-resistant and -sensitive ALLs. Consistent with this observation, PTEN expression was indistinguishable between 2 ALL subtypes (Figure 5B).
Given the range of upstream regulators that could potentially lead to PI3/Akt up-regulation, we compared phosphorylation patterns of 42 RTKs by using the Proteome Profiler Phospho RTK Array (R&D Systems) approach. We compared RTK phosphorylation between 4 representative apoptosis-resistant (ALLs 102,106, 111, and 141) and 2 apoptosis-sensitive (ALLs 112 and 114) tumors before and 1 hour after IR. Notably, resistant tumors revealed an individual pattern of differential phosphorylation, with untreated ALL 102 showing differential phosphorylation of ErbB4 and untreated ALL 106 differential phosphorylation of Flt3 (Figure S2A). In response to IR, however, tumor 102 exhibited induced phosphorylation of multiple kinase receptors (Figure S2B), whereas IR-treated ALL 111 exhibited moderate differential induction of FGFR3 phosphorylation (2C), and ALL 106 revealed differential phosphorylation of Mer, a proto-oncogene associated with development of lymphoid leukemia.47,48 Finally, resistant ALL 141 showed differential induction of both insulin R and IGF-I R phosphorylation (Figure S2C).
Taken together, our data suggest that both transcriptional and posttranscriptional up-regulation of prosurvival signals confers DNA damage–induced apoptosis resistance in ALL in a manner that might be individual to each resistant leukemia.
Suppression of prosurvival pathways restores sensitivity to IR induced DNA DSBs in vitro
In an attempt to sensitize apoptosis-resistant tumors to IR, we analyzed the impact of a panel of 5 inhibitors, against IGF-R1, MAPK MEK1, PI-3K, Akt and m-Tor, in 4 primary apoptosis-resistant ALL tumors. Consistent with a tumor-specific transcriptional and posttranscriptional deregulation of prosurvival pathways, each primary ALL responded to the panel of inhibitors in a unique manner. ALL 111 became IR sensitive after the addition of PI-3K inhibitor LY294002. Interestingly, and consistent with IR-induced differential phosphorylation of IGF-IR and insulin R, ALL 141 showed an increase in IR-induced apoptosis after preincubation with the IGF-R1 inhibitor AG1024. ALL 102 and ALL 106 both became IR sensitive following exposure to all the inhibitors except rapamycin (Figure 7A). The induction of IR-induced apoptosis in the presence of these inhibitors was confirmed by Western blotting in 2 representative apoptosis-resistant leukemias, ALL 111 and 141. These leukemias exhibited cleavage of caspase-3 and PARP1 8 hours after IR in the presence of LY294002 and AG1024, respectively, but not in the absence of the same inhibitors (Figure 7B).
Primary ALL tumors resistant to IR can be sensitized by targeted inhibition of prosurvival pathways. (A) Induction of IR-induced apoptosis measured by annexin V+/PI+ staining 24 hours after treatment with 20 μM UO126, 500 nM rapamycin, 30 μM LY294002, and 30 μM AG1024 in 4 apoptosis-resistant ALLs not previously included in the microarray experiments. The means and standard deviations from 3 experiments are shown as well as P values for each inhibitor relative to no treatment. The figure shows that the 4 patients with apoptosis-resistant ALL have individual patterns of response to inhibition of different prosurvival signals prior to IR. ALL 111 became IR sensitive after addition of PI-3 kinase inhibitor LY294002 only. ALL 141 showed an increase in IR-induced apoptosis after preincubation with IGFR inhibitor AG1024 only. ALL 102 and 106 are both IR sensitive following exposure to all inhibitors except rapamycin. (B) Western blotting showing post-IR–induced apoptosis after 8 hours of incubation with PI-3 inhibitor LY294002 in ALL 111 or IGFR inhibitor AG1024 in ALL 141 as measured by cleavage of PARP1 and appearance of the cleaved caspase-3. Both leukemias exhibit reduction in IR-induced phosphorylation of Akt in the presence of the respective inhibitors.
Primary ALL tumors resistant to IR can be sensitized by targeted inhibition of prosurvival pathways. (A) Induction of IR-induced apoptosis measured by annexin V+/PI+ staining 24 hours after treatment with 20 μM UO126, 500 nM rapamycin, 30 μM LY294002, and 30 μM AG1024 in 4 apoptosis-resistant ALLs not previously included in the microarray experiments. The means and standard deviations from 3 experiments are shown as well as P values for each inhibitor relative to no treatment. The figure shows that the 4 patients with apoptosis-resistant ALL have individual patterns of response to inhibition of different prosurvival signals prior to IR. ALL 111 became IR sensitive after addition of PI-3 kinase inhibitor LY294002 only. ALL 141 showed an increase in IR-induced apoptosis after preincubation with IGFR inhibitor AG1024 only. ALL 102 and 106 are both IR sensitive following exposure to all inhibitors except rapamycin. (B) Western blotting showing post-IR–induced apoptosis after 8 hours of incubation with PI-3 inhibitor LY294002 in ALL 111 or IGFR inhibitor AG1024 in ALL 141 as measured by cleavage of PARP1 and appearance of the cleaved caspase-3. Both leukemias exhibit reduction in IR-induced phosphorylation of Akt in the presence of the respective inhibitors.
Discussion
In this study, we investigated the clinical relevance of in vitro DNA DSB responses in pediatric B-precursor ALL. We demonstrated a significant correlation between the DNA DSB response in vitro and patients' response to treatment in vivo, as measured by blast clearance at days 8 and 15 of treatment and MRD at day 28. Resistance to apoptosis in vitro could be attributed to differential up-regulation of multiple prosurvival pathways in response to DNA DSBs and therefore may be one of the mechanisms underlying the poor clinical response in refractory ALL.
The notable correlation between the apoptosis response to IR in vitro and the clinical response to induction treatment suggests that alterations in the IR response pathways in ALL blasts are clinically relevant and may reflect responses to other DNA-damaging agents used in ALL treatment. This is not surprising, as many of the therapeutic agents currently used in ALL therapy are known to induce the same cellular responses observed after IR.15 Thus, IR response in vitro provides a relevant model system for the in vivo response to chemotherapy.
The major purpose of our transcriptional profiling was to elucidate the mechanisms behind resistance to DNA DSBs and potentially provide future therapeutic targets. Thus, unlike previous microarray studies that suggested a correlation between resistant disease and baseline expression of genes enrolled in pathways associated with cellular proliferation, cell-cycle regulation, and apoptosis,13,14,49-52 in this study we observed no differences in baseline transcription between IR-sensitive and IR-resistant ALL. This difference could be explained by the fact that we stratified our ALL cohort according to apoptosis response to IR-induced DNA DSB, unlike previous studies where cohorts were analyzed on the basis of MRD response in vivo or occurrence of relapse.49-52 Thus, it is probable that resistance defined by defective cellular response to DNA DSBs represents the mechanism of chemoresistance in only a subset of ALL.
Interestingly, in vitro resistance to apoptosis in pediatric ALL could not be attributed to the differential expression of a single set of IR-responsive genes. Instead, in our apoptosis-resistant group we identified coordinate up-regulation of genes involved in several pathways with prosurvival function, such as PI3K/Akt, MAPK, IGF, and inositol signaling, and the same pathways were also found to be activated at the posttranscriptional level. Importantly, the loss of integrity of the DNA DSB response was detectable at presentation in a significant subset of ALL, most likely representing an intrinsic feature of these tumors. Intriguingly, the defective response occurred in samples with a range of different recurrent cytogenetic abnormalities as well as those with no visible abnormalities, suggesting that no single genetic event was the likely cause of deregulation of survival response pathways. This was consistent with our inability to identify a single set of IR-responsive discriminating genes between apoptosis-resistant and -sensitive leukemias and with the variable pattern of RTK phosphorylation in resistant tumors. Also in support of this notion was our observation that RAS mutations did not segregate with either of the 2 ALL subtypes and the fact that no analyzed resistant tumor exhibited PTEN mutation. Thus, 2 alternative scenarios are likely to account for the presence of deregulated survival responses to DNA damage in a subset of pediatric ALL. First, differences in DNA damage–induced survival responses might reflect differences in cellular origin between apoptosis-resistant and -sensitive tumors. Second, and the more likely possibility, is that selective pressure for the loss of integrity of the DNA DSB response in ALL could incrementally lead to aberrant activation of several pathways with prosurvival function, the distribution of which is likely to be unique for each primary ALL.
It is intriguing that pediatric ALL does not appear to be associated with frequent defects in the ATM/p53 proapoptotic branch of DNA DSB responses. Indeed, in this study we have shown that that p53-dependent IR-responsive genes are uniformly up-regulated in both IR apoptosis-resistant and -sensitive cases, confirming our previous observations18 that pathogenic ATM or TP53 variants are exceptionally rare in pediatric ALL. This is in contrast with chronic lymphocytic leukemia (CLL), where ATM or TP53 mutations are the most common cause of treatment failure and where we have demonstrated a clear impact of presence of these mutations on survival.19,20 Thus, it appears that the nature of responses to DNA DSBs, which include both prosurvival and proapoptotic signals,16 provides the opportunity for selective pressure during leukemogenesis to target the DNA DSB response pathway at different levels in different types of leukemia.
Up-regulation of prosurvival pathways implicated in this study has been described in other forms of acute leukemias.53,54 Importantly, these pathways represent suitable therapeutic targets, and from that point of view, our observations have important clinical implications. A range of inhibitors targeting specific antiapoptotic pathways have recently become available, and their benefit in the induction of apoptosis in leukemic cells is beginning to emerge.55-66 Consistent with this, we have previously reported the positive effect of inhibition of NF-κB signaling by PARP inhibition on IR-induced ALL killing in vitro.18 Here, we show that modification of the response to IR in apoptosis-resistant ALL can be achieved by inhibition of other prosurvival molecules, including Akt, PI-3K, IGF-R1, and MEK, and that the mode of sensitization to DNA DSBs is likely to depend on the individual molecular make-up of each tumor. Our preliminary results suggest that PI-3/Akt activation may represent a converging point of deregulation in DNA damage response in resistant ALLs. Consequently, inhibition of the PI-3/Akt pathway might be a sensible initial approach toward sensitization of these tumors, a strategy inferred from our pharmacologic experiments. Furthermore, our in vitro DSB response stratification provides a valuable system to identify patients who will benefit from such treatment. Further investigations will be necessary, however, to establish prosurvival signaling molecule(s) whose inhibition will produce the most consistent sensitization of pediatric ALLs to DNA DSB–inducing agents.
In summary, we have shown that defective up-regulation of multiple prosurvival pathways during response to DNA DSBs in vitro represents a feature of a subset of pediatric ALL which is more likely to respond poorly to treatment. Thus, inhibition of prosurvival signaling could potentially improve the outcome in the subset of children with ALL defective in DNA DSB–induced apoptosis, and this approach warrants further investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Clemency Hawksley for technical assistance. Diagnosis of cytogenetic abnormalities was performed by the West Midlands Regional Genetics Laboratory at the Birmingham Women's Hospital. Patients' clinical and treatment details were provided by Sheila Parkes of the West Midlands Regional Children's Tumor Registry at the Birmingham Children's Hospital.
This study was supported by Leukaemia Research Fund, United Kingdom.
Authorship
Contribution: E. Marston and V.W. performed research, analyzed data, and wrote the paper; E. Maina, A.A., A.S., K.M., and J.J. performed research and analyzed data; C.M., K.S., J.E.P., and S.L. analyzed data; P.K. and F.F. analyzed data and wrote the paper; M.T. designed research and wrote the paper; and T.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tatjana Stankovic, CRUK Institute for Cancer Studies, Birmingham University, Vincent Drive, Edgbaston, B15 2TT United Kingdom; e-mail: t.stankovic@bham.ac.uk.