Abstract
The gamma isoform of PI3Kinase (PI3Kγ) controls leukocyte chemotaxis by participating in GPCR signaling, and by regulating cellular polarization. Here we show that PI3Kγ is required for efficient induction of CXC chemokine receptor 3 (CXCR3) on T cells upon activation. T cells from PI3Kγ−/− mice up-regulated CXCR3 less efficiently than wild-type controls both upon activation in vitro as well as during Leishmania mexicana infection. Inhibition of PI3Kinases using wortmannin and LY294002 or blockade of PI3Kγ activity using a selective inhibitor or PI3Kγ siRNA suppressed induction of CXCR3 on T cells following activation. Levels of CXCR3 and T-bet mRNA were significantly lower in PI3Kγ inhibitor–treated T cells, indicating that PI3Kγ may control CXCR3 expression in part through induction of T-bet. These results reveal a novel role for PI3Kγ in the induction of CXCR3 on T cells and suggest that PI3Kγ may regulate leukocyte chemotaxis by controlling the expression of chemokine receptors.
Introduction
The chemokine receptor CXCR3 is expressed on plasmacytoid dendritic cells (pDCs), natural killer (NK) cells, T cells, and microvascular endothelial cells.1 Three CXC chemokines, CXCL9, CXCL10, and CXCL11, signal via CXCR32 and mediate biologic functions such as cell migration and proliferation.3
CXCR3 mediates immunity against pathogens by regulating chemotaxis and function of T cells and other leukocytes,4-7 however, CXCR3 also contributes to pathogenesis of autoimmune diseases.8-11 Resting T cells express low levels of CXCR3. Upon activation, T cells up-regulate CXCR3 allowing for an increased responsiveness toward its ligands.12,13 Both IFN-γ and STAT1 are required for efficient induction of CXCR3 on CD4+ T cells but not CD8+ T cells.14
The phosphoinositide-3-kinases (PI3Ks) are required for generation of PIP3. Class I PI3Ks are dual-specificity lipid and protein kinases that control cell growth, proliferation, survival, adhesion, and motility. These enzymes include class IA (consisting of PI3Kα, PI3Kβ, and PI3Kδ) and class IB (PI3Kγ).15 Of these, PI3Kγ participates in leukocyte chemotaxis, mast cell degranulation, neutrophil respiratory burst, and TCR-induced T-cell activation.15-17
TCR-induced activation is essential for up-regulation of CXCR3 on T cells, suggesting that PI3Kγ may regulate CXCR3 expression on activated T cells.12,13 Therefore, we examined the role of PI3Kγ in regulating CXCR3 on T cells. Our findings show that PI3Kγ is critical for efficient induction of CXCR3 on activated T cells.
Methods
Mice
PI3Kγ−/− C57BL/6 mice were maintained at The Ohio State University. Wild-type C57BL/6 mice were purchased from Harlan (Madison, WI). IRB approval for this study was obtained from The Ohio State University.
In vitro stimulation of T cells
Cell suspensions were prepared from spleens of WT or PI3Kγ−/− mice, and T cells were isolated by immunomagnetic separation. Cells (90%-94% pure) were plated in 24-well plates at 106/mL and stimulated with plate-bound anti-CD3e (3 μg/mL) and anti-CD28 (4 μg/mL) at 37°C for 48 hours in the absence of PI3K inhibitor. Subsequently, cells were rested in their conditioned media for 24 hours without stimulation either in the presence of PI3K inhibitors (wortmannin, LY294002, or AS-605240) or DMSO as described previously.12,14
Generation of PI3Kγ siRNA and gene silencing
The Silencer Express Kit (Ambion, Austin, TX) was used to generate PI3Kγ siRNA-expressing vectors. Efficiency of gene silencing was confirmed by measuring PI3Kγ mRNA levels by real-time polymerase chain reaction (PCR). WT C57BL/6 T cells were stimulated with anti-CD3/CD28 for 48 hours and then transfected with PI3Kγ siRNA-expressing vector using a mouse T-cell Nucleofector kit (Amaxa, Gaithersburg, MD). CXCR3 expression was analyzed 24 hours after transfection using flow cytometry. As predicted by the manufacturer, T-cell transfection efficiency was 20% to 30% as determined by analyzing T cells transfected with a plasmid containing maxGFP.
Flow cytometry
Cells (1-2 × 105) were stained with PE-conjugated anti-CXCR3 antibodies (R&D Systems, Minneapolis, MN) and analyzed by flow cytometry.
Real-time PCR
Total RNA was extracted using TRIZOL Reagent (Invitrogen, Carlsbad, CA), and real-time PCR was performed as described previously.14 Primers and reaction conditions were found using the PRIMER BANK website.18 Data were normalized to the housekeeping gene gapdh and presented as fold induction over nonstimulated T cells using the delta-delta CT method.
Leishmania mexicana parasites and infections
WT and PI3Kγ−/− C57BL/6 mice were infected by inoculating 106Leishmania mexicana into footpads. Three weeks after infection, CXCR3-expressing T cells in the draining LNs were analyzed by flow cytometry.
Statistical analysis
Significant differences were determined by an unpaired Student t test. A value of P less than .05 was considered significant.
Results and discussion
T cells up-regulate CXCR3 upon activation, but the TCR-mediated signaling mechanisms that control CXCR3 expression are not clear. Class I PI3Ks are dual-specificity lipid and protein kinases that participate in numerous signaling pathways. Among those, PI3Kγ controls leukocyte migration and is expressed mainly in hematopoietic cells.15,17 PI3Kγ is activated primarily through GPCRs, which include chemokine receptors.15 However, PI3Kγ also participates in T-cell activation induced by TCRs.16
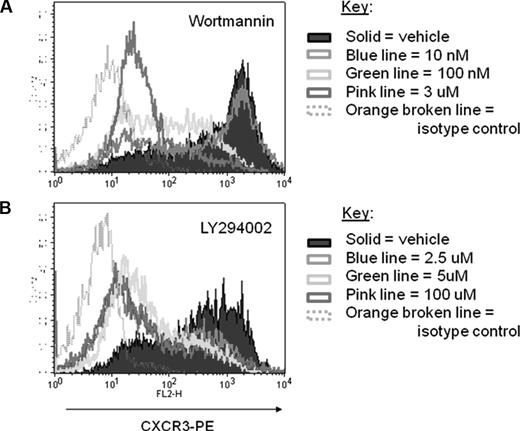
Our goal was to determine whether the PI3K pathway, and specifically PI3Kγ, controls CXCR3 expression by T cells following TCR-induced activation. Therefore, we analyzed CXCR3 levels on C57BL/6 T cells that were activated with anti-CD3/anti-CD28 antibodies in vitro and then treated with PI3K inhibitors as described in “In vitro stimulation of T cells.” Both wortmannin (Figure 1A) and LY294002 (Figure 1B) suppressed CXCR3 induction on T cells in a dose-dependent manner, suggesting that PI3K pathway regulates CXCR3 expression.
Inhibition of PI3K suppresses CXCR3 induction following in vitro T-cell activation. T cells from C57BL/6 mice were stimulated with anti-CD3/anti-CD28 antibodies for 48 hours and then rested with different concentrations of PI3K inhibitors: (A) wortmannin (10 nM, 100 nM, or 3 μM) or (B) LY294002 (2.5 μM, 5 μM, or 100 μM). Following 24-hour incubation with inhibitors, expression of CXCR3 on these cells was analyzed by flow cytometry. Isotype control–stained cells are represented by hollow, broken lines. Data are representative from 1 experiment of 3 independent experiments with similar results.
Inhibition of PI3K suppresses CXCR3 induction following in vitro T-cell activation. T cells from C57BL/6 mice were stimulated with anti-CD3/anti-CD28 antibodies for 48 hours and then rested with different concentrations of PI3K inhibitors: (A) wortmannin (10 nM, 100 nM, or 3 μM) or (B) LY294002 (2.5 μM, 5 μM, or 100 μM). Following 24-hour incubation with inhibitors, expression of CXCR3 on these cells was analyzed by flow cytometry. Isotype control–stained cells are represented by hollow, broken lines. Data are representative from 1 experiment of 3 independent experiments with similar results.
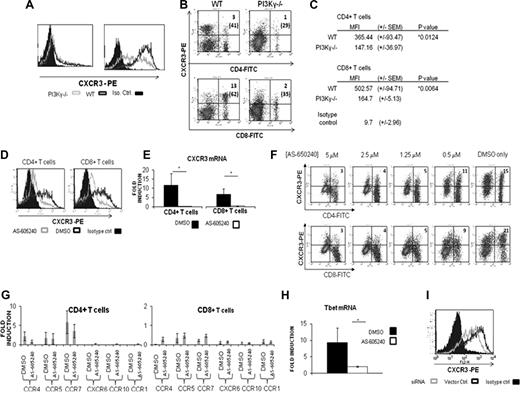
TCR-induced T-cell activation is essential for up-regulation of CXCR3, which mediates T-cell chemotaxis, likely through activating PI3Kγ. Because PI3Kγ participates in TCR-induced T-cell activation,16 we determined whether PI3Kγ regulates CXCR3 expression by comparing CXCR3 levels on anti-CD3/CD28–activated T cells from WT and PI3Kγ−/− C57BL/6 mice by flow cytometry. In addition, we analyzed CXCR3-expressing T cells in the lymph nodes of L mexicana–infected WT and PI3Kγ−/− mice. Basal levels of CXCR3 were comparably low on T cells of WT and PI3Kγ−/− mice (data not shown). However, activated PI3Kγ−/− T cells failed to up-regulate CXCR3 as efficiently as WT T cells (Figure 2A). Furthermore, lymph nodes of L mexicana–infected PI3Kγ−/− mice contained a lower proportion of CXCR3-expressing CD4+ and CD8+ T cells than WT mice (Figure 2B). Even within CD4+ and CD8+ compartments, PI3Kγ−/− mice contained a lower proportion of CXCR3+ T cells compared with WT mice with knockout T cells expressing less CXCR3 (Figure 2C).
PI3Kγ deficiency or selective blockade of PI3Kγ activity prevents efficient induction of CXCR3 on T cells upon activation. (A,B) T cells from C57BL/6 mice genetically deficient in functional PI3Kγ (PI3Kγ−/−) express less CXCR3 than wild-type C57BL/6 T cells. (A) Flow cytometric analysis of CXCR3 levels on anti-CD3/CD28–activated T cells from wild-type (black hollow peaks) and PI3Kγ−/− (gray hollow peaks) mice. (B) Flow cytometric analysis of lesion-draining LN cells excised from L mexicana–infected wild-type and PI3Kγ−/− C57BL/6 mice. Numbers outside parentheses indicate the percentage of LN cells, whereas numbers in parentheses represent the percentage of CXCR3+ T cells in the CD4+ or CD8+ compartment. Shown are representative results of 3 to 5 independent experiments. (C) Mean fluorescence intensity values (MFI) for CXCR3 staining on CD4+ and CD8+ LN T cells from L mexicana–infected mice. Numbers given are the mean MFI (± SEM) of CXCR3-PE staining on the surface of indicated T cells from 3 independent experiments. *P = .05. (D) Blockade of PI3Kγ activity suppresses efficient induction of CXCR3 on activated T cells. Activated T cells were treated with the PI3Kγ-selective inhibitor AS-605240 (1.25 μM; gray hollow peaks) or vehicle (black hollow peaks) as described before, and expression of CXCR3 was analyzed by flow cytometry. Isotype controls are solid peaks. (E) Analysis of CXCR3mRNA levels in AS-605240–treated versus vehicle-treated T cells by semiquantitative real-time PCR analysis. AS-605240–treated T cells (▭) showed significantly less induction of CXCR3 mRNA than vehicle-treated cells ( ). (F) The effect of AS-605240 on CXCR3 suppression was dose dependent. (G) AS-605240 treatment did not affect CCR1, CCR5, CCR4, CCR7, CXCR6, and CCR10 mRNA levels. (H) AS-605240–treated T cells (▭) displayed less induction of T-bet mRNA compared with vehicle controls (
). (F) The effect of AS-605240 on CXCR3 suppression was dose dependent. (G) AS-605240 treatment did not affect CCR1, CCR5, CCR4, CCR7, CXCR6, and CCR10 mRNA levels. (H) AS-605240–treated T cells (▭) displayed less induction of T-bet mRNA compared with vehicle controls ( ). Real-time PCR data for each group were normalized to the housekeeping gene gapdh and are expressed as fold induction over nonstimulated cells. (I) Effect of PI3Kγ gene silencing on induction of CXCR3. Activated T cells transfected with PI3Kγ siRNA expressed less CXCR3 compared with controls. Data in panels A-D and I are representative of at least 2 to 5 independent experiments with similar results. Panels F-H represent the mean results (± SEM) of 3 or more independent experiments. A P value from an unpaired Student t test less than .05 (*) was considered significant.
). Real-time PCR data for each group were normalized to the housekeeping gene gapdh and are expressed as fold induction over nonstimulated cells. (I) Effect of PI3Kγ gene silencing on induction of CXCR3. Activated T cells transfected with PI3Kγ siRNA expressed less CXCR3 compared with controls. Data in panels A-D and I are representative of at least 2 to 5 independent experiments with similar results. Panels F-H represent the mean results (± SEM) of 3 or more independent experiments. A P value from an unpaired Student t test less than .05 (*) was considered significant.
PI3Kγ deficiency or selective blockade of PI3Kγ activity prevents efficient induction of CXCR3 on T cells upon activation. (A,B) T cells from C57BL/6 mice genetically deficient in functional PI3Kγ (PI3Kγ−/−) express less CXCR3 than wild-type C57BL/6 T cells. (A) Flow cytometric analysis of CXCR3 levels on anti-CD3/CD28–activated T cells from wild-type (black hollow peaks) and PI3Kγ−/− (gray hollow peaks) mice. (B) Flow cytometric analysis of lesion-draining LN cells excised from L mexicana–infected wild-type and PI3Kγ−/− C57BL/6 mice. Numbers outside parentheses indicate the percentage of LN cells, whereas numbers in parentheses represent the percentage of CXCR3+ T cells in the CD4+ or CD8+ compartment. Shown are representative results of 3 to 5 independent experiments. (C) Mean fluorescence intensity values (MFI) for CXCR3 staining on CD4+ and CD8+ LN T cells from L mexicana–infected mice. Numbers given are the mean MFI (± SEM) of CXCR3-PE staining on the surface of indicated T cells from 3 independent experiments. *P = .05. (D) Blockade of PI3Kγ activity suppresses efficient induction of CXCR3 on activated T cells. Activated T cells were treated with the PI3Kγ-selective inhibitor AS-605240 (1.25 μM; gray hollow peaks) or vehicle (black hollow peaks) as described before, and expression of CXCR3 was analyzed by flow cytometry. Isotype controls are solid peaks. (E) Analysis of CXCR3mRNA levels in AS-605240–treated versus vehicle-treated T cells by semiquantitative real-time PCR analysis. AS-605240–treated T cells (▭) showed significantly less induction of CXCR3 mRNA than vehicle-treated cells ( ). (F) The effect of AS-605240 on CXCR3 suppression was dose dependent. (G) AS-605240 treatment did not affect CCR1, CCR5, CCR4, CCR7, CXCR6, and CCR10 mRNA levels. (H) AS-605240–treated T cells (▭) displayed less induction of T-bet mRNA compared with vehicle controls (
). (F) The effect of AS-605240 on CXCR3 suppression was dose dependent. (G) AS-605240 treatment did not affect CCR1, CCR5, CCR4, CCR7, CXCR6, and CCR10 mRNA levels. (H) AS-605240–treated T cells (▭) displayed less induction of T-bet mRNA compared with vehicle controls ( ). Real-time PCR data for each group were normalized to the housekeeping gene gapdh and are expressed as fold induction over nonstimulated cells. (I) Effect of PI3Kγ gene silencing on induction of CXCR3. Activated T cells transfected with PI3Kγ siRNA expressed less CXCR3 compared with controls. Data in panels A-D and I are representative of at least 2 to 5 independent experiments with similar results. Panels F-H represent the mean results (± SEM) of 3 or more independent experiments. A P value from an unpaired Student t test less than .05 (*) was considered significant.
). Real-time PCR data for each group were normalized to the housekeeping gene gapdh and are expressed as fold induction over nonstimulated cells. (I) Effect of PI3Kγ gene silencing on induction of CXCR3. Activated T cells transfected with PI3Kγ siRNA expressed less CXCR3 compared with controls. Data in panels A-D and I are representative of at least 2 to 5 independent experiments with similar results. Panels F-H represent the mean results (± SEM) of 3 or more independent experiments. A P value from an unpaired Student t test less than .05 (*) was considered significant.
Although these results suggest that PI3Kγ controls CXCR3 expression on T cells, it is possible that the failure of PI3Kγ−/− T cells to up-regulate CXCR3 is due to impaired TCR-induced activation.16 Therefore, we used an isoform-selective inhibitor of PI3Kγ (AS-605240) to determine whether PI3Kγ blockade after TCR activation prevents induction of CXCR3. AS-605240 competes with ATP for its binding pocket on PI3Kγ, effectively inhibiting the enzyme.17 T cells isolated from naive C57BL/6 mice were activated for 48 hours with anti-CD3/CD28 and then rested in the presence of AS-605240 for 24 hours. Expression and mRNA levels of CXCR3 were measured by flow cytometry and real-time reverse-transcription (RT)–PCR, respectively. Activated T cells rested with AS-605240 failed to up-regulate CXCR3 as efficiently as controls (Figure 2D). Low CXCR3 expression on AS-605240–treated T cells also correlated with low CXCR3 mRNA levels (Figure 2E). Furthermore, suppression of CXCR3 on T cells by AS-605240 was dose dependent (Figure 2F). Levels of IFN-γ, IL-4, and IL-2 were comparable in culture supernatants from AS-604850–treated and control T cells (data not shown). AS-605240 treatment had no effect on mRNA levels of CCR1, CCR5, CCR4, CCR7, CXCR6, or CCR10 (Figure 2G). These results demonstrate that PI3Kγ is required for efficient induction of CXCR3 on activated T cells and suggest that PI3Kγ controls CXCR3 levels by regulating CXCR3 gene transcription.
Expression of CXCR3 by T cells requires T-bet.19,20 SHP-1, which also suppresses PI3K activity,21 inhibits T-bet expression in T cells.22 We therefore measured T-bet mRNA levels in AS-605240–treated T cells. AS-605240–treated T cells showed decreases in both CXCR3 and T-bet mRNA compared with the controls (Figure 2E,H). These findings suggest that PI3Kγ controls T-bet induction in T cells and that T-bet may be involved in PI3Kγ-induced up-regulation of CXCR3.
Finally, to confirm the specificity of our findings, we examined the effect of PI3Kγ gene silencing on CXCR3 induction. Even with only 30% transfection efficiency, PI3Kγ siRNA-transfected T cells expressed significantly less CXCR3 than controls (Figure 2I).
In conclusion, our study has revealed a previously unknownfunction of PI3Kγ in CXCR3 induction on activated T cells. The findings suggest that since PI3Kγ may regulate T-cell chemotaxis by modulating CXCR3 levels, PI3Kγ-selective inhibitors may be useful in suppressing CXCR3 on pathogenic T cells to inhibit their action in chronic inflammatory diseases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work is supported by grants R01 AI51328 (A.R.S.) and R01 AI064320 (C.C.W.) from the National Institutes of Health (Bethesda, MD).
National Institutes of Health
Authorship
Contribution: J.B. performed experiments and prepared the paper; H.E.C. and S.O. performed experiments and reviewed the paper; B.L., T.R., W.L., C.W., and C.R. provided critical reagents and reviewed the paper; and A.R.S. provided laboratory space and funding, designed the experiments, and supervised paper preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Abhay R. Satoskar, Department of Microbiology, The Ohio State University, 484 W 12th Ave, Columbus, OH 43221; e-mail: satoskar.2@osu.edu.