Abstract
Robust T-cell responses without autoimmunity are only possible through a fine balance between activating and inhibitory signals. We have identified a novel modulator of T-cell expansion named proliferation-induced lymphocyte-associated receptor (PILAR). Surface PILAR is markedly up-regulated on CD4 and, to a lesser extent, on CD8 T cells on T-cell receptor engagement. In absence of CD28 costimulation, PILAR signaling through CD161 supports CD3 antibody-dependent and antigen-specificT-cell proliferation by increasing the expression of antiapoptotic Bcl-xL and induces secretion of T helper type 1 cytokines. These effects are abrogated by PILAR blockade with specific antibodies, which decrease surface levels of CD28. In contrast, PILAR induces apoptotic death on naive and early activated T cells if CD161 engagement is blocked. PILAR is expressed by approximately 7% to 10% of CD4 T cells in 2 samples of inflammatory synovial fluid, suggesting a potential role in the pathogenesis of joint inflammation. In addition, in the ovarian cancer microenvironment, effector T cells express PILAR, but not CD161, although expression of both can be augmented ex vivo. Our results indicate that PILAR plays a central role in modulating the extent of T-cell expansion. Manipulation of PILAR signaling may be important for treatment of autoimmune diseases and cancer.
Introduction
The balance between activating and inhibitory signals on T lymphocytes is crucial for maintaining an efficient adaptive immune response while preventing autoimmunity. T cells are activated by engagement of their T-cell receptor (TCR) by specific MHC-peptide complexes expressed on antigen-presenting cells (APCs), but other signals concurrently delivered determine the appropriate immune response. CD28 represents the archetype of an activating costimulatory receptor,1,2 but numerous molecules also contribute to maintain continued expansion and differentiation of T cells.3-8 Costimulatory molecules recognized by T cells are typically expressed by myeloid APCs, although recent reports indicate that T to T costimulatory interactions, mostly relevant for secondary responses,5,9 occur between CD27 and CD70.10 In addition, ectopic expression of CD80 and 4-1BBL costimulatory ligands on T lymphocytes has been recently proposed as a novel strategy for enhancing adoptive transfer therapy, because they induce potent auto-costimulation as well as activation of bystander T cells in vivo.11
Recent reports indicate that engagement of CD161, a C-type lectin receptor associated with natural killer (NK) cells, enhances IFN-γ and TNF-α production in the context of a TCR signal12,13 and induces the secretion of IL-12 by dendritic cells.14 Unlike in mouse lymphocytes, CD161 in humans is predominantly expressed by memory T cells and only by a small proportion of invariant NK T cells.12 The only known ligand for CD161 is CLEC2D/LLT1,13,15 another C-type lectin molecule that maps in the vicinity of NKG2D at the NK cluster. This narrow (≈2 Mb) region of human chromosome 12 encodes at least 18 different leukocyte receptors that exhibit a C-type lectin-like motif in their extracellular domain.
We have identified a novel human transmembrane molecule mapping at chromosome 12p13.31, named proliferation-induced lymphocyte-associated receptor (PILAR). PILAR is detected by 2 different antibodies on T lymphocytes, is up-regulated at the mRNA level on early TCR stimulation, and dramatically enhances T-cell expansion through CD161. Our results show a crucial role for PILAR in modulating the extent of cellular adaptive immune responses in humans and have implications for inflammation, cancer, and autoimmunity.
Methods
Tissues and cells
Tissues were obtained through Research Pathology (Dartmouth Medical School, Lebanon, NH), after institutional approval. Single-cell suspensions were obtained by gently forcing minced fresh specimens through a 70-μm pore mesh and subjected to Ficoll gradient centrifugation. Peripheral blood lymphocytes were obtained by leukapheresis/elutriation and Miltenyi bead–purified as described.3 Dendritic cells (DCs) were generated by incubating magnetically purified CD14+ cells for 7 days with granulocyte-macrophage colony stimulating factor (20 ng/mL; PeproTech, Rocky Hill, NJ) and IL-4 (50 ng/mL; R&D Systems, Minneapolis, MN). Synovial fluid specimens were obtained from inflamed knees in a patient with gout and rheumatoid arthritis, respectively.
Immunohistochemistry was performed using anti-CD161 monoclonal antibody (mAb; DX12 and B199.2) and the ABC kit (Chemicon International, Temecula, CA). Horse serum (1/10 dilution) was used as a negative control.
K562 cells were transduced with CD32 as described,16,17 to generate artificial antigen presenting cells (aAPCs). Expression of CD161 and PILAR was achieved with the pLENTI6/V5-D-TOPO lentiviral system (Invitrogen, Carlsbad, CA). 293 cells were stably transfected with a pCEFL-kz plasmid encoding hemagglutinin (HA)–PILAR. aAPCs were γ-irradiated (100 Gy), washed, and loaded with anti-CD3 (OKT3; eBioscience, San Diego, CA) or anti-CD3 plus anti-CD28 (15E8; Chemicon International) antibodies (Abs) at room temperature. T cells were added at a 10:1 ratio. A2+ aAPCs were generated by transducing K562 cells with lentiviral human CD80 and HLA-A*0201. Positive cells were selected by fluorescence-activated cell sorting (FACS).
Proliferation index of stimulated T cells, defined as the average number of cell divisions that a cell in the original population has undergone, was calculated with FlowJo software (TreeStar, Ashland, OR) using a model adjustment for 8 peaks for every sample.
Antibodies and cytokines
Flow cytometry was performed using anti-CD8 (OKT8), anti-CD28 (CD28.2), anti-CD45RA (HI100), anti-CD80 (2D10.4), and anti-CD62L (DREG56) Abs from eBioscience; anti-CCR7 (3D12) and anti-CD3 (SP34-2) Abs from BD Biosciences (San Jose, CA); anti-CD161 (B199.2) and anti–HLA-A2 (BB7.2) from Serotec (Oxford, United Kingdom); and custom rabbit polyclonal anti-PILAR Ab plus anti–rabbit Ig-FITC (Biomeda, Foster City, CA).
To generate PILAR-specific Abs, the structure of PILAR was predicted using RASMOL software v. 2.4 (http://openrasmol.org). On the basis of a putative structural similarity, a good matching of cysteines and the absence of high content of gaps, we assigned a CD69-like fold to the new sequence. We procured a rabbit polyclonal Ab against a fragment of PILAR located on a region of the extracellular domain not conserved on structurally related molecules and putatively forming a β-strand at the C-terminus (SFAFLSADGVHSSRGFIDIK). The same immunogen emulsified with adjuvant was used to immunize 5 female Balb/c mice by Covance Research Products (Denver, PA) to generate PILAR-specific monoclonal Abs through standard procedures.
Cytokines in supernatants from stimulated T cells were quantified in a Bio-Plex assay (Bio-Rad, Hercules, CA) using the Human-27-Plex panel. Plates were read in a Bio-Plex Array Reader (Bio-Rad).
Immunoprecipitation and immunoblotting
K562 cells transduced with CD161 or CLEC2D (variant 1) and 293 T cells stably transfected with HA-PILAR or a HA-tagged irrelevant protein (MGSTMYDVDYASRSCRYSHWRHASRGYSVSKC) were lysed on ice for 45 minutes at 4°C in 500 μL of lysis buffer (50 mM TrisCl, pH 7.5, 15 mM EDTA, 100 mM NaCl, 0.1% (wt/vol) Triton X-100, 1 mM dithiothreitol, and 1 mM Pefabloc SC [Roche Diagnostics, Mannheim, Germany]). The lysates were incubated on ice for 90 minutes with 20 μL of anti–CD-161 (DX12) or 3 μL of anti-HA (HA.11; Covance, Denver, PA) mAbs, or an irrelevant rabbit IgG (NeoMarkers, Fremont, CA). After centrifugation at 13 000g for 10 minutes at 4°C, the supernatants were incubated with 25 μL protein G/protein A–agarose beads (Calbiochem, San Diego, CA) for 45 minutes at 4°C with continuous rotation and washed (30 seconds, 1000g). Pellets were resuspended in 30 μL Laemmli buffer, boiled, loaded onto a 15% sodium dodecyl sulfate–polyacrylamide electrophoresis gel, transferred to a nitrocellulose membrane, blocked, and incubated with the indicated primary Ab. Immunoreactive bands were developed using horseradish peroxidase–conjugated secondary Abs (Bio-Rad) and chemiluminescent substrate (Pierce Chemical, Rockford, IL). CLEC2D was detected using a mouse anti–human mAb (clone no. 402659; R&D Systems).
Online supplemental material
The characterization of PILAR, generation of CD161 fusion proteins, T-cell nucleofection, and enzyme-linked immunospot (ELISPOT) analysis are detailed (Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The kinetics of PILAR up-regulation of CD4 T cells, the blockade of IFN-γ responses by PILAR, the phenotype of tumor-associated T cells, and the up-regulation of IL-10 on PILAR blockade are shown in Figure S1.
Results
Identification of a novel transmembrane molecule expressed in human spleen
We aligned the amino acid (AA) sequence of the known or predicted molecules mapping at chromosome 12p12-p13 and exhibiting a C-type lectin domain. The alignment showed a conserved pattern (C-P-x(2)-W-x(2)-[YF]-x(3)-C-Y-x(2)-S-x(5)-W-x(2)-S-x(3)-C), determined by the AAs encoded by uninterrupted genomic sequences. This motif was used to screen the genomic sequences (≈2 Mb) mapping at human chromosome 12p12-p13, translated in their 6 possible open reading frames. We identified a continuous genomic sequence encoding a putative protein with 74% identity with this aforementioned pattern (CSGDWLGVRDKCFYFSDDTRNWTASKIFC). This sequence was found on a GenBank mRNA entry identified as C-type lectin domain family 2, member A (NM_207375). However, we were unable to amplify the entire open reading frame from cDNA derived from 5 leukocyte subsets, including B cells, T cells, NK cells, monocyte-derived dendritic cells, and monocytes, indicating that this clone corresponds to a different variant. To generate the leukocyte-specific cDNA sequence, we performed a rapid amplification of cDNA ends–polymerase chain reaction (RACE-PCR) with cDNA from human spleen. The full-length cDNA sequence showed 5 exons that encode a transmembrane molecule containing a truncated C-type lectin-like domain (Figure 1A; GenBank no. EF127467). Sequence similarity searches identified CD69 and CLEC2D/LLT1 as the 2 closest human molecules (Figure 1B). After we publicly disclosed our molecule, a truncated sequence encoding 159 identical amino acids, identified as CLEC2A variant 1, was independently deposited (GenBank no. EU095394).
PILAR encodes a new lymphocyte transmembrane molecule. (A) PILAR gene spans 5 exons encoding a predicted transmembrane molecule that contains a truncated C-type lectin domain in the extracellular region. (B) PILAR alignment with EU095394 and the 2 closest human molecules: Leukocyte markers CD69 and CLEC2D. (C) TaqMan analysis of the expression of PILAR in 10 different human tissues. (D) PILAR is predominantly expressed by naive lymphocyte, but not by myeloid cells. These data are representative of DCs from 2 different donors.
PILAR encodes a new lymphocyte transmembrane molecule. (A) PILAR gene spans 5 exons encoding a predicted transmembrane molecule that contains a truncated C-type lectin domain in the extracellular region. (B) PILAR alignment with EU095394 and the 2 closest human molecules: Leukocyte markers CD69 and CLEC2D. (C) TaqMan analysis of the expression of PILAR in 10 different human tissues. (D) PILAR is predominantly expressed by naive lymphocyte, but not by myeloid cells. These data are representative of DCs from 2 different donors.
Novel molecule is a ligand for the CD161 immunoreceptor
Real-time quantitative PCR analysis showed that, in addition to the spleen, the novel sequence is constitutively expressed in the thymus and small intestine, suggesting that it may be present in lymphocytes (Figure 1C). Consequently, PCR analysis showed mRNA signal in T and B cells but not in epithelial tumor cells (J.R.C.-R., unpublished data, September 2005). We found the highest expression of the new molecule in CD8 T cells, B lymphocytes, and naive CD4 T cells, but no measurable mRNA expression was detected in myeloid leukocytes or NK cells (Figure 1D).
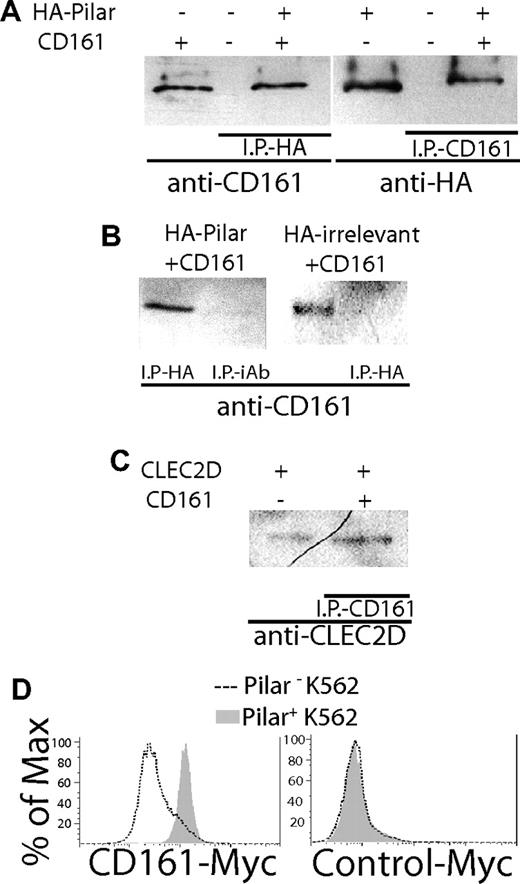
Because of the homology with CLEC2D/LLT1, we hypothesized that the new molecule could also bind to CD161. To determine this interaction, we stably transfected 293 cells with the novel sequence fused to a hemagglutinin (HA) epitope or with an HA-tagged irrelevant sequence. In parallel, we transduced CD161 into K562 cells. Lysates from CD161+ cells were then incubated for 2 hours with the HA+ lines ectopically expressing the new molecule or the irrelevant peptide, and immunoprecipitation was performed separately with anti-HA or anti-CD161 antibodies. Western blot analysis showed that immunoprecipitates from mixed lysates of CD161+K562 cells and 293 cells expressing the new molecule, performed with anti-HA antibodies, contained an approximately 40-kDa band that reacted with the anti-CD161 antibody. (Figure 2A left). As expected, immunoprecipitation with anti-CD161 antibodies showed that an approximately 20-kDa band reacted with the anti-HA antibody (Figure 2A right). In different experiments, we confirmed that the anti-CD161 Ab reacted with the expected approximately 40-kDa band when the lysates of CD161+ K562 cells and 293 cells expressing the new molecule were immunoprecipitated with the anti-HA Ab, but not when the immunoprecipitation was performed with an irrelevant Ab (Figure 2B left). Furthermore, no CD161 band was detected in HA-immunoprecipitates from mixed lysates of CD161+K562 cells and 293 cells expressing the irrelevant peptide (Figure 2B right). As expected, immunoprecipitates from lysates of 293 cells ectopically expressing the previously reported CD161 ligand LLT1/CLEC2D13,15 incubated with lysates from CD161+ cells produced an approximately 25-kDa single band reacting with anti-CLEC2D antibodies (Figure 2C). No band was detectable when CD161-expressing K562 cells were incubated with the parental 293 cells, or when 293 cells expressing the novel sequence were incubated with untransduced K562 cells (unpublished data).
PILAR is a ligand for CD161. (A) 293 cells transfected with HA-PILAR and K562 cells transduced with CD161 are identified as “+,” whereas “−” denotes the parental cell line. Cell extracts were incubated for 2 hours and immunoprecipitation (IP) was performed with either anti-HA Ab (HA11; left; Covance, Berkeley, CA) or anti-CD161 Ab (right), plus protein G/protein A agarose beads. The immunoprecipitates were run in a Western blot and analyzed for their reactivity against the opposite Ab. A positive control including either CD161+ cells (left) or PILAR+ cells (right) without IP was also included. (B) Coincubated PILAR+ and CD161+ cell extracts immunoprecipitated with an irrelevant Ab (iAb; left), or coincubated lysates from K562 cells ectopically expressing CD161 plus 293 cells expressing an irrelevant HA-tagged sequence (right) did not contain CD161. A positive control including CD161+ cells was included. (C) Immunoprecipitates of coincubated lysates from CLEC2D+ and CD161+ cells performed with an anti-CD161 Ab contained the expected approximately 25-kDa CLEC2D band. A positive control, including CLEC2D+ cells (left) without IP, was included. (D) Pilar transduced/untransduced (+/−) K562 cells were incubated with a CD161-Myc (left) or a control-Myc (GenBank no. NM_007048; right) protein (0.5 μg/mL, 45 minutes). Specific binding was confirmed 3 times with an anti–Myc-FITC Ab (Sigma-Aldrich, St Louis, MO).
PILAR is a ligand for CD161. (A) 293 cells transfected with HA-PILAR and K562 cells transduced with CD161 are identified as “+,” whereas “−” denotes the parental cell line. Cell extracts were incubated for 2 hours and immunoprecipitation (IP) was performed with either anti-HA Ab (HA11; left; Covance, Berkeley, CA) or anti-CD161 Ab (right), plus protein G/protein A agarose beads. The immunoprecipitates were run in a Western blot and analyzed for their reactivity against the opposite Ab. A positive control including either CD161+ cells (left) or PILAR+ cells (right) without IP was also included. (B) Coincubated PILAR+ and CD161+ cell extracts immunoprecipitated with an irrelevant Ab (iAb; left), or coincubated lysates from K562 cells ectopically expressing CD161 plus 293 cells expressing an irrelevant HA-tagged sequence (right) did not contain CD161. A positive control including CD161+ cells was included. (C) Immunoprecipitates of coincubated lysates from CLEC2D+ and CD161+ cells performed with an anti-CD161 Ab contained the expected approximately 25-kDa CLEC2D band. A positive control, including CLEC2D+ cells (left) without IP, was included. (D) Pilar transduced/untransduced (+/−) K562 cells were incubated with a CD161-Myc (left) or a control-Myc (GenBank no. NM_007048; right) protein (0.5 μg/mL, 45 minutes). Specific binding was confirmed 3 times with an anti–Myc-FITC Ab (Sigma-Aldrich, St Louis, MO).
In confirmatory experiments using a different approach, a soluble chimeric CD161-Myc epitope protein, but not an irrelevant-Myc protein, specifically bound to K562 cells ectopically expressing the entire open reading frame of the novel molecule.12,13 In contrast, no binding was observed to parental K562 cells (Figure 2D). Together, these data show that the novel transmembrane molecule interacts with CD161.
New CD161 ligand is transiently up-regulated in activated lymphocytes
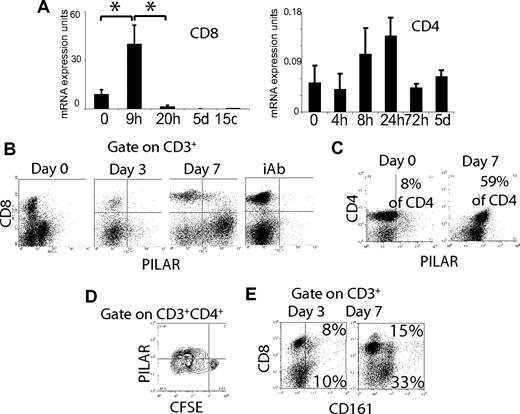
On the basis of the similarities with CD69, we postulated that the new CD161 ligand could be transiently up-regulated on lymphocyte activation.18 To test this hypothesis, we isolated peripheral CD8 T cells and incubated them with phytohemagglutinin (PHA) for different periods. The new CD161 ligand was maximally up-regulated 9 hours after activation of peripheral CD8 T cells (Figure S1A), with the mRNA levels rapidly decreasing thereafter.
To verify this pattern of mRNA up-regulation on CD3/CD28-dependent stimulation, we used a previously described artificial aAPC system.16,17 Briefly, (MHC-I–deficient) K562 cells that were transduced with CD32 were irradiated and coated with different concentrations of activating CD3 and CD28 antibodies. CD3/CD28 activation also induced a maximum mRNA overexpression of the CD161 ligand at 9 hours on CD8 T cells (Figure 3A left; P < .05, Mann-Whitney), although the up-regulation was lower than that induced by PHA. Lower but more persistent mRNA levels were found on CD4 T cells (Figure 3A right). This rapid up-regulation was followed by a decrease in expression below the constitutive levels found on naive lymphocytes after more than 20 hours. On the basis of this overexpression pattern and the presence of a transmembrane domain, we named the new CD161 ligand proliferation-induced lymphocyte-associated receptor (PILAR).
CD3/CD28 stimulation induces PILAR up-regulation. (A) CD3/CD28 (100 ng/mL each) stimulation of peripheral immunopurified CD8+ (left) or CD4+ (right) T cells (106/mL) resulted in a transient up-regulation of PILAR mRNA, with maximum levels between 9 and 24 hours. These data are representative of 3 independent experiments (total of 6 samples). Error bars indicate standard error. (B) Peripheral CD3+ T cells stimulated for different periods with aAPCs coated with agonistic anti-CD3/CD28 Abs (100 ng/mL) up-regulated PILAR at the protein level. This staining was performed with a PILAR-specific polyclonal Ab. These data, representative of 3 independent experiments, were confirmed using a monoclonal Ab (see also Figure S1). NTC indicates irrelevant rabbit Ab (Lab Vision, Freemont, CA) plus anti–rabbit Ig-FITC (Biomeda; top). (C) CD4 T cells contained in peripheral blood mononuclear cells stimulated for 7 days with aAPCs coated with agonistic CD3/CD28 Abs (100 ng/mL) up-regulated PILAR at the protein level in another independent experiment. (D) More than 40% of proliferating CD4 T cells from a different donor, but not cells that did not proliferate, expressed detectable levels of PILAR using the polyclonal Ab after 7 days of stimulation under identical conditions. (E) Up-regulation of CD161 on the peripheral T cells of a different donor after 3 and 7 days of CD3/CD28 stimulation using aAPCs (see also Figure S1F). Numbers in quadrants indicate percentage of gated cells in that quadrant (panels C and D, percentage of total CD4 T cells; panel E, percentage of total T cells).
CD3/CD28 stimulation induces PILAR up-regulation. (A) CD3/CD28 (100 ng/mL each) stimulation of peripheral immunopurified CD8+ (left) or CD4+ (right) T cells (106/mL) resulted in a transient up-regulation of PILAR mRNA, with maximum levels between 9 and 24 hours. These data are representative of 3 independent experiments (total of 6 samples). Error bars indicate standard error. (B) Peripheral CD3+ T cells stimulated for different periods with aAPCs coated with agonistic anti-CD3/CD28 Abs (100 ng/mL) up-regulated PILAR at the protein level. This staining was performed with a PILAR-specific polyclonal Ab. These data, representative of 3 independent experiments, were confirmed using a monoclonal Ab (see also Figure S1). NTC indicates irrelevant rabbit Ab (Lab Vision, Freemont, CA) plus anti–rabbit Ig-FITC (Biomeda; top). (C) CD4 T cells contained in peripheral blood mononuclear cells stimulated for 7 days with aAPCs coated with agonistic CD3/CD28 Abs (100 ng/mL) up-regulated PILAR at the protein level in another independent experiment. (D) More than 40% of proliferating CD4 T cells from a different donor, but not cells that did not proliferate, expressed detectable levels of PILAR using the polyclonal Ab after 7 days of stimulation under identical conditions. (E) Up-regulation of CD161 on the peripheral T cells of a different donor after 3 and 7 days of CD3/CD28 stimulation using aAPCs (see also Figure S1F). Numbers in quadrants indicate percentage of gated cells in that quadrant (panels C and D, percentage of total CD4 T cells; panel E, percentage of total T cells).
Independent FACS analysis performed with 2 different endotoxin-free (polyclonal and monoclonal) PILAR-specific Abs (Figure S1B) confirmed that PILAR surface expression was noticeable in less than 3% of CD4 naive T cells (Figure 3B; Figure S1C), became evident on approximately 15% to 18% of CD4 T cells after 5 days of CD3/CD28-stimulation (Figure S1C), and was detected on approximately 60% of CD4 but only on a minority of CD8 T cells after longer CD3/CD28 stimulation periods (Figure 3B,C). Of note, PILAR protein expression was restricted mostly to proliferating lymphocytes (Figure 3D). Similar results were obtained with polyclonal and monoclonal Abs in 3 independent experiments. Surprisingly, intracellular PILAR protein expression was clearly detectable after permeabilization on virtually all CD3/CD28-activated CD4 and CD8 T cells at day 3 (Figure S1D), indicating posttranslational regulation. Correspondingly, Psort-II software (http://psort.ims.u-tokyo.ac.jp/form2.html) predicted an endoplasmic reticulum membrane retention-like motif, PKYF, at the C-terminus of PILAR. Therefore, under physiologic conditions, a differential retardation of the transport of PILAR to the cell surface on CD4 and CD8 T cells might determine the timing of PILAR surface expression. Interestingly, most CD8 T cells but only half of CD4 T lymphocytes exhibited PILAR on their surface after only 5 days of stimulation with PHA (Figure S1C).
In addition, discordant protein versus mRNA expression is frequently observed on primed T cells, as initially they undergo translational attenuation.19-21 Correspondingly, we found a rapid PILAR mRNA degradation shortly after T-cell activation (Figure S1E), indicating that uncoupled mRNA and protein expression partially accounts for the disparity between mRNA and protein expression patterns with respect to time.
PILAR enhances TCR-mediated proliferation of human T cells
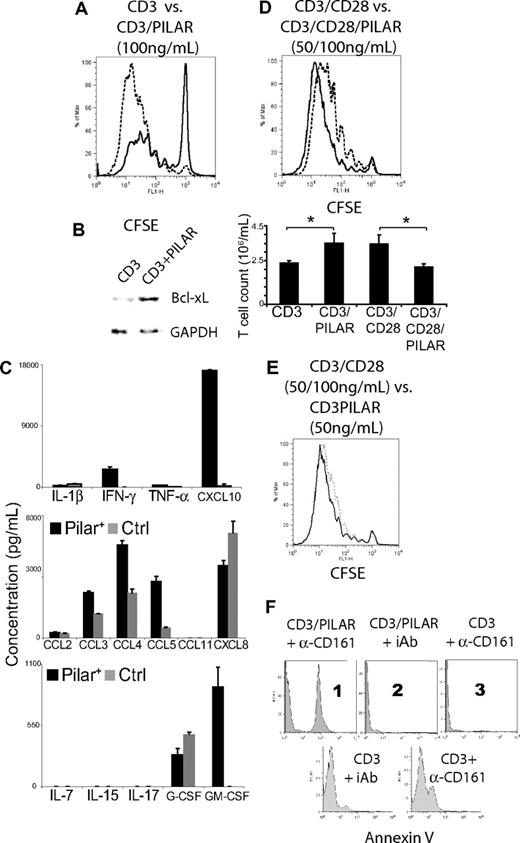
Because PILAR was rapidly up-regulated on activated T cells, we next tested the effect of increased PILAR availability on T-cell proliferation. We were not able to transfect naive T cells using multiple commercially available transfection reagents, which is consistent with previous reports.22 In addition, the use of nucleofection technology resulted in the nonspecific expression of surface PILAR in naive T cells nucleofected with a control plasmid. (Figure S1G). We therefore increased PILAR availability in our system by ectopically expressing PILAR on the PILAR− aAPCs. To define activating conditions of different stringency, we performed a proliferation assay using decreasing concentrations of anti-CD3 mAb (500 ng to 0.5 pg/mL), and a constant concentration of 100 ng/mL of anti-CD28 mAb. On the basis of 5-day proliferation analysis of naive T cells, we defined anti-CD3 concentrations of 100 ng/mL and 0.5 ng/mL as “optimal” and “limiting,” respectively. Hence, under optimal TCR-stimulation conditions but in the absence of CD28 signaling, ectopic PILAR dramatically enhanced the proliferation of naive T cells (53% increase in total proliferating cells; Figure 4A). Because costimulatory molecules prevent the induction of apoptosis by increasing Bcl-xL,23 we next compared its expression on T cells stimulated with agonistic CD3 antibodies in the presence or the absence of PILAR. As shown in Figure 4B, increasing PILAR availability augmented Bcl-xL levels and, to a much lesser extent, c-FLIP (not shown), after 24 hours of stimulation. Together, these data indicate that PILAR enhances the expansion of TCR-stimulated T cells by increasing their survival through enhanced expression of antiapoptotic proteins.
PILAR engagement of CD161 enhances T-cell proliferation. (A) CD3 (100 ng/mL)–mediated proliferation of naive T cells is dramatically enhanced at day 7 in the presence of ectopically expressed PILAR (dotted line), compared with mocked transduced aAPCs (thick line). (B) Ectopic expression of PILAR increases antiapoptotic Bcl-xL on T cells after 24 hours of stimulation with aAPCs coated with 100 ng/mL anti-CD3 Ab. (C) Comparison of the cytokine profiles of T cells activated for 7 days with 100 ng/mL anti-CD3 Ab, in the presence or the absence of ectopic PILAR. Levels are normalized by final cell numbers. Error bars indicate SE. (D top) Increasing PILAR availability by ectopic expression on the aAPCs in the presence of CD28 costimulation (100 ng/mL) resulted in a decrease of the CD3 (50 ng/mL)–mediated proliferation of naive T cells at day 7. Dotted line denotes cells incubated in the presence of ectopic PILAR, compared with mock transduced aAPCs (thick line). (D bottom) Final T-cell counts after 7 days of stimulation under the conditions detailed in panel A or D. Error bars indicate SE. (E) T cells expanded at a comparable rate in the presence of CD3 plus CD28 costimulation (50 and 100 ng/mL of each Ab) and in the presence of CD3 (50 ng/mL of Ab) plus ectopic PILAR expressed on the aAPCs. These results are representative of 2 independent experiments (total of 6 observations). (F) All data are representative of at least 3 experiments. (Top) Blockade of CD161 with 10 μg/mL Ab (B199.2) but not incubation with an identical concentration of an irrelevant IgG induces apoptosis on T cells activated with 100 ng/mL anti-CD3 Ab for 24 hours only in the presence of ectopic PILAR (1, 2, and 3). (Bottom) CD161-blocked T cells stimulated for 3 days also initiated apoptosis.
PILAR engagement of CD161 enhances T-cell proliferation. (A) CD3 (100 ng/mL)–mediated proliferation of naive T cells is dramatically enhanced at day 7 in the presence of ectopically expressed PILAR (dotted line), compared with mocked transduced aAPCs (thick line). (B) Ectopic expression of PILAR increases antiapoptotic Bcl-xL on T cells after 24 hours of stimulation with aAPCs coated with 100 ng/mL anti-CD3 Ab. (C) Comparison of the cytokine profiles of T cells activated for 7 days with 100 ng/mL anti-CD3 Ab, in the presence or the absence of ectopic PILAR. Levels are normalized by final cell numbers. Error bars indicate SE. (D top) Increasing PILAR availability by ectopic expression on the aAPCs in the presence of CD28 costimulation (100 ng/mL) resulted in a decrease of the CD3 (50 ng/mL)–mediated proliferation of naive T cells at day 7. Dotted line denotes cells incubated in the presence of ectopic PILAR, compared with mock transduced aAPCs (thick line). (D bottom) Final T-cell counts after 7 days of stimulation under the conditions detailed in panel A or D. Error bars indicate SE. (E) T cells expanded at a comparable rate in the presence of CD3 plus CD28 costimulation (50 and 100 ng/mL of each Ab) and in the presence of CD3 (50 ng/mL of Ab) plus ectopic PILAR expressed on the aAPCs. These results are representative of 2 independent experiments (total of 6 observations). (F) All data are representative of at least 3 experiments. (Top) Blockade of CD161 with 10 μg/mL Ab (B199.2) but not incubation with an identical concentration of an irrelevant IgG induces apoptosis on T cells activated with 100 ng/mL anti-CD3 Ab for 24 hours only in the presence of ectopic PILAR (1, 2, and 3). (Bottom) CD161-blocked T cells stimulated for 3 days also initiated apoptosis.
Significantly, PILAR overexpression profoundly increased the production of IFN-γ and the IFN-responsive chemokine CXCL10 by stimulated T cells (Figure 4C). Similarly, the inflammatory chemokines CCL3, CCL4, and CCL5, as well as GM-CSF, which increases the production of myeloid cells, were dramatically up-regulated by ectopic PILAR signaling. In contrast, PILAR had no effect on CXCL8 or G-CSF secretion.
PILAR/CD161 interactions are critical for CD3-mediated proliferation
Surprisingly, increased PILAR availability in the presence of CD3 plus CD28 signaling decreased T-cell proliferation (Figure 4D). The proliferation index (4 ± 0.2) and final cell count (3.4 ± 0.5 × 106 cells/mL) of CD3/CD28-stimulated T cells were significantly higher than that of CD3/CD28/PILAR-stimulated lymphocytes (3.2 ± 0.19 and 2.1 ± 0.1 × 106 cells/mL, respectively; n ≥ 6/group; P < .01 for both, Mann-Whitney). Interestingly, PILAR and CD28 were equipotent in our system, because both the proliferation indexes (3.8 ± 0.2) and final cell counts (3.5 ± 0.5 × 106 cells/mL) of CD3/PILAR-stimulated T cells were not statistically different from that of CD3/CD28-stimulated lymphocytes (Figure 4D top, E). However, the levels of PILAR on the surface of activated T cells may be significantly lower under physiologic conditions.
Consequently, we hypothesized that PILAR enhances T-cell proliferation through CD161 under suboptimal activating conditions and triggers an alternative regulatory pathway potentially through a different receptor in vigorously activated lymphocytes. To test this possibility, we activated human naive T cells under optimal CD3 stimulation conditions in the absence or the presence of ectopic PILAR and anti-CD161 Abs. Strikingly, in the presence of ectopic PILAR, more than 37% lymphocytes underwent apoptosis when an anti-CD161Ab, but not a control Ig, was added to the media, as indicated by annexin V staining at 24 hours (Figure 4F top, 1 and 2). The presence of the anti-CD161 Ab did not result in detectable apoptosis or cell death in the absence of ectopic PILAR at 24 hours in multiple independent experiments, ruling out a direct cytotoxic effect (Figure 4F top 3). However, on the initial expression of PILAR on the cell surface, approximately 24% of T cells activated for 3 days in the presence of the anti-CD161 Ab also started undergoing apoptosis (Figure 4F bottom).
PILAR blockade abrogates the expansion of both CD4 and CD8 T cells
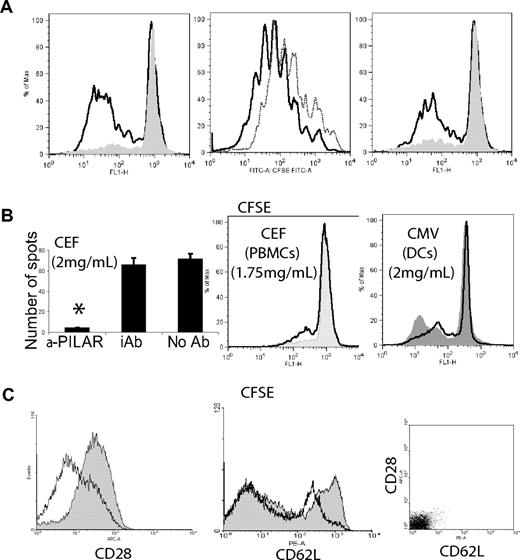
To confirm the critical role of PILAR on T-cell proliferation, we next examined whether PILAR blockade affected TCR-mediated signaling. Because the PILAR-specific rabbit polyclonal Ab showed its specificity by staining the same proportions of T cells as the mouse monoclonal Ab in stimulation experiments, we took advantage of the broader range of epitope coverage of the former for the neutralization of PILAR. In the presence of anti-PILAR polyclonal Abs, the CFSE dilution of magnetically purified T cells expanded under optimal conditions of CD3 stimulation was significantly lower than that from cells that were incubated with an irrelevant Ig. Furthermore, this signal was comparable to that of unstimulated lymphocytes (Figure 5A). A high level of costimulation (anti-CD28 at 100 ng/mL) was required to rescue the inhibition of proliferation mediated by PILAR blockade, although a decrease in the strong CD3/CD28-mediated expansion was still apparent. Comparable results were obtained with higher concentrations of plate-bound Ab. Under limiting conditions of TCR engagement, we observed a dramatic impairment of proliferation after the addition of an anti-PILAR Ab, even in the presence of costimulation (Figure 5A right). In addition, PILAR blockade abrogated the generation of IFN-γ–secreting, antigen-specific HLA-A2 CD8 T cells responding to a cocktail of 23 immunogenic, class I–restricted 8-11mers24 presented by A2+ aAPCs or autologous dendritic cells (Figure 5B left; Figure S1H; P = .02, Mann-Whitney test), as well as the proliferation of peripheral blood mononuclear cells (PBMCs) induced by these specific antigens (Figure 5B center). Addition of anti-PILAR Abs also impaired the proliferation of CD4 and CD8 T cells induced by autologous DCs pulsed with 138 different Cytomegalovirus 15-mers with 11 amino acid overlaps (Figure 5B right). These data confirm that endogenous PILAR, depending on the relative strength of the TCR signal and the degree of costimulation, is critical for the proliferation of naive T cells.
PILAR blockade impairs T-cell proliferation and IFN-γ production. (A) Miltenyi bead–purified, CFSE-labeled peripheral T cells were stimulated for 5 days with aAPCs coated with the following agonistic Abs: (left) αCD3 (100 ng/mL), (center) αCD3 plus αCD28 (100 ng/mL, each), (right) αCD3 plus αCD28 (0.5 and 100 ng/mL, respectively), in the presence of 20 μg/mL of either blocking anti-PILAR Abs (solid or dotted line) or an irrelevant rabbit Ab (open thick line). These results are representative of 4 independent experiments. (B) These results are representative of at least 2 independent experiments performed in quadruplicate. (left) IFN-γ ELISPOT analysis of PBMCs from an A2+ donor, stimulated for 7 days with A2+CD80+ aAPCs (10:1 ratio) pulsed with the CEF peptide pool (2 μg/mL of each peptide; Mabtech, Nacka Strand, Sweden), in the presence of20 μg/mL of anti-PILAR or an irrelevant Ab. (Center) CFSE-labeled PBMCs (2 × 106/mL) were incubated for 7 days with the CEF peptide pool in the presence of 20 μg/mL of anti-PILAR Abs (solid) or an irrelevant Ab (open). (Right) PBMCs stimulated for 7 days with autologous monocyte-derived dendritic cells (10:1 ratio, 106 total cells/mL), pulsed with pool of 138 Cytomegalovirus peptides (2 μg/mL; pp65 sequence, strain AD169; BD Bioscences), in the presence of 20 μg/mL of anti-PILAR (opened) or an irrelevant Ab (solid). Error bars indicate SE; *P = .02. (C) CD3/CD28 stimulation of peripheral T cells for 2 days in the presence of 20 μg/mL of anti-PILAR Abs (opened) resulted in a decrease in the expression of CD28 and CD62L, compared with the same lymphocytes stimulated with an irrelevant Ab (solid). CD28 and CD62L signal detected after staining with isotype control Ig is indicated. The experiment was repeated 3 times with similar results.
PILAR blockade impairs T-cell proliferation and IFN-γ production. (A) Miltenyi bead–purified, CFSE-labeled peripheral T cells were stimulated for 5 days with aAPCs coated with the following agonistic Abs: (left) αCD3 (100 ng/mL), (center) αCD3 plus αCD28 (100 ng/mL, each), (right) αCD3 plus αCD28 (0.5 and 100 ng/mL, respectively), in the presence of 20 μg/mL of either blocking anti-PILAR Abs (solid or dotted line) or an irrelevant rabbit Ab (open thick line). These results are representative of 4 independent experiments. (B) These results are representative of at least 2 independent experiments performed in quadruplicate. (left) IFN-γ ELISPOT analysis of PBMCs from an A2+ donor, stimulated for 7 days with A2+CD80+ aAPCs (10:1 ratio) pulsed with the CEF peptide pool (2 μg/mL of each peptide; Mabtech, Nacka Strand, Sweden), in the presence of20 μg/mL of anti-PILAR or an irrelevant Ab. (Center) CFSE-labeled PBMCs (2 × 106/mL) were incubated for 7 days with the CEF peptide pool in the presence of 20 μg/mL of anti-PILAR Abs (solid) or an irrelevant Ab (open). (Right) PBMCs stimulated for 7 days with autologous monocyte-derived dendritic cells (10:1 ratio, 106 total cells/mL), pulsed with pool of 138 Cytomegalovirus peptides (2 μg/mL; pp65 sequence, strain AD169; BD Bioscences), in the presence of 20 μg/mL of anti-PILAR (opened) or an irrelevant Ab (solid). Error bars indicate SE; *P = .02. (C) CD3/CD28 stimulation of peripheral T cells for 2 days in the presence of 20 μg/mL of anti-PILAR Abs (opened) resulted in a decrease in the expression of CD28 and CD62L, compared with the same lymphocytes stimulated with an irrelevant Ab (solid). CD28 and CD62L signal detected after staining with isotype control Ig is indicated. The experiment was repeated 3 times with similar results.
Notably, we observed a reduction in the surface expression of CD28 when naive T cells were stimulated with CD3/CD28 for 2 days in the presence of anti-PILAR Ab, compared with cells cultured with an irrelevant Ab (Figure 5C left). A major decrease in the expression of surface CD62L, which mediates lymphocyte homing during inflammation, was also observed on PILAR blockade (Figure 5C center). A comparable decrease was observed at day 7 and when lymphocytes were activated through PHA in the presence or the absence of anti-PILAR Ab (unpublished data). Taken together, these results suggest that, in vivo, PILAR may modulate the capacity of T lymphocytes to home to lymph nodes through CD62L, where they can undergo a robust expansion in the presence of increased CD28 signaling.
PILAR is expressed in different pathologic conditions
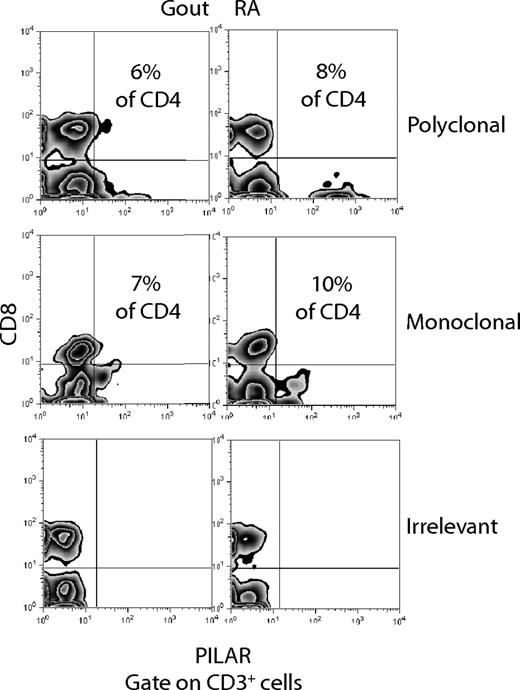
We next investigated a possible role for PILAR in the pathogenesis of different human diseases. To address this possibility, we first obtained 2 synovial fluid specimens from rheumatoid arthritis and gout patients. As expected, most cells exhibited FSC/SSC properties of neutrophils and macrophages, although we also found a noteworthy number of CD4 T cells (3% of total cells in rheumatoid arthritis; 4% of total cells in gout) and, to a lesser extent, of CD8 T cells (≈1% in of total cells both cases). As we observed previously on newly primed T cells, surface PILAR protein was found only on CD4 but not on CD8 T cells. Thus, approximately 10% of total CD4 T cells in rheumatoid synovial fluid and approximately 7% of CD4 T cells in the gout specimen expressed PILAR (Figure 6). These proportions are more than 3-fold higher than those found on naive CD4 T cells. Rabbit polyclonal and mouse monoclonal Abs produced similar results, corroborating the specificity of our analysis. Because PILAR is up-regulated on activated T cells and boosts robust T-cell proliferation, these results suggest a potential role for PILAR in the pathogenesis of joint inflammation. Ongoing studies will determine the extent of PILAR involvement in different autoimmune diseases.
PILAR is expressed on T cells in inflammatory synovial fluid. Proportions of PILAR+CD3+CD8− (CD4+) T cells in synovial fluid specimens from rheumatoid arthritis and gout patients using rabbit polyclonal or mouse monoclonal PILAR-specific Abs. Staining with an irrelevant mouse IgG (BD PharMingen, San Diego, CA) is additionally shown.
PILAR is expressed on T cells in inflammatory synovial fluid. Proportions of PILAR+CD3+CD8− (CD4+) T cells in synovial fluid specimens from rheumatoid arthritis and gout patients using rabbit polyclonal or mouse monoclonal PILAR-specific Abs. Staining with an irrelevant mouse IgG (BD PharMingen, San Diego, CA) is additionally shown.
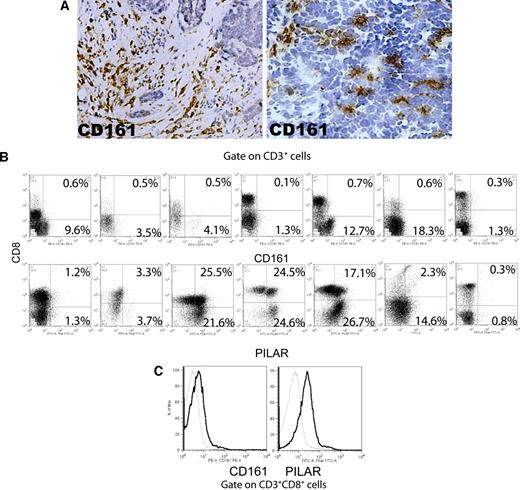
In addition, to define the expression of PILAR in human cancer, we procured single-cell suspensions from 7 unselected, mechanically dissociated human ovarian carcinoma specimens. More than 92% of CD8 T cells in these samples exhibited an effector memory (CCR7−CD69−CD45RA+/−) phenotype (Figure S1J). These cells did not express the invariant (Vα24-JαQ) TCR chain, suggesting that NK T cells do not play a significant role in the physiopathology of established ovarian carcinomas. Although we observed a significant infiltration of CD161+ cells in both the stroma and the tumor islets of most specimens analyzed (Figure 7A), CD161 was found on less than 1% of CD8 T cells by flow cytometry (Figure 7B top). In contrast, we found variable levels of CD161 on autologous tumor-derived CD4 T cells. Because CD161 expression in healthy donors is mostly found on effector memory T cells,12 these data imply that CD161-mediated stimulation may be specifically abrogated by the tumor microenvironment on effector T cells. Finally, most tumors contained CD4 and CD8 tumor-infiltrating T cells that expressed PILAR (Figure 7B top), suggesting that CD4+ lymphocytes, which include CD25+Foxp3+ regulatory T cells,25 may be more susceptible to PILAR-mediated expansion. Importantly, CD3/CD28-mediated T-cell stimulation resulted in a significant up-regulation of both surface CD161 and PILAR on CD8 T cells (Figure 7C), suggesting that antitumor T cells could be recovered from tumor-induced immunosuppression and expanded ex vivo through PILAR/CD161 manipulation.
PILAR, but not CD161, is expressed by a proportion of both CD4+ and CD8+ tumor-infiltrating T cells. (A) CD161 is expressed by individual cells in the ovarian carcinoma microenvironment. Data are representative of 20 immunostained specimens. Original magnification, ×100 (left); ×200 (right). Photomicrographs were taken with a Nikon Eclipse 801 (Nikon, Melville, NY) at 10× (left) or 20× (right) 0.30 plan Fluor objective. Images were acquired using Micropublisher v. 5.0 (QImaging, Surrey, BC) and processed using Image ProPlus v. 5.1.1 (Media Cybernetics, Bethesda, MD). (B) PILAR (polyclonal Ab) is expressed by tumor-associated CD4 and CD8 T cells, whereas CD161 is only found on CD4 T lymphocytes. Gated on viable cells. Numbers in quadrants indicate percentage of CD3+ T cells in that quadrant. (C) Expression of both PILAR and CD161 can be recovered on tumor-associated cytotoxic T cells ex vivo by CD3/CD28 (100 ng/mL each) stimulation. Dotted, tumor-derived unstimulated CD8+ T cells; thick, the same cells after stimulation with CD3/CD28 for 5 days. Gated on viable cells.
PILAR, but not CD161, is expressed by a proportion of both CD4+ and CD8+ tumor-infiltrating T cells. (A) CD161 is expressed by individual cells in the ovarian carcinoma microenvironment. Data are representative of 20 immunostained specimens. Original magnification, ×100 (left); ×200 (right). Photomicrographs were taken with a Nikon Eclipse 801 (Nikon, Melville, NY) at 10× (left) or 20× (right) 0.30 plan Fluor objective. Images were acquired using Micropublisher v. 5.0 (QImaging, Surrey, BC) and processed using Image ProPlus v. 5.1.1 (Media Cybernetics, Bethesda, MD). (B) PILAR (polyclonal Ab) is expressed by tumor-associated CD4 and CD8 T cells, whereas CD161 is only found on CD4 T lymphocytes. Gated on viable cells. Numbers in quadrants indicate percentage of CD3+ T cells in that quadrant. (C) Expression of both PILAR and CD161 can be recovered on tumor-associated cytotoxic T cells ex vivo by CD3/CD28 (100 ng/mL each) stimulation. Dotted, tumor-derived unstimulated CD8+ T cells; thick, the same cells after stimulation with CD3/CD28 for 5 days. Gated on viable cells.
Discussion
Here we report the identification of a novel modulator of T-cell expansion named PILAR. PILAR is markedly up-regulated on the surface of CD4 T cells and supports CD3 antibody-dependent and antigen-specific T-cell proliferation through CD161 engagement. Blockade of endogenous PILAR with specific Abs completely abrogates the proliferation of human T cells induced by CD3 agonistic Abs and decreases CD3/CD28-induced T-cell expansion. The requirement for PILAR-induced signaling is not restricted to Ab-induced activation, because PILAR neutralization also prevents T-cell expansion when the proliferating signal is triggered by specific antigens presented by autologous dendritic cells. In addition, PILAR overexpression profoundly enhances T-cell expansion, which is at least partially mediated by the up-regulation of antiapoptotic Bcl-xL. PILAR blockade decreases the surface expression of CD28 and CD62L at early stages of T-cell activation, when proliferation is still unappreciable. In contrast, reflecting a lower level of activation and proliferation, the levels of both CD62L and CD28 on proliferating cells are significantly lower after 7 days of CD3/CD28 stimulation in the absence of PILAR blockade (not shown). These results suggest a mechanism whereby PILAR predominantly contributes to a robust (CD28-mediated) expansion of human T cells anchored through CD62L at lymphatic locations.
Although PILAR exhibits an overall stimulatory effect on T-cell expansion, we found that blocking CD161 engagement results in apoptotic death on naive and early activated T cells within 24 hours in the presence of increased PILAR availability. This was not caused by a direct cytotoxic effect of the anti-CD161 Ab, because there was no apoptosis on primed lymphocytes in the absence of ectopic PILAR. It is possible that PILAR induces T-cell apoptosis through a second receptor present on naive T cells that is up-regulated shortly after TCR stimulation and that this apoptotic cell death is prevented by the engagement of CD161. Because PILAR decreases the proliferation of CD3/CD28 costimulated T cells, it is tempting to speculate that this putative alternative receptor is also up-regulated on CD28 signaling. Collectively, these results point to a model whereby PILAR, depending on the receptor preferentially expressed at different stages of T-cell activation, either increases T-cell survival through CD161 or induces apoptotic death through a different mediator.
Importantly, we found significantly higher levels of PILAR expression on T cells compared with any other cell type. Using different polyclonal and monoclonal Abs, we found intracellular PILAR on virtually all T cells after CD3/CD28 activation for 3 days, although surface expression was mostly restricted to proliferating CD4 T cells and became evident on most cells 4 days later. We were not able to ectopically express PILAR on human T cells without inducing their activation or the unspecific up-regulation of surface PILAR, but we could express PILAR ectopically on the surface of the same cells that bear the CD3/CD28 activating antibodies. Although this represents a safe system to investigate the effect of increasing PILAR availability in a human system without preactivating T cells, PILAR is not endogenously expressed on myeloid APCs. Consequently, in vivo, T cells (predominantly CD4 lymphocytes) must present PILAR to themselves and to each other on neighbor T cells within the same cluster for engagement to CD161 expressed in a reciprocal manner. Therefore, our results suggest that a finely regulated differential retardation of the transport of PILAR to the cell surface determines the timing of productive PILAR surface expression on activated CD4 T helper cells, but not on CD8 lymphocytes. T to T costimulatory interactions have previously been described for the CD27-CD70 pair.10 However, unlike CD70-CD27 interactions, which affect predominantly secondary responses,5,9 PILAR engagement of CD161 should predominantly affect the initial priming. Interestingly, a recent report described how the ectopic expression of costimulatory ligands on T lymphocytes dramatically enhanced auto- and trans-costimulation of T cells in vivo in a similar manner.11 The confirmation of these biologic effects through the manipulation of PILAR expression in animal models may be complicated by the different expression patterns of CD161 in humans and mice.12 In addition, we did not find any mouse equivalent located in the counterpart gene locus. Nevertheless, at least 3 mouse genes (Clec2ag, Clec2af, and Clec2ai) exhibit a high degree of similarity with both PILAR and human CLEC2D, suggesting that several functionally related molecules may also cooperatively modulate T-cell expansion in mice.
In agreement with previous reports about posttranscriptional regulation on primed T cells,19-21 PILAR mRNA and protein (confirmed with monoclonal and polyclonal Abs) exhibited discordant expression. Uncoupled mRNA and protein expression are frequently observed on primed T cells, which use rapid mRNA degradation for turning gene expression off in an activation-dependent manner.26 We found a more than 90-fold decrease in the PILAR mRNA levels after only 45 minutes of transcriptional blockade, significantly shorter than the usual half-life of standard mRNA transcripts. Primed T cells, unlike restimulated T cells,20 undergo transient translational attenuation.21 These effects are mediated by the phosphorylation of eIF2, which alters its capacity to be “recharged” by the nucleotide-exchange factor eIF2B. This leads to a decrease in the availability of active initiation complexes.20 Correspondingly, a recent report showed that approximately 50% of genes expressed by primed CD8 T cells exhibited discordant protein versus mRNA expression.19 When the researchers compared primed CD8 T cells to the naive parental population, they found that effector CD8 T cells yield approximately 4 times more total RNA than naive cells, whereas total protein content in effector cells was increased only 1.5 times more than that of naive cells. In independent studies, critical cytokines for T-cell differentiation, including IL-4, also showed uncoupled mRNA and protein levels.20
We found that PILAR is present in different pathologic conditions. First, we observed that, up to 10% of synovial fluid CD4 T cells exhibited surface PILAR. Because PILAR enhances T-cell activation and proliferation, these data suggest a possible role for PILAR in the pathogenesis of joint inflammation. Interestingly, a recent genome-wide association study identified a significant accumulation of single-nucleotide polymorphisms in patients with type 1 diabetes in the narrow region on chromosome 12p13 to which PILAR maps.27 In addition, PILAR was expressed on both ovarian cancer–infiltrating CD4 and CD8 T cells. However, the expression of the stimulatory receptor CD161 was restricted to CD4 lymphocytes, which include CD25+Foxp3+ regulatory T cells,25 suggesting that they may be more susceptible to PILAR-mediated expansion. In contrast, CD8+ lymphocytes, the only cell type known to exert immune pressure against ovarian cancer growth,25 may be prone to PILAR-mediated apoptotic death because of their lack of CD161 expression. Importantly, CD3/CD28-mediated T-cell stimulation resulted in a significant up-regulation of surface CD161 on CD8 T cells, suggesting that ovarian cancer–specific lymphocytes may be recovered from tumor-induced immunosuppression ex vivo and possibly converted into activated cytotoxic cells with stronger proliferative capacity. Our observation that the expression of PILAR on CD8 T cells increased dramatically on activation supports this conclusion.
In summary, PILAR represents a novel and significant modulator of cellular immune responses in humans. Our results provide a mechanistic rationale for the manipulation of PILAR signaling, which may unveil new interventions against autoimmunity, inflammation, and cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Immune Monitoring Laboratory for performing the Bioplex experiments.
This work was supported by a 2006 Liz-Tilberis Award from the Ovarian Cancer Research Fund (New York, NY), by grants from the American Cancer Society (IRG-82-003-22), the National Center for Research Resources (2P20RR016437-06); the National Cancer Institute (R01CA124515), and the National Institutes of Health (AI51547 and AI51877); and by the “Ramon Areces” Foundation (E.H.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: J.R.C.-G. and E.H. conceptualized and designed the experiments; E.H., J.C-R., Y.C.N., U.S., and D.M. did the experiments and provided intellectual contributions; X.A.E. performed the confirmatory T-cell expansion experiments and quantified cytokine productions; W.F.R., P.P., and P.G. provided leukocyte and tissue samples, analyzed the data, and contributed to the preparation of the manuscript.
Conflict-of-interest disclosure: Several of the authors (J.R.C-G., E.H., J.C-R., Y.C.N., and D.M.) have filed a patent application related to the work described in the present study. The remaining authors declare no competing financial interests.
Correspondence: Jose R. Conejo-Garcia, 640W Borwell, HB 7556, 1 Medical Center Dr, Lebanon, NH 03756; e-mail: jose.r.conejo-garcia@dartmouth.edu.
References
Author notes
*E.H., J.R.C-R., and Y.C.N. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal