In this issue of Blood, Waning and colleagues investigate the function of the ubiquitin ligase Cul4A, known for its involvement in protein degradation and histone modification. The present study reveals an essential requirement for Cul4A in hematopoiesis.
The continuous generation of short-lived blood cells depends on tightly controlled proliferation and differentiation of hematopoietic progenitors. Changes in gene expression are part of this developmental program, orchestrated by a combination of epigenetic, transcriptional, and RNA-based events. However, the aforementioned molecular processes only affect transcription and translation; they cannot eliminate already synthesized gene products. So, how does the counterpoint to gene expression—the regulation of protein destruction—contribute to hematopoiesis?
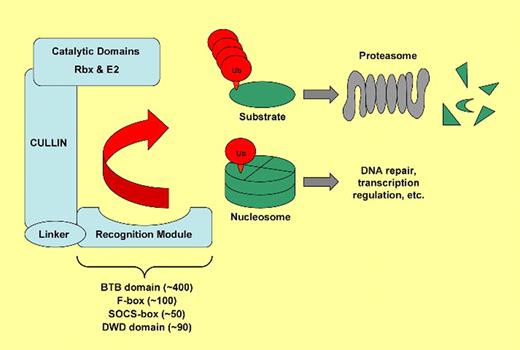
To address this question, Waning and colleagues at Kristin Chun's laboratory have generated an inducible knockout of Cullin 4 (Cul4A), a master regulator of protein degradation. Originally named for their ability to select proteins for removal,1 cullins form the backbone of modular complexes that selectively transfer ubiquitin onto target proteins (see figure). Once modified by multiple ubiquitin molecules, the substrate proteins are then recognized and destroyed by the proteasome. Cullins are critical for several cellular pathways, and Cul4A has been linked to DNA synthesis and repair as well as to transcriptional regulation. A previous study has indicated that Cul4A influences differentiation of hematopoietic progenitors.3 However, with the embryonic lethality of the homozygous Cul4A knockout and only mild signs of haploinsufficiency, the specific contribution of this mol-ecule to adult hematopoiesis has remained elusive.4 Here, the authors have generated a conditional knockout of Cul4A driven by interferon-inducible Cre recombination. Upon deletion, animals develop a striking pancytopenia. They die within a few days, with widespread signs of apoptosis in rapidly proliferating tissues such as the intestine and the bone marrow.
Simplified model of cullin-based ubiquitin ligases. Mammals possess 7 cullins (Cul1, 2, 3, 4A, 4B, 5, and 7) that each form a scaffold allowing substrate-specificity modules to connect to domains with ubiquitin ligase activity. Target proteins (green) are either modified by a chain containing multiple ubiquitin molecules (red)—a prerequisite for degradation by the proteasome—or they are monoubiquitinated. Each cullin uses a certain type of recognition module for substrate binding. Cul4A uses specificity modules that contain DWD domains, of which there are up to 90 in humans, to target a diverse set of proteins for ubiquitination. Substrates of Cul4A include Cdt1 and p27, both involved in cell-cycle regulation, and the histones H2A, H3, and H4.2
Simplified model of cullin-based ubiquitin ligases. Mammals possess 7 cullins (Cul1, 2, 3, 4A, 4B, 5, and 7) that each form a scaffold allowing substrate-specificity modules to connect to domains with ubiquitin ligase activity. Target proteins (green) are either modified by a chain containing multiple ubiquitin molecules (red)—a prerequisite for degradation by the proteasome—or they are monoubiquitinated. Each cullin uses a certain type of recognition module for substrate binding. Cul4A uses specificity modules that contain DWD domains, of which there are up to 90 in humans, to target a diverse set of proteins for ubiquitination. Substrates of Cul4A include Cdt1 and p27, both involved in cell-cycle regulation, and the histones H2A, H3, and H4.2
The described phenotype following Cul4A excision is impressive, but we still lack a molecular and cellular explanation for the rapid loss of hematopoietic cells. Areas for future research therefore include the identification of the substrate protein(s) of Cul4A, whose lack of ubiquitination in the knockout mice is responsible for the observed bone marrow failure. The authors propose that p27 and Cdt1 accumulation in Cul4A knockout cells are to blame for induction of apoptosis, but this point remains unproven. In particular, the possibility of a nonproteolytic function of Cul4A (eg, histone modification) has not been addressed. A second avenue for further research would pursue the precise definition of those cells affected most by Cul4A deletion. The data provided by Waning and colleagues suggest that rapidly proliferating progenitor cells are more susceptible to loss of Cul4A than quiescent stem cells. Third, given that it is ubiquitously expressed, how does the deletion of Cul4A affect other organs? The tissue-specific consequences of protein degradation have recently been exemplified by the observation that cullin-mediated removal of a particular transcription factor promotes differentiation in neurons, whereas the very same elimination process induces malignant transformation in the epithelial lineage.5 In addition to its physiological function, Cul4A has also been implicated in oncogenesis; its gene is amplified in several cancers, and mutations in the substrate recognition module DDB2 cause xeroderma pigmentosum, a disease characterized by DNA-repair defects.
In conclusion, the conditional Cul4A mouse model provides a powerful tool for studying the role of cullin-mediated ubiquitination in vivo. The investigations of Waning et al have already revealed essential pathways in hematopoietic and other regenerating tissues; there are likely more to be discovered.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal