Abstract
Inappropriate expression of EVI1 (ecotropic virus integration-1), in particular splice form EVI1-1D, through chromosome 3q26 lesions or other mechanisms has been implicated in the development of high-risk acute myeloid leukemia (AML). To validate the clinical relevance of EVI1-1D, as well as of the other EVI1 splice forms and the related MDS1/EVI1 (ME) gene, real-time quantitative polymerase chain reaction was performed in 534 untreated adults with de novo AML. EVI1-1D was highly expressed in 6% of cases (n = 32), whereas 7.8% were EVI1+ (n = 41) when all splice variants were taken into account. High EVI1 predicted a distinctly worse event-free survival (HR = 1.9; P = .002) and disease-free survival (HR = 2.1, P = .006) following multivariate analysis. Importantly, we distinguished a subset of EVI1+ cases that lacked expression of ME (EVI1+ME−; n = 17) from cases that were ME+ (EVI1+ME+; n = 24). The atypical EVI1+ME− expression pattern exhibited cytogenetically detectable chromosomal 3q26 breakpoints in 8 cases. Fluorescence in situ hybridization revealed 7 more EVI1+ME− cases that carried cryptic 3q26 breakpoints, which were not found in the EVI1+ME+ group. EVI1+ME− expression predicts an extremely poor prognosis distinguishable from the general EVI1+ AML patients (overall survival [OS]: P < .001 and event-free survival [EFS]: P = .002). We argue that EVI1/ME quantitative expression analysis should be implemented in the molecular diagnostic procedures of AML.
Introduction
Inappropriate expression of EVI1, through chromosome 3q26 lesions (eg, t(3;3)(q21;q26) or inv3(q21;q26)) has been implicated in the development or progression of high-risk acute myeloid leukemia (AML).1,2 Importantly, EVI1 is also highly expressed in a subgroup of AML without 3q26 rearrangements.3,4 High expression of EVI1 (ie, splice form EVI1-1D) is an independent negative prognostic indicator of survival in AML irrespective of the presence of 3q26 rearrangements.3 At least 4 additional splice variants of EVI1 were recently identified (ie, EVI1-1A, -1B, -1C, and -3L5 ), differing mainly in their 5′ untranslated regions. Since, we previously determined only the relative expression of EVI1-1D,3 it is feasible that EVI1+ AML cases have been underestimated. The prognostic value of EVI1 in AML, taking into account the distinct EVI1 splice forms, has not been evaluated yet.
Myeloid cells may also express MDS1/EVI1 (ME), an EVI1 fusion variant generated through intergenic splicing with MDS1,6 a gene located approximately 140 kb upstream of EVI1 with a currently unknown function. Among EVI1+ AML patients, leukemias with selective ME expression (ME+) can be distinguished from those that are ME negative (ME−).3,4 Currently, it is unclear whether EVI1+ME− and EVI1+ME+ leukemias are clinically and biologically different. Normal CD34+ bone marrow cells express EVI1 as well as ME,7,8 suggesting that an EVI1+ME− expression pattern in AML is abnormal. In fact, chromosomal breaks in 3q26 may occur between MDS1 and EVI1,9 thereby preventing ME fusion but instigating transcriptional activation of EVI1 alone. How frequently ME negativity in EVI1+ leukemias (EVI1+ME− genotype) is the result of genetic alterations in this locus is unknown.
It has also remained unexplained why certain leukemias express high levels of EVI1 without carrying a 3q26 abnormality. It is conceivable that these AML cells represent normal marrow CD34+ precursors, which have been shown to express EVI1 as well as ME.7,8 Another explanation could be that EVI1 and ME expression is the result of defects in other genes, which function upstream and cause high EVI1 and ME levels by elevating their transcription. In this study, we examined another possibility, that is, whether hidden 3q26 lesions exist in EVI1+AML cases without cytogenetically detectable aberrations in this locus.
We demonstrate in a cohort of 534 AML cases that high EVI1 expression, considering the various currently known EVI1 splice variants, is an independent predictor of poor survival. Of the EVI1+ AMLs, a considerable number of patients could be identified by real-time quantitative polymerase chain reaction (RQ-PCR) detecting only alternative EVI1 splice forms, but not EVI1-1D. The EVI1+ME− subgroups of AMLs often carry chromosome 3q26 lesions, some cryptic and only recognizable by fluorescence in situ hybridization. Importantly, among the EVI1+ AMLs, the EVI1+ME− leukemia subtype showed an extremely poor treatment outcome. Finally, the results reveal a positive correlation between EVI1+ME+ overexpression and 11q23 chromosomal abnormalities, suggesting a possible role for MLL fusion proteins in the regulation of EVI1 and ME expression.
Methods
Patients and molecular analyses
Leukemic blast cells were isolated from bone marrow or blood of 534 patients with AML, enrolled in the Dutch-Belgian Hemato-Oncology Cooperative Group (HOVON)–04,10-12 -29, -32, -42, or -43 protocols (available at http://www.hovon.nl/hovon/main.asp? M = 4&M = 5&M = 1&L = 1&P = true&C = 135&LNK = T). The control group contained 7 healthy bone marrow specimens. Blasts and mononuclear cells form healthy bone marrow specimens and AML samples were purified as previously reported.13 Reverse-transcription polymerase chain reaction (RT-PCR) and sequence analyses for mutations in FLT3-ITD, FLT3-TKD, NPM1, N-RAS, K-RAS, and CEBPA were performed as described previously.14-17
All subjects provided written informed consent in accordance with the Declaration of Helsinki. This research has been approved by the Institutional Review Board of Erasmus University Medical Center.
Real-time quantitative PCR and Northern blot analyses
RNA isolation, cDNA synthesis, and real-time quantitative RT-PCR (RQ-PCR) were performed as described.3,5,13 EVI1-1D splice form and ME expression levels were determined using probes,3 whereas the other EVI1 splice variants5 (-1A, -1B, -1C, and -3L) were analyzed using SYBR green (Applied Biosystems, Foster City, CA). A systematic overview of the EVI1 splice forms and the primer/probe localizations are shown in Figure 1. Primer and probe sequences are shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). EVI1 expression levels were determined using the ΔΔCt method.18 The average expression of each EVI1 splice variant and ME in 7 bone marrow samples from healthy volunteers was used as calibrator. The mean Ct values in the normal bone marrow samples were 29.6 (± 1.1) for EVI1-1A, 29.1 (± 0.82) for EVI1-1B, 37.2 (± 1.7) for EVI1-1C, 38.6 (± 1.2) for EVI1-1D, 32.8 (± 0.92) EVI1-3L, and 35.9 (± 1.9) for ME. The Ct values obtained were normalized for the internal reference,3 porphobilinogen deaminase (PBGD). The mean PBGD Ct value for normal bone marrow samples was 27.8 (± 1.0). For the ΔΔCt calculation to be valid,18 the absolute value of the slope in the plot of the log cDNA dilution versus ΔCt was determined for all primer combinations and was close to zero.
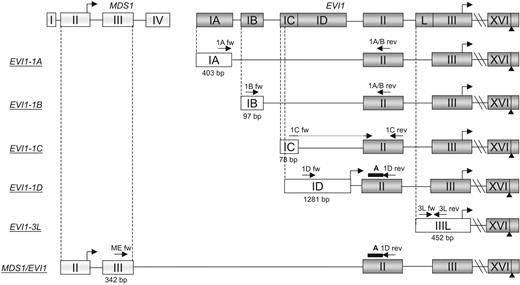
Gene structure and primer/probe locations of EVI1 splice variants -1A, -1B, -1C, -1D, -3L, and MDS1/EVI1 (ME). The exons, introns, and translational starts are depicted in boxes, connective lines, and standup arrows, respectively. The first exon's size in base pairs (bp), primers (arrows), and probes (bold line) are shown. Nucleotide sequences of primer/probe are presented in Table S1.
Gene structure and primer/probe locations of EVI1 splice variants -1A, -1B, -1C, -1D, -3L, and MDS1/EVI1 (ME). The exons, introns, and translational starts are depicted in boxes, connective lines, and standup arrows, respectively. The first exon's size in base pairs (bp), primers (arrows), and probes (bold line) are shown. Nucleotide sequences of primer/probe are presented in Table S1.
All samples were tested in duplicate and the average values were used for quantification. The amplification efficiency of each primer combination using 5 different dilutions (equal to 1.25 ng to 20 ng total RNA) was determined using mRNA isolated from 4 EVI1+ samples. The mean amplification efficiencies of EVI1-1A, -1B, -1C, -1D, -3L, and ME were, respectively, 1.00, 0.99, 0.90, 1.00, 0.94, and 0.95. Northern blot analyses for EVI1 expression were carried out as described previously.3
Fluorescence in situ hybridization
Dual-color fluorescence in situ hybridization (FISH) was performed with BAC clones located on chromosome 3q26, the EVI1 and/or the MDS1 locus, RP11-82C9 (EVI1), RP11-672P8 (EVI1), RP11-141C22 (MDS1), and RP11-250A4 (3q26; MDS1). Furthermore, BAC clones RP11-456K4 and RP11-912D21 located on chromosome 3q21, the ribophorin I (RPN1) locus, and RP1-196F4 located on 3q telomere were used. Clone isolation and labeling were performed using biotin-16-dUTP and digoxigenin-11-dUTP (Roche Diagnostics Belgium, Vilvoorde, Belgium) according to the manufacturer's protocol. The FISH analysis was performed as previously described.19 Each sample was analyzed by 2 different hybridizations. The evaluation of the hybridization pattern and signal intensity served as a reciprocal internal control. A minimum of 100 interphase cells and 10 metaphases were analyzed for each probe combination. Threshold values for true positivity were calculated from the average percentages, plus 3 times the standard deviations of nuclei falsely positive for each of the aberrant hybridization patterns in the control group. The control group consists of 4 healthy control and 14 EVI1+ME+ samples that yielded more than 90% normal metaphases, that is, 2 pairs of red and green fusion signals in both hybridizations with BAC clones RP11-82C9 plus RP11-141C22 and RP11-672P8 plus RP11-250A4.
Statistical analysis
Statistical analysis was performed with Stata Statistical Software, Release 9.2 (Stata, College Station, TX). Spearman rank test, Fisher exact test, chi-square test, and Student t test were calculated using Excel software (Microsoft, Redmond, WA). All patients received induction therapy and were included in the survival analysis. Actuarial probabilities of overall survival (OS, with death due to any cause), event-free survival (EFS, with failure in case of no complete remission at day 1 [CR1] or relapse or death), and disease-free survival (DFS; with death in CR1 or relapse) were estimated by the method of Kaplan and Meier.20 The Cox proportional hazards analysis21 was applied to determine the association of high total EVI1 expression (as a binary and as a continues variable) with OS, EFS, and DFS without and with adjustment for age, cytogenetic risk (ie, favorable, intermediate, or unfavorable22 ), and FLT3 internal tandem duplication (FLT3 ITD) together with known important poor prognostic AML markers (ie, monosomy 7 and MLL translocations). All tests were 2 tailed, and a P value of less than .05 was considered statistically significant.
Results
The predictive value of EVI1-1D validated in an independent cohort of 272 AML patients
We have previously demonstrated in patients with newly diagnosed AML that high EVI1 mRNA levels (ie, splice form 1D [EVI1-1D]; Figure 1) significantly predict for poor survival.3 Here, we show in an independent cohort of 272 cases of newly diagnosed AML (cohort A; Table S2) high EVI1-1D levels in 6.2% (n = 17) patients (Table 1). Importantly, high EVI1-1D expression again correlated with significantly reduced event-free survival (EFS) and overall survival (OS; P < .001; Figure S1). In the following experiments, we combined cohort A with samples from the previously investigated patient cohort3 (cohort B [n = 262]; Table S2). Only patients who were treated according to the HOVON cooperative group protocols were included. In cohort B, 5.8% of cases (n = 15) were EVI1-1D+ (Table 1). Hence, among the combined 534 AMLs, 6.0% (n = 32) of cases were EVI1-1D+.
Relative expression of EVI1 splice variants (-1D, -1A, -1B, -1C, -3L) and MDS1/EVI1 (ME), a priori karyotype and final FISH results in EVI1+ patients from cohorts A and B
| No. . | 1D . | 1A . | 1B . | 1C . | 3L . | ME . | Karyotype (ISCN 1995) . | FISH . |
|---|---|---|---|---|---|---|---|---|
| Cohort A | ||||||||
| 1 | 850 | 1634 | 199 | 80 | 819 | 0,1 | 45,XY,inv(3)(p12p2?4),−7[16]/46,XY[8] | inv(3) |
| 2 | 540 | 2159 | 75 | 73 | 157 | 0,2 | 45,XY,t(3;3)(q21;q26),−7[20] | ND |
| 3 | 254 | 1473 | 42 | 6 | 479 | 0,1 | 46,XY,t(3;3)(q21;q26)[51] | ND |
| 4 | 265 | 1405 | 67 | 58 | 76 | 0,0 | 45,XX,−7[24] | inv(3) |
| 5 | 352 | 88 | 33 | 4 | 65 | 0,5 | 47,XX,del(5)(q23q34),+del(21)(q21q22)[4]/47,idem,t(2;3)(p2?2;q2?7)[20]/46,XX[3] | t(2;3) |
| 6 | 796 | 943 | 160 | 225 | 131 | 3 | 45,XY,inv(3)(q21q26.2),−7[13] | ND |
| 7 | 68 | 74 | 47 | 5 | 43 | 8 | 45,XY,−7[16] | inv(3) |
| 8 | 166 | 177 | 42 | 5 | 43 | 25 | 46,XY[14] | NA |
| 9 | 50 | 84 | 21 | 3 | 137 | 89 | 48,XY,+9,+21[9]/49,idem,+21[14] | NA |
| 10 | 686 | 408 | 209 | 57 | 361 | 95 | 47,XX,del(3)(q25) or del(3)(q21q26),+mar | NA |
| 11 | 54 | 163 | 50 | 1 | 212 | 145 | 46,XX,t(6;11)(q27;q23)[36] | NN |
| 12 | 217 | 174 | 73 | 10 | 327 | 224 | 53,XY,+6,+8,+9,t(11;16)(q23;p13),+13,+14,+19,+21[15] | NN |
| 13 | 227 | 694 | 88 | 3 | 855 | 269 | 46,XX | NA |
| 14 | 120 | 380 | 85 | 6 | 527 | 289 | 46,XX,t(6;11)(q27;q23)[28] | NN |
| 15 | 94 | 494 | 119 | 4 | 680 | 437 | 46,XY[30] | NA |
| 16 | 183 | 1013 | 136 | 30 | 628 | 934 | 47,XY,+?8(9%)/46,XY | NA |
| 17 | 54 | 3 | 2 | 5 | 2185 | 1048 | 46,XY,t(3;21)(q26;q22),del(12)(p12p13)[20] | ND |
| 18 | 13 | 132 | 7 | 0,0 | 21 | 0,2 | 46,XY[38] | inv(3) |
| 19 | 21 | 89 | 11 | 0,5 | 90 | 24 | 46,XY,?der(11)(q2?)[3]/46,XY[18] | NN |
| 20 | 14 | 67 | 16 | 0,5 | 21 | 32 | 46,XY,inv(3)(q21q26)[32] | inv(3) |
| 21 | 26 | 99 | 27 | 1 | 135 | 86 | 46,XY,t(11;19)(q23;p13.1)[15]/46,XY[5] | NN |
| 22 | 7 | 27 | 9 | 2 | 77 | 216 | NA | NA |
| Cohort B | ||||||||
| 23 | 1624 | 1256 | 363 | 59 | 834 | 19 | 45,XX,inv(3)(q22q26),−7[29]/46,XX[1] | ND |
| 24 | 584 | 417 | 116 | 74 | 37 | 4 | 45,XY,inv(3)(q22q26),−7[25] | ND |
| 25 | 506 | 845 | 119 | 37 | 350 | 0,2 | 45,XY,inv(3)(q12q26.2),−7[20] | ND |
| 26 | 700 | 1121 | 161 | 76 | 244 | 0,0 | 45,XX,−7[27]/46,XX[3] | inv(3) |
| 27 | 214 | 1074 | 44 | 9 | 541 | 0,1 | NA | NN |
| 28 | 173 | 172 | 28 | 10 | 46 | 0,0 | 46,XX[68] | inv(3) |
| 29 | 196 | 239 | 69 | 6 | 167 | 0,0 | 46,XX,t(1;6)(p32;q24∼25),del(2)(q34)[33]/46,XX[1] | inv(3) |
| 30 | 829 | 1258 | 132 | 68 | 333 | 63 | NA | NA |
| 31 | 745 | 2354 | 147 | 23 | 1309 | 286 | 45,XY,−7,t(9;11)(p21;q23)[33] | NN |
| 32 | 315 | 641 | 174 | 9 | 2532 | 532 | 46,XX,t(11;19)(q23;p13)[21] | NN |
| 33 | 207 | 186 | 118 | 5 | 373 | 168 | 46,XY,t(9;22)(q34;q11)[22] | NN |
| 34 | 192 | 897 | 112 | 8 | 3266 | 539 | 46,XX,t(2;9;11)(p13;p22;q23)[20] | NA |
| 35 | 83 | 306 | 43 | 3 | 517 | 448 | 46,XY[40] | NN |
| 36 | 79 | 67 | 36 | 3 | 204 | 1065 | 46,XX,del(7)(q22)[41]/46,XX[1] | NN |
| 37 | 52 | 157 | 27 | 0,9 | 605 | 429 | 46,XY,t(6;11)(q25;q23)[22] | NN |
| 38 | 14 | 7 | 6 | 0,1 | 95 | 139 | 47,XY,+13[11]/46,XY[28] | NN |
| 39 | 16 | 39 | 9 | 0,9 | 33 | 223 | 45,XY,−7,t(7;8)(q22;p11)[21] | NN |
| 40 | 11 | 6 | 6 | 1,7 | 144 | 397 | 47,XX,+13[27]/46,XX[1] | NN |
| 41 | 14 | 13 | 5 | 2,8 | 234 | 1830 | 46,XY,−7,add(12)(p12),+mar[46]/46,XY[54] | NN |
| No. . | 1D . | 1A . | 1B . | 1C . | 3L . | ME . | Karyotype (ISCN 1995) . | FISH . |
|---|---|---|---|---|---|---|---|---|
| Cohort A | ||||||||
| 1 | 850 | 1634 | 199 | 80 | 819 | 0,1 | 45,XY,inv(3)(p12p2?4),−7[16]/46,XY[8] | inv(3) |
| 2 | 540 | 2159 | 75 | 73 | 157 | 0,2 | 45,XY,t(3;3)(q21;q26),−7[20] | ND |
| 3 | 254 | 1473 | 42 | 6 | 479 | 0,1 | 46,XY,t(3;3)(q21;q26)[51] | ND |
| 4 | 265 | 1405 | 67 | 58 | 76 | 0,0 | 45,XX,−7[24] | inv(3) |
| 5 | 352 | 88 | 33 | 4 | 65 | 0,5 | 47,XX,del(5)(q23q34),+del(21)(q21q22)[4]/47,idem,t(2;3)(p2?2;q2?7)[20]/46,XX[3] | t(2;3) |
| 6 | 796 | 943 | 160 | 225 | 131 | 3 | 45,XY,inv(3)(q21q26.2),−7[13] | ND |
| 7 | 68 | 74 | 47 | 5 | 43 | 8 | 45,XY,−7[16] | inv(3) |
| 8 | 166 | 177 | 42 | 5 | 43 | 25 | 46,XY[14] | NA |
| 9 | 50 | 84 | 21 | 3 | 137 | 89 | 48,XY,+9,+21[9]/49,idem,+21[14] | NA |
| 10 | 686 | 408 | 209 | 57 | 361 | 95 | 47,XX,del(3)(q25) or del(3)(q21q26),+mar | NA |
| 11 | 54 | 163 | 50 | 1 | 212 | 145 | 46,XX,t(6;11)(q27;q23)[36] | NN |
| 12 | 217 | 174 | 73 | 10 | 327 | 224 | 53,XY,+6,+8,+9,t(11;16)(q23;p13),+13,+14,+19,+21[15] | NN |
| 13 | 227 | 694 | 88 | 3 | 855 | 269 | 46,XX | NA |
| 14 | 120 | 380 | 85 | 6 | 527 | 289 | 46,XX,t(6;11)(q27;q23)[28] | NN |
| 15 | 94 | 494 | 119 | 4 | 680 | 437 | 46,XY[30] | NA |
| 16 | 183 | 1013 | 136 | 30 | 628 | 934 | 47,XY,+?8(9%)/46,XY | NA |
| 17 | 54 | 3 | 2 | 5 | 2185 | 1048 | 46,XY,t(3;21)(q26;q22),del(12)(p12p13)[20] | ND |
| 18 | 13 | 132 | 7 | 0,0 | 21 | 0,2 | 46,XY[38] | inv(3) |
| 19 | 21 | 89 | 11 | 0,5 | 90 | 24 | 46,XY,?der(11)(q2?)[3]/46,XY[18] | NN |
| 20 | 14 | 67 | 16 | 0,5 | 21 | 32 | 46,XY,inv(3)(q21q26)[32] | inv(3) |
| 21 | 26 | 99 | 27 | 1 | 135 | 86 | 46,XY,t(11;19)(q23;p13.1)[15]/46,XY[5] | NN |
| 22 | 7 | 27 | 9 | 2 | 77 | 216 | NA | NA |
| Cohort B | ||||||||
| 23 | 1624 | 1256 | 363 | 59 | 834 | 19 | 45,XX,inv(3)(q22q26),−7[29]/46,XX[1] | ND |
| 24 | 584 | 417 | 116 | 74 | 37 | 4 | 45,XY,inv(3)(q22q26),−7[25] | ND |
| 25 | 506 | 845 | 119 | 37 | 350 | 0,2 | 45,XY,inv(3)(q12q26.2),−7[20] | ND |
| 26 | 700 | 1121 | 161 | 76 | 244 | 0,0 | 45,XX,−7[27]/46,XX[3] | inv(3) |
| 27 | 214 | 1074 | 44 | 9 | 541 | 0,1 | NA | NN |
| 28 | 173 | 172 | 28 | 10 | 46 | 0,0 | 46,XX[68] | inv(3) |
| 29 | 196 | 239 | 69 | 6 | 167 | 0,0 | 46,XX,t(1;6)(p32;q24∼25),del(2)(q34)[33]/46,XX[1] | inv(3) |
| 30 | 829 | 1258 | 132 | 68 | 333 | 63 | NA | NA |
| 31 | 745 | 2354 | 147 | 23 | 1309 | 286 | 45,XY,−7,t(9;11)(p21;q23)[33] | NN |
| 32 | 315 | 641 | 174 | 9 | 2532 | 532 | 46,XX,t(11;19)(q23;p13)[21] | NN |
| 33 | 207 | 186 | 118 | 5 | 373 | 168 | 46,XY,t(9;22)(q34;q11)[22] | NN |
| 34 | 192 | 897 | 112 | 8 | 3266 | 539 | 46,XX,t(2;9;11)(p13;p22;q23)[20] | NA |
| 35 | 83 | 306 | 43 | 3 | 517 | 448 | 46,XY[40] | NN |
| 36 | 79 | 67 | 36 | 3 | 204 | 1065 | 46,XX,del(7)(q22)[41]/46,XX[1] | NN |
| 37 | 52 | 157 | 27 | 0,9 | 605 | 429 | 46,XY,t(6;11)(q25;q23)[22] | NN |
| 38 | 14 | 7 | 6 | 0,1 | 95 | 139 | 47,XY,+13[11]/46,XY[28] | NN |
| 39 | 16 | 39 | 9 | 0,9 | 33 | 223 | 45,XY,−7,t(7;8)(q22;p11)[21] | NN |
| 40 | 11 | 6 | 6 | 1,7 | 144 | 397 | 47,XX,+13[27]/46,XX[1] | NN |
| 41 | 14 | 13 | 5 | 2,8 | 234 | 1830 | 46,XY,−7,add(12)(p12),+mar[46]/46,XY[54] | NN |
Relative expression of each of the distinct EVI1 transcripts was determined as explained in “Real-time quantitative PCR and Northern blot analysis.” A positive EVI1 or ME signal (ie, >30) is indicated by italics.
ME indicates MDS1/EVI1; ISCN, International System for Human Cytogenetic Nomenclature; FISH, fluorescent in situ hybridization; NN, normal 3q21, 3q26, and 3q telomere locus; inv(3), inversion of chromosomal band 3q21 and 3q26; NA, not available; NN, normal at the 3q21, 3q26, and 3q telomere locus as determined by FISH; and ND, not determined.
EVI1+ cases identified by expression analysis of alternative EVI1 splice forms
Four other EVI1 splice forms have been reported, which differ mainly in their 5′-untranslated region (ie, EVI1-1A, -1B, -1C, and -3L; Figure 1). To investigate for the frequency of expression of each of these EVI1 splice variants in AML and to verify whether EVI1-1D− AMLs might express other EVI1 splice forms, we determined the relative expression by splice form–specific RQ-PCRs (Figure 1). A sample was considered EVI1+ if the relative expression was more than 30 for one or more EVI1 splice variants. Different cutoff points (ie, 50, 30, 20, and 10) were tested based on event-free survival and showed minor differences in survival and in EVI1+ patients (40, 41, 52, and 57, respectively). Differences in survival between EVI1+ versus EVI1− cases appeared to be significant in each situation (data not shown). To prevent including false-positive patients we chose a cutoff level of 30 for further analysis and calculations.
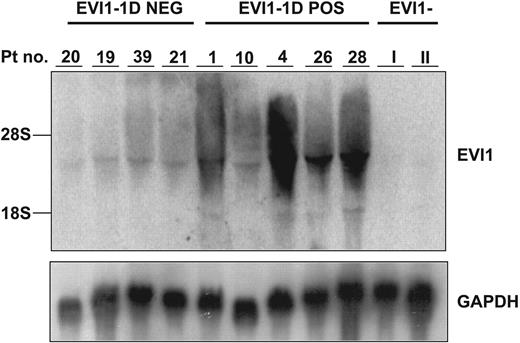
Although Spearman correlation coefficients comparing expression levels of distinct splice forms were high (Figure S2), indicative of frequent coexpression, 9 EVI1-1D− cases were identified that were positive for one or more other EVI1 splice forms (Table 1). The total EVI1+ fraction (EVI1+) was increased to 7.8% (n = 41 cases). EVI1-1A or EVI1-3L was found to be most frequently expressed in those cases. Northern blot analysis performed on samples from selected patients that expressed distinct EVI1 splice variants (Figure 2), revealed bands of the expected size, confirming the true identity of EVI1.
EVI1 mRNA expression levels in EVI1-1D+ and EVI1-1D− AML samples in cohort A and B determined by Northern blot. Human 600-bp EVI1 probe and as control a murine GAPDH fragment were used. Patients I and II represent AML samples without EVI1 expression. The patient numbers correspond to those in Table 1.
EVI1 mRNA expression levels in EVI1-1D+ and EVI1-1D− AML samples in cohort A and B determined by Northern blot. Human 600-bp EVI1 probe and as control a murine GAPDH fragment were used. Patients I and II represent AML samples without EVI1 expression. The patient numbers correspond to those in Table 1.
EVI1 expression and clinical characteristics
No differences in age and sex distributions, FAB classifications, pretreatment white blood cell (WBC) counts, or percentages of bone marrow blasts were observed between patients with EVI1+ and EVI1− de novo AML. Clinical characteristics of EVI1+ versus EVI1− patients within the cohort of 534 AML patients are depicted in Table 2. Chromosomal aberrations in the EVI1 locus (ie, 3q26 abnormalities) were seen in 8 (20%) of the 41 EVI1+ cases, whereas only 2 (0.4%) of 493 EVI1− AMLs showed a 3q26 abnormality (Tables 1,2). Possibly, in those latter 2 cases another gene present in the 3q26 locus may have been affected. Other cytogenetic lesions that are frequently seen in association with EVI1 positivity are −7/7q− deletions and translocations involving 11q23. Deletions −7/7q− were found in 13 (38%) of 41 EVI1+ leukemias and 34 (7%) of 493 EVI1− cases. Translocations involving 11q23 were observed in 8 (20%) of 41 EVI1+ versus 8 (1.6%) of 493 EVI1− AMLs. Furthermore, an inverse correlation was seen between EVI1+ patients and NPM1 mutations (P < .001).
Clinical characteristics of EVI1+ patients in relation to clinical parameters, morphology, cytogenetics, and molecular characteristics of 534 patients with newly diagnosed AML
| . | No. of EVI1− patients . | No. of EVI1+ patients (%) . | P . |
|---|---|---|---|
| Sex* | .19 | ||
| Male | 242 | 25 (11) | — |
| Female | 251 | 16 (17) | — |
| Age, y* | .22 | ||
| Younger than 35 | 117 | 14 (11) | — |
| Between 35 and 50 | 166 | 12 (15) | — |
| Older than 50 | 210 | 15 (15) | — |
| HOVON protocol* | |||
| 04A | 49 | 3 (6) | >.99 |
| 04 | 40 | 8 (17) | .50 |
| 29 | 203 | 12 (6) | .32 |
| 32 | 5 | 0 (0) | >.99 |
| 42 | 140 | 11 (7) | .85 |
| 43 | 55 | 7 (11) | .54 |
| FAB* | |||
| M0 | 16 | 2 (11) | .64 |
| M1 | 104 | 6 (5) | .42 |
| M2 | 123 | 6 (5) | .18 |
| M3 | 24 | 1 (4) | .71 |
| M4 | 82 | 13 (14) | .11 |
| M5 | 104 | 10 (9) | .85 |
| M6 | 7 | 1 (12) | .47 |
| Mx | 33 | 2 (6) | >.99 |
| Cytogenetic abnormalities*† | |||
| −5/5q− | 16 | 3 (16) | .23 |
| −7/7q− | 21 | 13 (38) | <.001 |
| 3q26 | 2 | 8 (80) | <.001 |
| t(9;22)(q34;q11) | 1 | 1 (50) | .17 |
| t(11q23) | 8 | 8 (50) | <.001 |
| t(15;17)(q22;q21) | 21 | 1 (5) | .71 |
| t(8;21)(q22;q22) | 39 | 0 (0) | .039 |
| inv(16)/t(16;16) | 37 | 0 (0) | .065 |
| +8 | 22 | 2 (8) | .71 |
| +21 | 3 | 2 (40) | .05 |
| t(6;9)(p23;q34) | 6 | 0 (0) | >.99 |
| Complex | 20 | 1 (5) | >.99 |
| Other | 65 | 6 (8) | >.99 |
| Normal | 218 | 6 (3) | <.001 |
| ND | 20 | 2 (9) | >.99 |
| Cytogenetic risk* | |||
| Favorable | 89 | 1 (1) | .23 |
| Intermediate | 347 | 17 (5) | <.001 |
| Unfavorable | 57 | 23 (29) | <.001 |
| Molecular abnormalities* | |||
| FLT3 ITD | 135 | 5 (4) | .027 |
| FLT3 TKD | 43 | 1 (2) | .23 |
| KRAS | 5 | 0 (0) | >.99 |
| NRAS | 40 | 5 (11) | .57 |
| CEBPA | 43 | 1 (2) | .23 |
| NPM1 | 158 | 2 (1) | <.001 |
| WBC count, ×109/L,‡ mean; SD | 52; 63 | 45; 49 | .18 |
| Platelet count, ×109/L,‡ mean; SD | 72; 101 | 165; 241 | .01 |
| Blast % in BM,‡ mean; SD | 61; 27 | 61; 24 | .38 |
| . | No. of EVI1− patients . | No. of EVI1+ patients (%) . | P . |
|---|---|---|---|
| Sex* | .19 | ||
| Male | 242 | 25 (11) | — |
| Female | 251 | 16 (17) | — |
| Age, y* | .22 | ||
| Younger than 35 | 117 | 14 (11) | — |
| Between 35 and 50 | 166 | 12 (15) | — |
| Older than 50 | 210 | 15 (15) | — |
| HOVON protocol* | |||
| 04A | 49 | 3 (6) | >.99 |
| 04 | 40 | 8 (17) | .50 |
| 29 | 203 | 12 (6) | .32 |
| 32 | 5 | 0 (0) | >.99 |
| 42 | 140 | 11 (7) | .85 |
| 43 | 55 | 7 (11) | .54 |
| FAB* | |||
| M0 | 16 | 2 (11) | .64 |
| M1 | 104 | 6 (5) | .42 |
| M2 | 123 | 6 (5) | .18 |
| M3 | 24 | 1 (4) | .71 |
| M4 | 82 | 13 (14) | .11 |
| M5 | 104 | 10 (9) | .85 |
| M6 | 7 | 1 (12) | .47 |
| Mx | 33 | 2 (6) | >.99 |
| Cytogenetic abnormalities*† | |||
| −5/5q− | 16 | 3 (16) | .23 |
| −7/7q− | 21 | 13 (38) | <.001 |
| 3q26 | 2 | 8 (80) | <.001 |
| t(9;22)(q34;q11) | 1 | 1 (50) | .17 |
| t(11q23) | 8 | 8 (50) | <.001 |
| t(15;17)(q22;q21) | 21 | 1 (5) | .71 |
| t(8;21)(q22;q22) | 39 | 0 (0) | .039 |
| inv(16)/t(16;16) | 37 | 0 (0) | .065 |
| +8 | 22 | 2 (8) | .71 |
| +21 | 3 | 2 (40) | .05 |
| t(6;9)(p23;q34) | 6 | 0 (0) | >.99 |
| Complex | 20 | 1 (5) | >.99 |
| Other | 65 | 6 (8) | >.99 |
| Normal | 218 | 6 (3) | <.001 |
| ND | 20 | 2 (9) | >.99 |
| Cytogenetic risk* | |||
| Favorable | 89 | 1 (1) | .23 |
| Intermediate | 347 | 17 (5) | <.001 |
| Unfavorable | 57 | 23 (29) | <.001 |
| Molecular abnormalities* | |||
| FLT3 ITD | 135 | 5 (4) | .027 |
| FLT3 TKD | 43 | 1 (2) | .23 |
| KRAS | 5 | 0 (0) | >.99 |
| NRAS | 40 | 5 (11) | .57 |
| CEBPA | 43 | 1 (2) | .23 |
| NPM1 | 158 | 2 (1) | <.001 |
| WBC count, ×109/L,‡ mean; SD | 52; 63 | 45; 49 | .18 |
| Platelet count, ×109/L,‡ mean; SD | 72; 101 | 165; 241 | .01 |
| Blast % in BM,‡ mean; SD | 61; 27 | 61; 24 | .38 |
FAB indicates French-American-British classification; BM, bone marrow; FLT3 ITD, internal tandem duplication of the FLT3 gene; FLT3 TKD, a mutation in tyrosine kinase domain of the FLT3 gene; Mx, FAB not available; ND, not determined; —, not applicable; and SD, standard deviation.
P values were calculated using the 2-tailed chi-square test.
All patients with a specific abnormality were considered irrespective of the presence of additional abnormalities.
P values were calculated using 2-tailed t test.
High EVI1 expression is an independent prognostic marker in AML
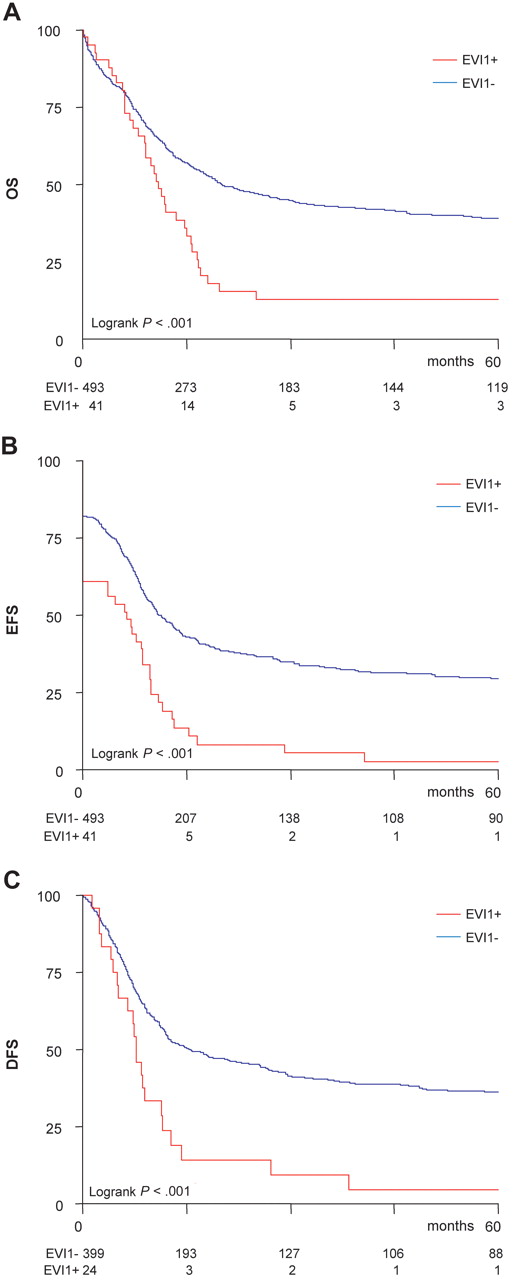
We next investigated the prognostic impact of EVI1 positivity (ie, EVI1-1A, -1B, -1C, -1D, or -3L positive) in the total cohort of 534 patients with AML. Patients with EVI1+ AML less often attained a complete remission (61% vs 82%; χ2P = .001) and the probability of relapse was considerably higher compared with EVI1− AMLs (51% vs 41%; χ2P = .04). Survival analysis revealed a severe disadvantage for EVI1+ AML patients regarding the 5-year overall survival (OS) (13% ± 5% vs 39% ± 2%; Figure 3A), event-free survival (EFS) (3% ± 3% vs 29% ± 2%; Figure 3B), and disease-free survival (DFS; probability of relapse; 5% ± 4 vs 32% ± 3%; Figure 3C). Table 3 describes the increased hazard ratios for EVI1+AML for OS, EFS, and DFS following univariate analysis and multivariate analysis.
High EVI1 expression associates with poor survival outcome in AML. Kaplan Meier analysis of (A) overall survival (OS), (B) event-free survival (EFS), and (C) disease-free survival (DFS) shows an inferior outcome for EVI1+ patients in comparison with patients without EVI1 overexpression in a total cohort of 534 AML patients.
High EVI1 expression associates with poor survival outcome in AML. Kaplan Meier analysis of (A) overall survival (OS), (B) event-free survival (EFS), and (C) disease-free survival (DFS) shows an inferior outcome for EVI1+ patients in comparison with patients without EVI1 overexpression in a total cohort of 534 AML patients.
Univariate and multivariate analysis of high EVI1 expression as prognostic factor for survival
| . | EFS . | DFS . | OS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | (95% CI) . | P . | HR . | (95% CI) . | P . | HR . | (95% CI) . | P . | |
| Univariable analysis, high EVI1 expression* | 2.17 | (1.55-3.03) | <.001 | 2.26 | (1.46-3.50) | <.001 | 1.91 | (1.34-2.72) | <.001 |
| Multivariable analysis | |||||||||
| Intermediate† | 1.62 | (1.17-2.26) | .004 | 1.91 | (1.30-2.82) | .001 | 1.84 | (1.27-2.68) | .001 |
| Unfavorable† | 2.90 | (1.88-4.49) | <.001 | 3.8 | (2.27-6.36) | <.001 | 3.46 | (2.14-5.60) | <.001 |
| Age | |||||||||
| 35 to 50 y | 1.03 | (0.78-1.35) | .84 | 1.10 | (0.81-1.52) | .53 | 1.22 | (0.90-1.65) | .20 |
| Older than 50 y | 1.23 | (0.94-1.60) | .13 | 1.17 | (0.86-1.60) | .33 | 1.48 | (1.11-1.98) | .007 |
| FLT3 ITD‡ | 1.40 | (1.11-1.77) | .005 | 1.25 | (0.93-1.67) | .13 | 1.55 | (1.21-1.99) | <.001 |
| Monosomy 7§ | 1.37 | (0.89-2.11) | .15 | 1.05 | (0.58-1.89) | .87 | 1.35 | (0.86-2.13) | .20 |
| MLL translocation‖ | 0.68 | (0.35-1.29) | .24 | 0.67 | (0.32-1.39) | .28 | 0.79 | (0.39-1.56) | .49 |
| High EVI1 expression* | 1.88 | (1.29-2.76) | .002 | 2.14 | (1.29-3.54) | .006 | 1.47 | (0.98-2.21) | .072 |
| . | EFS . | DFS . | OS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | (95% CI) . | P . | HR . | (95% CI) . | P . | HR . | (95% CI) . | P . | |
| Univariable analysis, high EVI1 expression* | 2.17 | (1.55-3.03) | <.001 | 2.26 | (1.46-3.50) | <.001 | 1.91 | (1.34-2.72) | <.001 |
| Multivariable analysis | |||||||||
| Intermediate† | 1.62 | (1.17-2.26) | .004 | 1.91 | (1.30-2.82) | .001 | 1.84 | (1.27-2.68) | .001 |
| Unfavorable† | 2.90 | (1.88-4.49) | <.001 | 3.8 | (2.27-6.36) | <.001 | 3.46 | (2.14-5.60) | <.001 |
| Age | |||||||||
| 35 to 50 y | 1.03 | (0.78-1.35) | .84 | 1.10 | (0.81-1.52) | .53 | 1.22 | (0.90-1.65) | .20 |
| Older than 50 y | 1.23 | (0.94-1.60) | .13 | 1.17 | (0.86-1.60) | .33 | 1.48 | (1.11-1.98) | .007 |
| FLT3 ITD‡ | 1.40 | (1.11-1.77) | .005 | 1.25 | (0.93-1.67) | .13 | 1.55 | (1.21-1.99) | <.001 |
| Monosomy 7§ | 1.37 | (0.89-2.11) | .15 | 1.05 | (0.58-1.89) | .87 | 1.35 | (0.86-2.13) | .20 |
| MLL translocation‖ | 0.68 | (0.35-1.29) | .24 | 0.67 | (0.32-1.39) | .28 | 0.79 | (0.39-1.56) | .49 |
| High EVI1 expression* | 1.88 | (1.29-2.76) | .002 | 2.14 | (1.29-3.54) | .006 | 1.47 | (0.98-2.21) | .072 |
P values were calculated using the Cox regression model.
HR indicates hazard ratio with high EVI1 expression; CI, confidence interval; EFS, event-free survival; DFS, disease-free survival; OS, overall survival; and FLT3 ITD, FLT3 gene internal tandem duplication.
High EVI1 expression versus no EVI1 expression.
Cytogenetic risk versus favorable cytogenetic risk.
FLT3 ITD versus no FLT3 ITD.
Monosomy 7 versus no monosomy 7.
MLL translocation versus no MLL translocation.
Multivariate analysis established high EVI1 expression as an independent prognostic marker in relation to FLT3-ITD mutations and other prognostic cytogenetic abnormalities with hazard ratios for EFS and DFS of 1.88 (95% CI, 1.29-2.76; P = .002) and 2.14 (95% CI, 1.29-3.54; P = .006), respectively (Table 3), whereas for OS a hazard ratio with a decreasing trend of 1.47 (95% CI, 0.98-2.21, P = .073) was established.
We performed a Cox regression analysis with expression of EVI1-1A, 1B, 1D, and 3L as a continuous variable. The data were log-transformed to guarantee an equal distribution. The results shown in Table S3 demonstrate that the relative expression of variants EVI1-1A, 1B, and 3L, but not EVI1-1D, was of significant influence in the EFS, DFS, and OS of the AMLs studied.
We also performed a Cox regression analysis where EVI1 expression as a time-dependent covariate was added to the original multivariate model for OS. High EVI1 expression as a time-dependent covariant was not of significant influence in this model (HR = 1.0, 95% CI; 0.98-1.04, P = .54).
EVI1 positivity and MDS1/EVI1 negativity associate with 3q26 lesions
Since EVI1 is normally coexpressed with the intergenic splice form MDS1/EVI1 (ME; Figure 1), we argued that the atypical dissociated expression pattern EVI1+ME− that is noted in a proportion of cases of AML3 might be caused by chromosome 3q26 lesions disrupting the MDS1/EVI1 locus. We estimated ME levels in the EVI1+ AML subgroup and observed absence of ME expression in 17 of 41 cases. Six of these (6/17, 36%) carried a cytogenetically detectable 3q26 abnormality, while the remaining leukemias carried other cytogenetic aberrations or had a normal karyotype (Table 1). ME was coexpressed with EVI1 in 24 of 41 cases. Only 2 (8%) of 24 EVI1+ME+ cases showed a 3q26 lesion. In fact, in one case (no. 20) ME expression levels were borderline (Table 1), while in the other case (no. 17) the translocation t(3;21) was apparent, which is known to result in an AML1/MDS1/EVI1 fusion.23
Frequent hidden 3q26 lesions in EVI1+ME− AML patients
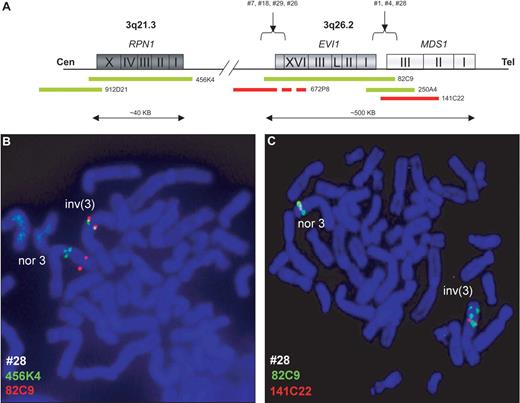
The positive association between EVI1+ME− expression and cytogenetically detectable 3q26 abnormalities prompted us to investigate whether hidden 3q26 chromosomal lesions might be present in the EVI1+ME− subgroup of AML. Dual-color FISH analyses using BAC clones covering EVI1 and MDS1 (Figure 4A) were carried out on metaphase spreads of 10 cases of EVI1+ME− AML without any a priori cytogenetically detectable 3q26 abnormality. In 8 of 10 patients (nos. 1, 4, 5, 7, 18, 26, 28, and 29), split signals were observed in metaphase and interphase nuclei, indicating the presence of hidden 3q26 lesions (Figure 4B; Table 1; and Figure S3A-C). In addition, the FISH analysis redefined the breakpoint of the cytogenetically identified t(2;3)(p2?;q2?7) into t(2;3)(p2?;q26) (no. 5; Figure S3D-E). Next, FISH analysis using BAC clones covering RPN1 (3q21) and EVI1 revealed involvement of the RPN1 locus in the remaining 7 patients (Figure 4C; Table 1). The complete FISH results for each patient analyzed are shown in Table 1. FISH experiments using the same BAC clones covering EVI1, MDS1, and RPN1 were carried out in EVI1+ME+ leukemias but did not demonstrate the existence of inv3(q21;q26) or t(3;3)(q21;q26) in any of the 14 cases studied. Other chromosome 3 translocations were excluded in these samples by FISH using a BAC clone located telomeric on 3q (Figure S3F).
Fluorescent in situ hybridization (FISH) of chromosome 3q26 and 3q21 loci reveal hidden 3q26 aberrations. BAC clone localization from centromere (Cen) to telomere (Tel) (A). A metaphase from EVI1+ patient no. 28 revealed a cryptic inv(3)(q21q26) (inv3) and a normal chromosome 3 (nor3) using EVI1 (RP11-82C9) and MDS1 (RP11-141C22) (B) and RPN1 (RP11-456K4) BAC clones (C). Micrographs after FISH were acquired by imaging with a fluorescence microscope (Axio-Imager Z1; Zeiss, Sliedrecht, The Netherlands) fitted with a Plan-Apochromat at 100x/1.40 numeric aperture oil objective, a CCD video camera (Metasystems, Altlussheim, Germany), and using Isis software for capturing and processing fluorescent images (v5.1.7, Metasystems).
Fluorescent in situ hybridization (FISH) of chromosome 3q26 and 3q21 loci reveal hidden 3q26 aberrations. BAC clone localization from centromere (Cen) to telomere (Tel) (A). A metaphase from EVI1+ patient no. 28 revealed a cryptic inv(3)(q21q26) (inv3) and a normal chromosome 3 (nor3) using EVI1 (RP11-82C9) and MDS1 (RP11-141C22) (B) and RPN1 (RP11-456K4) BAC clones (C). Micrographs after FISH were acquired by imaging with a fluorescence microscope (Axio-Imager Z1; Zeiss, Sliedrecht, The Netherlands) fitted with a Plan-Apochromat at 100x/1.40 numeric aperture oil objective, a CCD video camera (Metasystems, Altlussheim, Germany), and using Isis software for capturing and processing fluorescent images (v5.1.7, Metasystems).
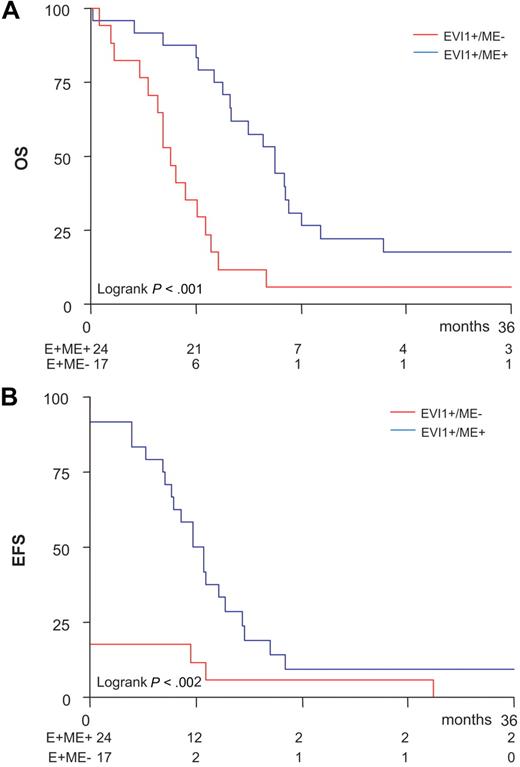
EVI1+ME− and EVI1+ME+ AMLs are cytogenetically and clinically different
No significant differences in sex and age distributions, FAB classification, WBC counts, bone marrow blast percentages, karyotype risk classification, and molecular abnormalities were observed between the EVI1+ME− (n = 17) and the EVI1+ME+ (n = 24) subsets of patients with AML (Table S4). Besides the frequent occurrence of 3q26 lesions, patients with EVI1+ME− AML also frequently presented a −7/7q− deletion (n = 9; P = .11). Interestingly, the platelet count, which was significantly elevated in the total EVI1+ group compared with the EVI1− AML patients (Table 1), was elevated only in the EVI1+ME− subgroup but not in the EVI1+ME+ subgroup (P = .024; Table S4). EVI1+ME+ cases frequently carried 11q23 abnormalities (n = 8, P = .013). While EVI1+ME+ and EVI1+ME− subgroups both showed a poor treatment outcome (Figure 5), EVI1+ME− cases showed an even worse prognosis. This was evident from the fact that they less often achieved complete remission than patients with EVI1+ME+ AML (18% vs 92%; χ2P = .001) and showed a significantly reduced OS (9% ± 3% vs 24% ± 6%; Figure 5A) and EFS (3% ± 3% vs 29% ± 2%; Figure 5B) at 1.5 years. In fact, a significantly worse OS, EFS, and DFS (OS: HR = 2.45, P = .002; EFS: HR = 3.21, P < .001; DFS: HR = 2.44, P = .13) for EVI1+ME− cases compared with EVI1+ME+ AML patients were also evident from multivariate analysis, although one should keep in mind that patients numbers were relatively small for such subdividing and this requires further validation in other cohorts.
Inferior outcome for EVI1+ME− patients in comparison with EVI1+ME+patients. Kaplan Meier analysis of (A) overall survival (OS) and (B) event-free survival (EFS) shows an inferior outcome for EVI1+ME− patients in comparison with EVI1+ME+ patients in the cohort of 41 EVI1+ AML patients.
Inferior outcome for EVI1+ME− patients in comparison with EVI1+ME+patients. Kaplan Meier analysis of (A) overall survival (OS) and (B) event-free survival (EFS) shows an inferior outcome for EVI1+ME− patients in comparison with EVI1+ME+ patients in the cohort of 41 EVI1+ AML patients.
Discussion
Molecular markers are of utmost importance in resolving the genetic heterogeneity of AML and deriving discriminative prognostic algorithms. Recently, markers such as expression levels of EVI1,3 BAALC,24 and ERG25 have been demonstrated to be powerful indicators of prognostically distinct AML subsets. High expression of EVI1 (ie, in particular splice variant EVI1-1D) has been shown to be a strong negative indicator for treatment response in AML.3 Here, we confirmed the adverse prognostic significance of EVI1-1D in an independent cohort of 272 AML patients. In addition, we demonstrate in a large cohort of 534 adults with pretreatment AML that 41 (7.8%) cases expressed high levels of EVI1 mRNA, which significantly predicted for an unfavorable prognosis. Importantly, 22% of these cases (9/41) were EVI1-1D−, but could be designated as EVI1+ when the expression of the other recently identified EVI1 splice forms5 was determined. Our findings inquire for the development of a reproducible assay, which discriminates EVI1+ from EVI1− AML for diagnostic purposes.
A diagnostic assay should be rapid and quantitative and it should take into account each of the different EVI1 splice forms. A multiplex PCR assay, by which the expression levels of each of the different EVI1 splice variants26,27 will be determined separately, is feasible but may be complicated to interpret. It may also be possible to develop primer probe combinations, which recognize all splice forms in one single EVI1-specific Q-PCR. Such an assay has the advantage that “high” versus “low” EVI1 cases may be determined in one easy-to-use test and it may provide an even better possibility to study EVI1 expression level as a continuous rather than a categoric variable. This would allow us to study more precisely whether a correlation exists between actual EVI1 expression levels and survival time of AML patients.
Although, a correlation analysis as discussed above would be helpful to further substantiate that high EVI1 levels associate with poor treatment response, a diagnostic assay should provide binary results and allows a reliable discrimination between EVI1+ and EVI1− cases. To avoid the inclusion of false-positive cases, we scored patients EVI1+ with expression levels of 30 or higher compared with 6 healthy bone marrow controls. Consequently, it may be possible that, by choosing such a high cutoff level, EVI1+ cases have been missed. Since important decisions will be made based on the outcome of a diagnostic EVI1 assay, it will be essential to include proper reproducible positive and negative controls, and develop algorithms by which EVI1+ cases will be indisputably identified.
AMLs with 3q26 lesions are among the most aggressive forms of human leukemia.3,28 An important message of this study is that a large fraction of AML cases that carried a 3q26 aberration was missed by standard karyotyping but was found positive by FISH. The abnormal pattern of elevated EVI1 levels but absence of MDS1/EVI1 expression (EVI1+ME−) provided the clue to these cryptic 3q26 abnormalities. Since EVI1 and ME are normally coexpressed, we hypothesized that a cryptic break within the 3q locus between these 2 closely related genes might cause their dissociated expression. EVI1+ME− leukemias that we recognized represent the most unfavorable subgroup of AML and they include most of the patients who carry 3q26 lesions. This provides another argument to implement an EVI1/ME multiplex RQ-PCR in the molecular diagnostic procedures of AML. Applying EVI1/ME RQ-PCR and karyotyping in combination with MDS1/EVI1- and RPN1-specific FISH on selected cases will disclose AMLs that belong to this distinctly unfavorable subgroup.
In only 2 EVI1+ME− cases, we did not find 3q26 lesions by FISH, suggesting another mechanism for aberrant EVI1 expression. BLIMP, an EVI1 homologue, which is a frequent target for chromosomal breaks (1p36) in B-cell lymphomas, has been shown to carry BLIMP point mutations in certain cases without 1p36 translocations,29 giving rise to short BLIMP (like EVI1) forms in favor of the long (like ME) product. Whether within the small EVI1+ME− group without major 3q26 lesions mutations at the molecular level have occurred in the MDS1/EVI1 locus remains to be investigated.
Using FISH, we did not detect hidden 3q26 aberrations in EVI1+ME+ AMLs. We carried out nucleotide sequencing in all EVI1+ME+ AML patients available but have not found any lesions at the molecular level in EVI1 or ME (data not shown). It is possible that excessive EVI1 and ME levels observed in those AML samples are caused by genetic defects in disease genes acting upstream of ME and EVI1. Interestingly, in a significant number of EVI1+ME+ leukemias, 11q23 chromosomal alterations were found. Kumar et al have reported that hematopoietic stem cells from MLL-AF9 knockin mice express high levels of EVI1,30 which may suggest that enforced expression of MLL-fusion genes directly or indirectly affect EVI1 and ME transcription. Transduction experiments in hematopoietic precursor cells may shed light on a putative effect of MLL-fusion proteins on transcription of ME and EVI1. Another subset of EVI1+ME+ leukemias did not carry cytogenetically identifiable 11q23 lesions. We ruled out hidden 11q23 translocations in those patients using Southern blot analyses and MLL-specific FISH (data not shown). Neither did we observe the existence of MLL-PTD by applying genomic PCR on those patients31 (data not shown), so that for the time being there is no suggestive clue as regards possible mechanisms for the excessively high levels of EVI1 and ME in this particular patient group.
Assessment of EVI1/ME not only provides a useful diagnostic and prognostic marker, but at least in EVI1+ME− AML with coexistent 3q26 chromosomal lesions, it pinpoints AML cases in which EVI1 may be the major disease gene playing a critical role in leukemic transformation. It remains a major challenge to unravel the mechanisms of transformation by EVI1 protein. EVI1 encodes a nuclear protein, which interacts with several proteins important in transcriptional control (eg, CtBp1,32 HDAC,33 SMAD3,34 P/CAF,32 and GATA135 ). Studies unraveling how these interactions exactly take place and as to whether those associations are crucial in leukemic transformation may provide insight that could be valuable for developing tools to specifically target EVI1+ leukemia cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Egied Simons for the graphic assistance. We are indebted to our colleagues from the bone marrow transplantation group and molecular diagnostics laboratory of the Department of Hematology for storage of the samples and molecular analyses, respectively.
This work was supported by grants from the Dutch Cancer Society Koningin Wilhelmina Fonds and the Association for International Cancer Research.
Authorship
Contribution: S.L., H.B.B., and R.D. designed the study; S.L., E.D., A.H., and C.A.J.E. performed research; S.L., Y.N., H.B.B., and R.D. analyzed data, P.J.M.V., H.B.B., and B.L revised the paper; S.L. and R.D. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruud Delwel, Erasmus University Medical Center, Department of Hematology, room Ee1392, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: h.delwel@erasmusmc.nl.