To the editor:
Three related articles in Blood have recently highlighted an issue with a significant impact on the study of regulatory T cells (Tregs), namely the specificity and affinity of the antibodies used to detect forkhead box P3 (FOXP3).1-3 There is phenomenal interest in Tregs and controversial hypotheses derived from conflicting data have been relatively common. This often reflects differences between model systems, diseases, and species, variability between individual patients, and the choice of experimental reagents.
The retention of only a partial clone name on commercialization of our anti-FOXP3 monoclonal antibodies, and the tendency not to sufficiently emphasize the original publication describing antibody production and characterization in product literature, may have caused some confusion. While the authors1-3 purchased 236A/E7, 206D, and 259D from different companies, each of these 3 different hybridoma cell lines was originally generated from the same fusion.4 Pillai and Karandikar's2 suggestion that 259D is conclusively a less sensitive detector of total FOXP3 conflicts not only with the data from Tran and Shevach,1,3 but also with our own data.4 Our initial experience was that 259D/C7 exhibited strong reactivity with FOXP3, both by immunohistochemistry and flow cytometry of FOXP3 transfectants.4 Here we present immunolabeling data that demonstrates that antibody 259D/C7 detects tonsillar FOXP3+ Tregs in routinely fixed tissue as effectively as any of our FOXP3 antibodies (Figure 1A). Antibodies 150D/E4 and 206D/B1 showed less labeling, as previously reported,4 although no immunolabeling data were originally presented. Furthermore, using Biolegend's (San Diego, CA) commercially prepared Alexa 488 direct fluorochrome conjugates, 259D exhibited comparable labeling of CD4+ peripheral blood cells to 206D and 150D, by intracellular flow cytometric staining.5
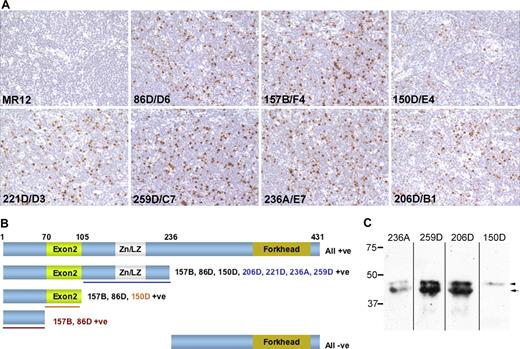
Comparative reactivity of “in house” hybridoma supernatants containing our FOXP3 monoclonal antibodies. Panel A illustrates immunoperoxidase labeling of the same area from sequentially cut sections from a single, paraffin-fixed and formalin-embedded tonsil, using previously described methodology.4 Tonsil tissue was collected with informed patient consent under ethical approval from the Oxfordshire Clinical Research Ethics Committee in accordance with the Declaration of Helsinki. Cells were counterstained with hematoxylin Gill No. 2 (Sigma-Aldrich, St Louis, MO), mounted in Aquatex (VWR International, Poole, United Kingdom) and viewed using a 40×/0.65 numerical aperture objective (Zeiss) on a bright field Axioskop microscope (Zeiss, Welwyn Garden City, United Kingdom). Images were captured using a Micropublisher 5MP RTV camera (QImaging, Surrey, BC). In terms of both frequency and intensity of FOXP3+ cell labeling, antibody 259D/C7 was as effective as the other clones. Panel B illustrates the full-length and truncated recombinant FOXP3 proteins that were expressed in COS-1 cells and their reactivity with the panel of FOXP3 antibodies by immunoperoxidase labeling. The expression of each truncated FOXP3 protein was also verified by detection of the Xpress epitope tag. The clone names in colored fonts indicate those antibodies with an epitope in the region of FOXP3 indicated by underlining in the same color. Numbers above the full-length protein give amino acid coordinates for FOXP3 truncations. Panel C shows Western blotting detection of FOXP3 expression in a lysate prepared from CD4+CD25+ peripheral blood mononuclear cells that were purified using a Regulatory T-Cell Isolation Kit (Miltenyi Biotech, Bisley, United Kingdom) from a blood buffy coat preparation obtained from the National Blood Service (Bristol, United Kingdom). Antibodies 259D/C7 and 206D/B1 strongly labeled both FOXP3 isoforms, while weaker labeling was observed with 236A/E7. As predicted, the smaller FOXP3 isoform lacking exon 2 was not detected by 150D/A3.
Comparative reactivity of “in house” hybridoma supernatants containing our FOXP3 monoclonal antibodies. Panel A illustrates immunoperoxidase labeling of the same area from sequentially cut sections from a single, paraffin-fixed and formalin-embedded tonsil, using previously described methodology.4 Tonsil tissue was collected with informed patient consent under ethical approval from the Oxfordshire Clinical Research Ethics Committee in accordance with the Declaration of Helsinki. Cells were counterstained with hematoxylin Gill No. 2 (Sigma-Aldrich, St Louis, MO), mounted in Aquatex (VWR International, Poole, United Kingdom) and viewed using a 40×/0.65 numerical aperture objective (Zeiss) on a bright field Axioskop microscope (Zeiss, Welwyn Garden City, United Kingdom). Images were captured using a Micropublisher 5MP RTV camera (QImaging, Surrey, BC). In terms of both frequency and intensity of FOXP3+ cell labeling, antibody 259D/C7 was as effective as the other clones. Panel B illustrates the full-length and truncated recombinant FOXP3 proteins that were expressed in COS-1 cells and their reactivity with the panel of FOXP3 antibodies by immunoperoxidase labeling. The expression of each truncated FOXP3 protein was also verified by detection of the Xpress epitope tag. The clone names in colored fonts indicate those antibodies with an epitope in the region of FOXP3 indicated by underlining in the same color. Numbers above the full-length protein give amino acid coordinates for FOXP3 truncations. Panel C shows Western blotting detection of FOXP3 expression in a lysate prepared from CD4+CD25+ peripheral blood mononuclear cells that were purified using a Regulatory T-Cell Isolation Kit (Miltenyi Biotech, Bisley, United Kingdom) from a blood buffy coat preparation obtained from the National Blood Service (Bristol, United Kingdom). Antibodies 259D/C7 and 206D/B1 strongly labeled both FOXP3 isoforms, while weaker labeling was observed with 236A/E7. As predicted, the smaller FOXP3 isoform lacking exon 2 was not detected by 150D/A3.
Pillai and Karandikar2 propose that differences may arise through FOXP3 antibodies recognizing different epitopes. We agree that this is a valid issue, particularly because human Tregs express an additional, smaller isoform of the FOXP3 protein that derives from a transcript lacking the second coding exon.6 Immunohistochemical labeling of truncated recombinant FOXP3 proteins demonstrated that all 7 antibodies recognized epitopes within the first 235 amino acids (aa) of the FOXP3 protein (Figure 1B). The absence of reactivity to the highly conserved FOXP3 forkhead domain is consistent with their lack of cross-reactivity with the related FOXP proteins.4 Antibodies157B/F4 and 86D/D6 recognized an epitope encoded by exon 1 (aa 1-70), 150D/A3 an epitope encoded by exon 2 (aa 70-105) and thus, 206D/B1, 221D/D3, 236A/E7 and 259D/C7 bind within the remaining N-terminus (aa 105-235). Antibodies 259D/C7 and 206D/B1 both labeled FOXP3 from CD4+CD25+ Tregs equally effectively by Western blotting. As expected, the smaller FOXP3 isoform lacking exon 2 was not recognized by 150D/A3 (Figure 1C). We therefore conclude that 259D/C7 represents a sensitive detector of FOXP3 protein expression.
We strongly advocate the use of well-validated mono-clonal antibodies to FOXP3 and the confirmation of unexpected or conflicting data with additional antibody reagents. Furthermore, as FOXP3 mRNA can be detected in the absence of FOXP3 protein expression,7,8 we recommend caution in interpreting data where FOXP3 gene expression is presented as the sole corroborating evidence to support discordant protein data.
Authorship
This work was supported by funding from the Leukaemia Research Fund (United Kingdom) and Cancer Research UK.
Contribution: B.C.F., P.A.B., and P.J.B. designed and performed experiments, and contributed to writing the manuscript. A.H.B. designed research and wrote the manuscript.
Conflict-of-interest disclosure: A.H.B. receives royalty payments from the commercialization of the FOXP3 antibodies described in Roncador et al.4 The other authors declare no competing financial interests.
Correspondence: Dr Alison H. Banham, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, Level 4 Academic Block, John Radcliffe Hospital, Headington, Oxford, Oxfordshire, OX3 9DU, United Kingdom; e-mail: alison.banham@ndcls.ox.ac.uk.