Abstract
Minor histocompatibility (H) antigens are the molecular targets of allo-immunity responsible both for the development of antitumor effects and for graft-versus-host disease (GVHD) in allogeneic hematopoietic stem cell transplantation (allo-HSCT). However, despite their potential clinical use, our knowledge of human minor H antigens is largely limited by the lack of efficient methods of their characterization. Here we report a robust and efficient method of minor H gene discovery that combines whole genome association scans (WGASs) with cytotoxic T-lymphocyte (CTL) assays, in which the genetic loci of minor H genes recognized by the CTL clones are precisely identified using pooled-DNA analysis of immortalized lymphoblastoid cell lines with/without susceptibility to those CTLs. Using this method, we have successfully mapped 2 loci: one previously characterized (HMSD encoding ACC-6), and one novel. The novel minor H antigen encoded by BCL2A1 was identified within a 26 kb linkage disequilibrium block on chromosome 15q25, which had been directly mapped by WGAS. The pool size required to identify these regions was no more than 100 individuals. Thus, once CTL clones are generated, this method should substantially facilitate discovery of minor H antigens applicable to targeted allo-immune therapies and also contribute to our understanding of human allo-immunity.
Introduction
Currently, allogeneic hematopoietic stem cell transplantation (allo-HSCT) has been established as one of the most effective therapeutic options for hematopoietic malignancies1 and is also implicated as a promising approach for some solid cancers.2 Its major therapeutic benefits are obtained from allo-immunity directed against patients' tumor cells (graft-versus-tumor [GVT] effects). However, the same kind of allo-immune reactions can also be directed against normal host tissues resulting in graft-versus-host disease (GVHD). In HLA-matched transplants, both GVT and GVHD are initiated by the recognition of HLA-bound polymorphic peptides, or minor histocompatibility (H) antigens, by donor T cells. Minor H antigens are typically encoded by dichotomous single nucleotide polymorphism (SNP) alleles, and may potentially be targeted by allo-immune reactions if the donor and recipient are mismatched at the minor H loci. Identification and characterization of minor H antigens that are specifically expressed in hematopoietic tissues, but not in other normal tissues, could contribute to the development of selective antileukemic therapies while minimizing unfavorable GVHD reactions, one of the most serious complications of allo-HSCT.3,4 Unfortunately, the total number of such useful minor H antigens that are currently molecularly characterized is still disappointingly small, including HA-1,5 HA-2,6 ACC-1Y and ACC-2,7 DRN-7,8 ACC-6,9 LB-ADIR-1F,10 HB-1,11 LRH-1,12 and 7A7-PANE1,13 limiting the number of patients eligible for such GVT-oriented immunotherapy.
Several techniques have been developed to identify novel minor H antigens targeted by CTLs generated from patients who have undergone transplantation. Among these, linkage analysis based on the cytotoxicity of the CTL clones against panels of lymphoblastoid cell lines (B-LCLs) from large pedigrees was proposed as a novel genetic approach,14 and has been successfully applied to identify novel minor H epitopes encoded by the BCL2A1 and P2RX5 genes.7,12 Nevertheless, the technology is still largely limited by its resolution, especially when large segregating families are not available. Linkage analysis using B-LCL panels from the Centre d'Etude du Polymorphisme Humain (CEPH) could only localize minor H loci within a range of 1.64 Mb to 5.5 Mb, which still contained 11 to 46 genes,7,12,14 thus requiring additional selection procedures to identify the actual minor H genes.
On the other hand, clinically relevant minor H antigens might be associated with common polymorphisms within the human population, and therefore could be ideal targets of genetic association studies, considering recent advances of large-scale genotyping technologies and the assets of the International HapMap Project.15,16 In this alternative genetic approach using the extensive linkage disequilibrium (LD) found within the human genome, target loci can be more efficiently localized within relatively small haplotype blocks without depending on limited numbers of recombination events, given the large number of genotyped genetic markers.17 Moreover, since the presence of a target minor H allele in individual target cells can be determined by ordinary immunologic assays using minor H antigen-specific CTLs, the characterization of minor H antigens should be significantly more straightforward than identifying alleles associated with typical common complex diseases, for which typically weak-to-moderate genetic effects have been assumed.18
In this report, we describe a high-performance, cost-effective method for the identification of minor H antigens, in which whole genome association scans (WGASs) are performed based on SNP array analysis of pooled DNA samples constructed from cytotoxicity-positive (CTX+) and cytotoxicity-negative (CTX−) B-LCLs as determined by their susceptibility to CTL clones. Based on this method, termed WGA/CTL, we were able to map the previously characterized ACC-6 minor H locus to a 115-kb block containing only 4 genes, including HMSD.9 Moreover, using the same approach, a novel minor H antigen encoded by the BCL2A1 gene was identified within a 26-kb block containing only BCL2A1 on chromosome 15q25. Surprisingly, the pool size required to identify these regions was no more than 100 individuals. Thus, this WGA/CTL method has significant potential to accelerate the discovery of minor H antigens that could be used in more selective, and thus more effective, allo-immune therapies in the near future.
Methods
Cell isolation and cell cultures
This study was approved by the institutional review board of the Aichi Cancer Center and the University of Tokyo. All blood or tissue samples were collected after written informed consent was obtained in accordance with the Declaration of Helsinki. B-LCLs were derived from allo-HSCT donors, recipients, and healthy volunteers. B-LCLs were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 1 mM sodium pyruvate.
Generation of CTL lines and clones
CTL lines were generated from peripheral blood mononuclear cells (PBMCs) obtained after transplantation by stimulation with irradiated (33 Gy) recipient PBMCs harvested before HSCT, thereafter stimulated weekly in RPMI 1640 supplemented with 10% pooled human serum and 2 mM l-glutamine. IL-2 was added on days 1 and 5 after the second and third stimulations. CTL clones were isolated by standard limiting dilution and expanded as previously described.7 CTL-1B9 was isolated from PBMCs harvested on day 30 after transplantation from a patient receiving a marrow graft from his HLA-identical sibling (HLA A11, A24, B39, B51, Cw7, Cw14), and CTL-2A12 has been described recently.9
Chromium release assay
Target cells were labeled with 0.1 mCi (3.7 MBq) of 51Cr for 2 hours, and 103 target cells/well were mixed with CTL at the effector-to-target (E/T) ratio indicated in a standard 4-hour cytotoxicity. All assays were performed at least in duplicate. Percent specific lysis was calculated as follows: ((Experimental cpm − Spontaneous cpm) / (Maximum cpm − Spontaneous cpm)) × 100.
Immunophenotyping by enzyme-linked immunosorbent assay
B-LCL cells (20 000 per well, which had been retrovirally transduced with restriction HLA cDNA for individual CTLs, if necessary) were plated in each well of 96-well round-bottomed plates, and corresponding CTL clones (10 000 per well) were added to each well. After overnight incubation at 37°C, 50 μL supernatant was collected and released IFN-γ was measured by standard enzyme-linked immunosorbent assay (ELISA).
Construction of pooled DNA and microarray experiments
Genomic DNA was individually extracted from immunophenotyped B-LCLs. After DNA concentrations were measured and adjusted to 50 μg/mL using the PicoGreen dsDNA Quantitation Reagent (Molecular Probes, Eugene, OR), the DNA specimens from CTX+ and CTX− B-LCLs were separately combined to generate individual pools. DNA pools were analyzed in pairs using Affymetrix GeneChip SNP-genotyping microarrays (Affymetrix, Tokyo, Japan) according to the manufacturer's protocol,19,20 where 2 independent experiments were performed for each array type (for more detailed statistical analysis for generated microarray data, see Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Estimation of LD blocks
LD structures of the candidate loci were evaluated based on empirical data from the International Hap Map Project (http://www.hapmap.org/).15 LD data for the relevant HapMap panels were downloaded from the HapMap web site and further analyzed using Haploview software (http://www.broad.mit.edu/mpg/haploview/).21
Transfection of 293T cells and ELISA
Twenty thousand 293T cells retrovirally transduced with HLA-A*2402 were plated in each well of 96-well flat-bottomed plates, cultured overnight at 37°C, then transfected with 0.12 μg of plasmid containing full-length BCL2A1 cDNA generated from either the patient or his donor using Trans IT-293 (Mirus, Madison, WI). B-LCLs of the recipient and his donor were used as positive and negative controls, respectively. Ten thousand CTL-1B9 cells were added to each well 20 hours after transfection. After overnight incubation at 37°C, 50 μL of supernatant was collected and IFN-γ was measured by ELISA.
SNP identification by direct sequencing
Complementary DNA prepared from B-LCLs was polymerase chain reaction (PCR) amplified for the coding region of BCL2A1 using the following primers: sense: 5′-AGAAGATGACAGACTGTGAATTTGG -3′; antisense: 5′-TCAACAGTATTGCTTCAGGAGAG-3′.
PCR products were purified and directly sequenced with the same primer and BigDye Terminator kit (version 3.1) by using ABI PRISM 3100 (Applied Biosystems, Foster City, CA).
Confirmatory SNP genotyping
Genotyping was carried out using fluorogenic 3′-minor groove binding (MGB) probes in a PCR assay. PCR was conducted in 10-μL reactions containing both allelic probes, 500 nM each of the primers, 1 × TaqMan Universal PCR Master Mix (Applied Biosystems), and 1 μL (100 ng) DNA. PCR cycling conditions were as follows: predenature, 50°C for 2 minutes, 95°C for 10 minutes, followed by 35 cycles of 92°C for 15 seconds and 60°C for 1 minute in a GeneAmp PCR System 9700 (Applied Biosystems). The PCR products were analyzed on an ABI 7900HT with the aid of SDS 2.2 software (Applied Biosystems).
Epitope reconstitution assay
The candidate BCL2A1-encoded minor H epitope and its allelic counterpart (DYLQYVLQI) peptides were synthesized by standard Fmoc chemistry. 51Cr-labeled CTX− donor B-LCLs were incubated with graded concentrations of the peptides and then used as targets in standard cytotoxicity assays.
Results
CTL-based typing and SNP array analysis of pooled DNA
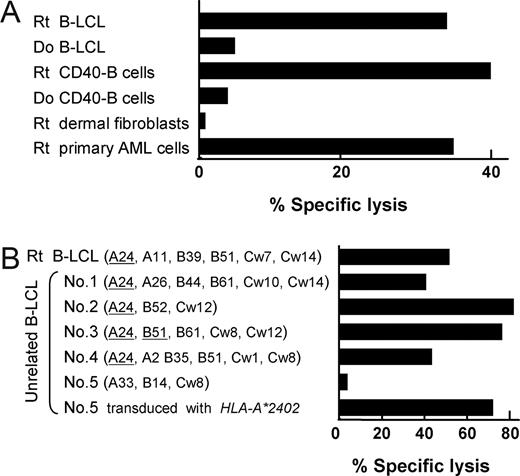
CTL-2A12 and CTL-1B9 are CTL clones established from the peripheral blood of 2 patients with leukemia who had received HLA-identical sibling HSCTs. Each clone demonstrated specific lysis against the B-LCLs of the recipient but not against donor B-LCLs, indicating recognition of minor H antigen (Figure 1A and Kawase et al9 ). The minor H antigen for CTL-2A12 had been previously identified by expression cloning9 ; on the other hand, the target minor H antigen for the HLA-A24–restricted CTL-1B9 clone, which was apparently hematopoietic lineage–specific (Figure 1A) and present in approximately 80% of the Japanese population (data not shown), had not yet been determined. Using these CTL clones, a panel of B-LCLs expressing the restriction HLA (HLA-B44 for CTL-2A12 and HLA-A24 for CTL-1B9) endogenously or retrovirally transduced, were subjected to “immunophenotyping” for the presence or absence of the minor H antigen by ELISA and, if necessary, by standard chromium release assay (CRA). Based on the assay results, for CTL-2A12 we initially collected 44 cytotoxicity-positive (CTX+) and 44 cytotoxicity-negative (CTX−) B-LCLs after screening 132 B-LCLs, while 57 CTX+ and 38 CTX− B-LCLs were obtained from 121 B-LCLs for CTL-1B9. From these sets of B-LCL panels, pools of DNA were generated and subjected to analysis on Affymetrix GeneChip 100 K and 500 K microarrays in duplicate.19,20
Specificity of CTL-1B9 against hematopoietic cells and its restriction HLA. (A) The cytolytic activity of CTL-1B9 was evaluated in a standard 4-hour 51Cr release assay (E/T ratio, 20:1). Targets used were B-LCL, CD40-activated (CD40-B) B cells, dermal fibroblasts, and primary acute myeloid leukemia cells from the recipient (Rt), and B-LCL and CD40-B cells from his donor (Do). Rt dermal fibroblasts were pretreated with 500 U/mL IFN-γ and 10 ng/mL TNF-α for 48 hours before 51Cr labeling. (B) Cytolytic activity of CTL-1B9 against a panel of B-LCLs derived from unrelated individuals, each of whom shared 1 or 2 class I MHC allele(s) with the recipient from whom the CTL-1B9 was generated. The shared HLA allele(s) with the recipient are underlined. B-LCLs (no. 5) which did not share any HLA alleles with the recipient, were retrovirally transduced with HLA-A*2402 cDNA and included to confirm HLA-A*2402 restriction by CTL-1B9. Results are typical of 2 experiments and data are the mean plus or minus the standard deviation (SD) of triplicates.
Specificity of CTL-1B9 against hematopoietic cells and its restriction HLA. (A) The cytolytic activity of CTL-1B9 was evaluated in a standard 4-hour 51Cr release assay (E/T ratio, 20:1). Targets used were B-LCL, CD40-activated (CD40-B) B cells, dermal fibroblasts, and primary acute myeloid leukemia cells from the recipient (Rt), and B-LCL and CD40-B cells from his donor (Do). Rt dermal fibroblasts were pretreated with 500 U/mL IFN-γ and 10 ng/mL TNF-α for 48 hours before 51Cr labeling. (B) Cytolytic activity of CTL-1B9 against a panel of B-LCLs derived from unrelated individuals, each of whom shared 1 or 2 class I MHC allele(s) with the recipient from whom the CTL-1B9 was generated. The shared HLA allele(s) with the recipient are underlined. B-LCLs (no. 5) which did not share any HLA alleles with the recipient, were retrovirally transduced with HLA-A*2402 cDNA and included to confirm HLA-A*2402 restriction by CTL-1B9. Results are typical of 2 experiments and data are the mean plus or minus the standard deviation (SD) of triplicates.
Detection of association between minor H phenotypes and marker SNPs
Genetic mapping of the minor H locus was performed by identifying marker SNPs that showed statistically significant deviations in allele-frequencies between CTX+ and CTX− pools based on the observed allele-specific signals in the microarray experiments. For this purpose, we evaluated the deviations of observed allele ratios between CTX+ and CTX− pools for each SNP on a given array (Document S1). An SNP was considered as positive for association if its test statistic exceeded an empirically determined threshold that provided a “genome-wide” P value of .05 in duplicate experiments (Document S1, Figures S1,S2, and Table S1). Threshold values for different pool sizes are also provided in Table S2 for further experiments. The positive SNPs eventually obtained for both CTLs are summarized in Table 1, where the 10 SNPs showing the highest test statistics are listed for individual experiments.
Positive SNPs from pooled DNA analysis
| CTL-2A12, Exp 1 . | CTL-2A12, Exp 2 . | CTL-1B9, Exp 1 . | CTL-1B9, Exp 2 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID . | Chr . | Position . | ΔRAΔRB . | rsID . | Chr . | Position . | ΔRAΔRB . | rsID . | Chr . | Position . | ΔRAΔRB . | rsID . | Chr . | Position . | ΔRAΔRB . |
| 50K X bal | |||||||||||||||
| rs10513933 | 18 | 59699669 | 0.366* | rs10513933 | 18 | 59699669 | 0.511† | rs1363258 | 5 | 103297593 | 0.239 | rs10499174 | 6 | 131209689 | 0.352* |
| rs9320028 | 18 | 59668150 | 0.255‡ | rs9320028 | 18 | 59668150 | 0.360* | rs726083 | 3 | 67093729 | 0.203 | rs30058 | 5 | 122325602 | 0.240 |
| rs6102 | 18 | 59721450 | 0.221 | rs10485873 | 7 | 3503743 | 0.157 | rs639243 | 5 | 31392931 | 0.198 | rs150724 | 16 | 61960443 | 0.213 |
| rs724533 | 23 | 116440574 | 0.137 | rs219323 | 14 | 59510440 | 0.150 | rs1936461 | 10 | 56519024 | 0.186 | rs1993129 | 8 | 63618836 | 0.208 |
| rs1341112 | 6 | 104919391 | 0.136 | rs10506892 | 12 | 82478539 | 0.147 | rs763876 | 12 | 94922502 | 0.186 | rs356946 | 13 | 69066751 | 0.201 |
| rs470490 | 18 | 61182216 | 0.136 | rs10492269 | 12 | 97786333 | 0.144 | rs958404 | 7 | 133054441 | 0.179 | rs2869268 | 4 | 86421898 | 0.184 |
| rs2826718 | 21 | 21471423 | 0.134 | rs10483466 | 14 | 35986827 | 0.139 | rs10486727 | 7 | 41672315 | 0.178 | rs287002 | 12 | 40312537 | 0.183 |
| rs10506697 | 12 | 73241741 | 0.128 | rs5910124 | 23 | 116408616 | 0.137 | rs2833488 | 21 | 32010112 | 0.176 | rs1146808 | 13 | 67688608 | 0.182 |
| rs10506891 | 12 | 82393029 | 0.127 | rs10512545 | 17 | 66337079 | 0.134 | rs379212 | 5 | 60977687 | 0.172 | rs10501287 | 11 | 42446011 | 0.180 |
| rs308995 | 14 | 59657919 | 0.125 | rs295678 | 5 | 58186928 | 0.131 | rs1954004 | 14 | 58627872 | 0.170 | rs564993 | 5 | 31393476 | 0.177 |
| 50KHindIII | |||||||||||||||
| rs9320032 | 18 | 59712191 | 0.486† | rs9320032 | 18 | 59712191 | 0.506† | rs1842353 | 8 | 63617543 | 0.244* | rs9300692 | 13 | 101216476 | 0.225‡ |
| rs8090046 | 18 | 59773066 | 0.207‡ | rs8090046 | 18 | 59773066 | 0.245* | rs10521202 | 17 | 12755289 | 0.201‡ | rs1842353 | 8 | 63617543 | 0.210‡ |
| rs1474220 | 2 | 108525317 | 0.193‡ | rs10498752 | 6 | 41876488 | 0.210‡ | rs7899961 | 10 | 59696431 | 0.198‡ | rs10520983 | 5 | 31314700 | 0.195‡ |
| rs10498752 | 6 | 41876488 | 0.178 | rs1941538 | 18 | 37994337 | 0.176 | rs9320974 | 6 | 124421441 | 0.197‡ | rs1334375 | 13 | 80897038 | 0.173 |
| rs2298578 | 21 | 21632551 | 0.167 | rs7682770 | 4 | 152748018 | 0.174 | rs10520983 | 5 | 31314700 | 0.179 | rs10519164 | 15 | 75412758 | 0.163 |
| rs7516032 | 1 | 91618962 | 0.165 | rs1445862 | 5 | 3675257 | 0.169 | rs1862446 | 5 | 147460749 | 0.170 | rs9322063 | 6 | 146852196 | 0.152 |
| rs5030938 | 10 | 70645922 | 0.164 | rs4696976 | 4 | 21058616 | 0.167 | rs1358778 | 20 | 13266796 | 0.169 | rs8067384 | 17 | 37926265 | 0.150 |
| rs1883041 | 21 | 44921845 | 0.158 | rs5030938 | 10 | 70645922 | 0.165 | rs1873790 | 4 | 83422480 | 0.166 | rs10521202 | 17 | 12755289 | 0.147 |
| rs3902916 | 4 | 189045176 | 0.155 | rs3902916 | 4 | 189045176 | 0.165 | rs1220724 | 4 | 70888705 | 0.162 | rs7914904 | 10 | 62749969 | 0.141 |
| rs1000551 | 20 | 58709208 | 0.154 | rs1883041 | 21 | 44921845 | 0.164 | rs9300692 | 13 | 101216476 | 0.157 | rs1220724 | 4 | 70888705 | 0.141 |
| 250K NspI | |||||||||||||||
| rs9950903 | 18 | 59781783 | 0.534† | rs9950903 | 18 | 59781783 | 1.036† | rs1879894 | 15 | 78055874 | 0.846† | rs1879894 | 15 | 78055874 | 1.072† |
| rs1463835 | 3 | 23539615 | 0.532† | rs8090586 | 18 | 59781864 | 0.518† | rs9646294 | 16 | 6110019 | 0.484† | rs6771859 | 3 | 190642054 | 0.387† |
| rs16975459 | 18 | 37802275 | 0.383* | rs6473170 | 8 | 80664840 | 0.338* | rs17734332 | 5 | 134945240 | 0.365† | rs10512261 | 9 | 98804394 | 0.299* |
| rs8090586 | 18 | 59781864 | 0.367* | rs4510128 | 18 | 59782312 | 0.310‡ | rs566619 | 7 | 41381538 | 0.345* | rs12122772 | 1 | 60384564 | 0.287* |
| rs16872621 | 4 | 22081055 | 0.312‡ | rs1006755 | 18 | 59782026 | 0.300‡ | rs17737566 | 6 | 50345280 | 0.310* | rs2153155 | 4 | 26034162 | 0.248‡ |
| rs870582 | 6 | 125097114 | 0.301‡ | rs7039378 | 9 | 118735938 | 0.258 | rs3849955 | 9 | 28350374 | 0.285* | rs17126896 | 14 | 53320494 | 0.246‡ |
| rs1015416 | 18 | 59720363 | 0.270‡ | rs1860563 | 16 | 6418899 | 0.258 | rs4616156 | 13 | 86581518 | 0.273* | rs1328652 | 13 | 35607527 | 0.240 |
| rs2155907 | 11 | 97599883 | 0.227 | rs4699126 | 4 | 105709109 | 0.212 | rs2484698 | 1 | 217474460 | 0.263* | rs7021551 | 9 | 27446645 | 0.237 |
| rs2112948 | 5 | 50994294 | 0.222 | rs10275055 | 7 | 156212079 | 0.204 | rs17139603 | 11 | 79638632 | 0.262* | rs252817 | 5 | 106752487 | 0.237 |
| rs2919747 | 2 | 129681506 | 0.217 | rs1526411 | 7 | 124658309 | 0.201 | rs2156737 | 4 | 100642529 | 0.246‡ | rs10772587 | 12 | 12681356 | 0.235 |
| 250K StyI | |||||||||||||||
| rs6102 | 18 | 59721450 | 0.597† | rs6102 | 18 | 59721450 | 0.495† | rs9383925 | 6 | 151975774 | 0.819† | rs201204 | 6 | 104842863 | 0.688† |
| rs9951512 | 18 | 59690885 | 0.374* | rs9945924 | 18 | 59771746 | 0.407* | rs6497397 | 16 | 19646258 | 0.311‡ | rs12556155 | 23 | 108836419 | 0.442† |
| rs6496897 | 15 | 90493249 | 0.320‡ | rs9951512 | 18 | 59690885 | 0.317‡ | rs917252 | 7 | 22219990 | 0.289‡ | rs4791422 | 17 | 10605304 | 0.435† |
| rs9945924 | 18 | 59771746 | 0.315‡ | rs1983205 | 3 | 157782892 | 0.314‡ | rs1019403 | 3 | 7823997 | 0.260‡ | rs7749012 | 6 | 106459559 | 0.336* |
| rs12707805 | 8 | 107404746 | 0.303‡ | rs950865 | 5 | 2720684 | 0.307‡ | rs17053134 | 5 | 155373544 | 0.259‡ | rs509951 | 5 | 31385483 | 0.308‡ |
| rs10971778 | 9 | 33893184 | 0.296‡ | rs2278179 | 18 | 59715512 | 0.292‡ | rs11710880 | 3 | 72214965 | 0.246 | rs16879024 | 8 | 32225711 | 0.256‡ |
| rs6565076 | 16 | 81487818 | 0.294‡ | rs10427722 | 22 | 36417752 | 0.289‡ | rs17167866 | 7 | 13919264 | 0.237 | rs2100054 | 15 | 75293482 | 0.252 |
| rs2278179 | 18 | 59715512 | 0.291‡ | rs17156659 | 7 | 82046820 | 0.271 | rs10867062 | 9 | 137935241 | 0.237 | rs11811023 | 1 | 143805934 | 0.240 |
| rs7806238 | 7 | 29906442 | 0.290‡ | rs4502324 | 18 | 4811261 | 0.262 | rs5925800 | 23 | 23278707 | 0.235 | rs17382798 | 15 | 75256074 | 0.231 |
| rs965888 | 18 | 38062658 | 0.283‡ | rs1348428 | 2 | 225927288 | 0.260 | rs2255831 | 4 | 146614313 | 0.234 | rs2030302 | 17 | 12526591 | 0.231 |
| CTL-2A12, Exp 1 . | CTL-2A12, Exp 2 . | CTL-1B9, Exp 1 . | CTL-1B9, Exp 2 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID . | Chr . | Position . | ΔRAΔRB . | rsID . | Chr . | Position . | ΔRAΔRB . | rsID . | Chr . | Position . | ΔRAΔRB . | rsID . | Chr . | Position . | ΔRAΔRB . |
| 50K X bal | |||||||||||||||
| rs10513933 | 18 | 59699669 | 0.366* | rs10513933 | 18 | 59699669 | 0.511† | rs1363258 | 5 | 103297593 | 0.239 | rs10499174 | 6 | 131209689 | 0.352* |
| rs9320028 | 18 | 59668150 | 0.255‡ | rs9320028 | 18 | 59668150 | 0.360* | rs726083 | 3 | 67093729 | 0.203 | rs30058 | 5 | 122325602 | 0.240 |
| rs6102 | 18 | 59721450 | 0.221 | rs10485873 | 7 | 3503743 | 0.157 | rs639243 | 5 | 31392931 | 0.198 | rs150724 | 16 | 61960443 | 0.213 |
| rs724533 | 23 | 116440574 | 0.137 | rs219323 | 14 | 59510440 | 0.150 | rs1936461 | 10 | 56519024 | 0.186 | rs1993129 | 8 | 63618836 | 0.208 |
| rs1341112 | 6 | 104919391 | 0.136 | rs10506892 | 12 | 82478539 | 0.147 | rs763876 | 12 | 94922502 | 0.186 | rs356946 | 13 | 69066751 | 0.201 |
| rs470490 | 18 | 61182216 | 0.136 | rs10492269 | 12 | 97786333 | 0.144 | rs958404 | 7 | 133054441 | 0.179 | rs2869268 | 4 | 86421898 | 0.184 |
| rs2826718 | 21 | 21471423 | 0.134 | rs10483466 | 14 | 35986827 | 0.139 | rs10486727 | 7 | 41672315 | 0.178 | rs287002 | 12 | 40312537 | 0.183 |
| rs10506697 | 12 | 73241741 | 0.128 | rs5910124 | 23 | 116408616 | 0.137 | rs2833488 | 21 | 32010112 | 0.176 | rs1146808 | 13 | 67688608 | 0.182 |
| rs10506891 | 12 | 82393029 | 0.127 | rs10512545 | 17 | 66337079 | 0.134 | rs379212 | 5 | 60977687 | 0.172 | rs10501287 | 11 | 42446011 | 0.180 |
| rs308995 | 14 | 59657919 | 0.125 | rs295678 | 5 | 58186928 | 0.131 | rs1954004 | 14 | 58627872 | 0.170 | rs564993 | 5 | 31393476 | 0.177 |
| 50KHindIII | |||||||||||||||
| rs9320032 | 18 | 59712191 | 0.486† | rs9320032 | 18 | 59712191 | 0.506† | rs1842353 | 8 | 63617543 | 0.244* | rs9300692 | 13 | 101216476 | 0.225‡ |
| rs8090046 | 18 | 59773066 | 0.207‡ | rs8090046 | 18 | 59773066 | 0.245* | rs10521202 | 17 | 12755289 | 0.201‡ | rs1842353 | 8 | 63617543 | 0.210‡ |
| rs1474220 | 2 | 108525317 | 0.193‡ | rs10498752 | 6 | 41876488 | 0.210‡ | rs7899961 | 10 | 59696431 | 0.198‡ | rs10520983 | 5 | 31314700 | 0.195‡ |
| rs10498752 | 6 | 41876488 | 0.178 | rs1941538 | 18 | 37994337 | 0.176 | rs9320974 | 6 | 124421441 | 0.197‡ | rs1334375 | 13 | 80897038 | 0.173 |
| rs2298578 | 21 | 21632551 | 0.167 | rs7682770 | 4 | 152748018 | 0.174 | rs10520983 | 5 | 31314700 | 0.179 | rs10519164 | 15 | 75412758 | 0.163 |
| rs7516032 | 1 | 91618962 | 0.165 | rs1445862 | 5 | 3675257 | 0.169 | rs1862446 | 5 | 147460749 | 0.170 | rs9322063 | 6 | 146852196 | 0.152 |
| rs5030938 | 10 | 70645922 | 0.164 | rs4696976 | 4 | 21058616 | 0.167 | rs1358778 | 20 | 13266796 | 0.169 | rs8067384 | 17 | 37926265 | 0.150 |
| rs1883041 | 21 | 44921845 | 0.158 | rs5030938 | 10 | 70645922 | 0.165 | rs1873790 | 4 | 83422480 | 0.166 | rs10521202 | 17 | 12755289 | 0.147 |
| rs3902916 | 4 | 189045176 | 0.155 | rs3902916 | 4 | 189045176 | 0.165 | rs1220724 | 4 | 70888705 | 0.162 | rs7914904 | 10 | 62749969 | 0.141 |
| rs1000551 | 20 | 58709208 | 0.154 | rs1883041 | 21 | 44921845 | 0.164 | rs9300692 | 13 | 101216476 | 0.157 | rs1220724 | 4 | 70888705 | 0.141 |
| 250K NspI | |||||||||||||||
| rs9950903 | 18 | 59781783 | 0.534† | rs9950903 | 18 | 59781783 | 1.036† | rs1879894 | 15 | 78055874 | 0.846† | rs1879894 | 15 | 78055874 | 1.072† |
| rs1463835 | 3 | 23539615 | 0.532† | rs8090586 | 18 | 59781864 | 0.518† | rs9646294 | 16 | 6110019 | 0.484† | rs6771859 | 3 | 190642054 | 0.387† |
| rs16975459 | 18 | 37802275 | 0.383* | rs6473170 | 8 | 80664840 | 0.338* | rs17734332 | 5 | 134945240 | 0.365† | rs10512261 | 9 | 98804394 | 0.299* |
| rs8090586 | 18 | 59781864 | 0.367* | rs4510128 | 18 | 59782312 | 0.310‡ | rs566619 | 7 | 41381538 | 0.345* | rs12122772 | 1 | 60384564 | 0.287* |
| rs16872621 | 4 | 22081055 | 0.312‡ | rs1006755 | 18 | 59782026 | 0.300‡ | rs17737566 | 6 | 50345280 | 0.310* | rs2153155 | 4 | 26034162 | 0.248‡ |
| rs870582 | 6 | 125097114 | 0.301‡ | rs7039378 | 9 | 118735938 | 0.258 | rs3849955 | 9 | 28350374 | 0.285* | rs17126896 | 14 | 53320494 | 0.246‡ |
| rs1015416 | 18 | 59720363 | 0.270‡ | rs1860563 | 16 | 6418899 | 0.258 | rs4616156 | 13 | 86581518 | 0.273* | rs1328652 | 13 | 35607527 | 0.240 |
| rs2155907 | 11 | 97599883 | 0.227 | rs4699126 | 4 | 105709109 | 0.212 | rs2484698 | 1 | 217474460 | 0.263* | rs7021551 | 9 | 27446645 | 0.237 |
| rs2112948 | 5 | 50994294 | 0.222 | rs10275055 | 7 | 156212079 | 0.204 | rs17139603 | 11 | 79638632 | 0.262* | rs252817 | 5 | 106752487 | 0.237 |
| rs2919747 | 2 | 129681506 | 0.217 | rs1526411 | 7 | 124658309 | 0.201 | rs2156737 | 4 | 100642529 | 0.246‡ | rs10772587 | 12 | 12681356 | 0.235 |
| 250K StyI | |||||||||||||||
| rs6102 | 18 | 59721450 | 0.597† | rs6102 | 18 | 59721450 | 0.495† | rs9383925 | 6 | 151975774 | 0.819† | rs201204 | 6 | 104842863 | 0.688† |
| rs9951512 | 18 | 59690885 | 0.374* | rs9945924 | 18 | 59771746 | 0.407* | rs6497397 | 16 | 19646258 | 0.311‡ | rs12556155 | 23 | 108836419 | 0.442† |
| rs6496897 | 15 | 90493249 | 0.320‡ | rs9951512 | 18 | 59690885 | 0.317‡ | rs917252 | 7 | 22219990 | 0.289‡ | rs4791422 | 17 | 10605304 | 0.435† |
| rs9945924 | 18 | 59771746 | 0.315‡ | rs1983205 | 3 | 157782892 | 0.314‡ | rs1019403 | 3 | 7823997 | 0.260‡ | rs7749012 | 6 | 106459559 | 0.336* |
| rs12707805 | 8 | 107404746 | 0.303‡ | rs950865 | 5 | 2720684 | 0.307‡ | rs17053134 | 5 | 155373544 | 0.259‡ | rs509951 | 5 | 31385483 | 0.308‡ |
| rs10971778 | 9 | 33893184 | 0.296‡ | rs2278179 | 18 | 59715512 | 0.292‡ | rs11710880 | 3 | 72214965 | 0.246 | rs16879024 | 8 | 32225711 | 0.256‡ |
| rs6565076 | 16 | 81487818 | 0.294‡ | rs10427722 | 22 | 36417752 | 0.289‡ | rs17167866 | 7 | 13919264 | 0.237 | rs2100054 | 15 | 75293482 | 0.252 |
| rs2278179 | 18 | 59715512 | 0.291‡ | rs17156659 | 7 | 82046820 | 0.271 | rs10867062 | 9 | 137935241 | 0.237 | rs11811023 | 1 | 143805934 | 0.240 |
| rs7806238 | 7 | 29906442 | 0.290‡ | rs4502324 | 18 | 4811261 | 0.262 | rs5925800 | 23 | 23278707 | 0.235 | rs17382798 | 15 | 75256074 | 0.231 |
| rs965888 | 18 | 38062658 | 0.283‡ | rs1348428 | 2 | 225927288 | 0.260 | rs2255831 | 4 | 146614313 | 0.234 | rs2030302 | 17 | 12526591 | 0.231 |
Significant SNPs that appeared on both experiments are underlined.
Genomewide P < .01.
Genomewide P < .001.
Genomewide P < .05.
Mapping of the minor H loci by WGASs
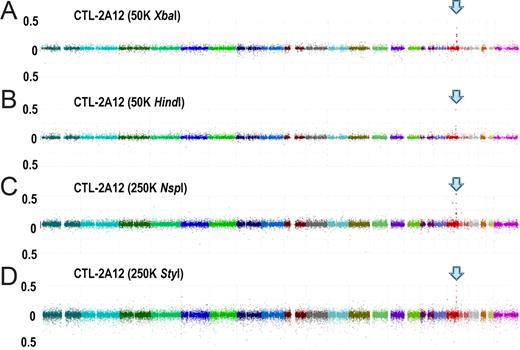
All the SNPs significantly associated with susceptibility to CTL-2A12 were correctly mapped within a single 115 kb LD block at chromosome 18q21 containing the HMSD gene (Figures 2 and 3A), which had been previously shown to encode the ACC-6 minor H antigen recognized by CTL-2A12.9 According to the above criteria, no false-positive SNPs were reported in any array types (Table 1). Confirmation genotyping of individual B-LCLs from both panels revealed none of the 44 that had been immunophenotyped as CTX− were misjudged, while 8 of the 44 CTX+ B-LCLs were found to actually carry no minor H-positive allele for ACC-6, which was likely due to the inclusion of individual B-LCLs showing borderline cytotoxicity (data not shown).
Whole genome association scans performed with pooled DNA generated based on immunophenotyping with CTL-2A12. Pooled DNAs generated from 44 CTX+ and 44 CTX− B-LCLs were analyzed with 50 K XbaI (A), 50 K HindIII (B), 250 K NspI (C), and 250 K StyI (D) arrays. Test statistics were calculated for all SNPs and plotted in the chromosomal order. In all SNP array types, a common association peak is observed at 18q21, to which the minor H antigen for CTL-2A12, encoded by the HMSD gene, had been mapped based on expression cloning9 (arrows).
Whole genome association scans performed with pooled DNA generated based on immunophenotyping with CTL-2A12. Pooled DNAs generated from 44 CTX+ and 44 CTX− B-LCLs were analyzed with 50 K XbaI (A), 50 K HindIII (B), 250 K NspI (C), and 250 K StyI (D) arrays. Test statistics were calculated for all SNPs and plotted in the chromosomal order. In all SNP array types, a common association peak is observed at 18q21, to which the minor H antigen for CTL-2A12, encoded by the HMSD gene, had been mapped based on expression cloning9 (arrows).
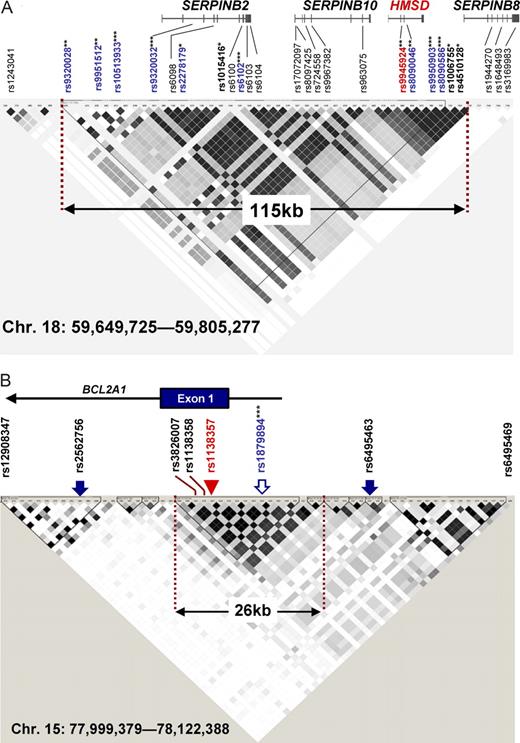
Linkage disequilibrium (LD) block mapped by CTL-2A12 and CTL-1B9. (A) An LD block map identified by pairwise r2 plot from HapMap CEU data are overlayed with SNPs available from Affymetrix GeneChip SNP-genotyping microarrays (arrows) and 4 genes in the 115 kb block. SNPs that emerged repeatedly in the 2 independent experiments are indicated in blue. The genomewide P values for positive SNPs are shown as follows: *P < .05; **P < .01; ***P < .001. The intronic SNP (rs9945924) controlling the alternative splicing of HMSD transcripts and expression of encoded ACC-6 minor H antigen is indicated in red. (B) LD blocks identified by pairwise r2 plot from HapMap JPT data are overlayed with SNPs available from Affymetrix GeneChip SNP-genotyping microarrays (arrows) and exon 1 of the BCL2A1 gene. The only SNP showing a high association with CTL-1B9 immunophenotypes (rs1879894) is shown as an open arrow. The nonsynonymous SNP (rs1138357) controlling the expression of the minor H antigen recognized by CTL-1B9 is indicated by a red arrowhead. ***SNP with genomewide P < .001. The 2 SNPs adjacent to the 26 kb LD block (rs2562756 and rs6495463) never gave a significant genomewide P value.
Linkage disequilibrium (LD) block mapped by CTL-2A12 and CTL-1B9. (A) An LD block map identified by pairwise r2 plot from HapMap CEU data are overlayed with SNPs available from Affymetrix GeneChip SNP-genotyping microarrays (arrows) and 4 genes in the 115 kb block. SNPs that emerged repeatedly in the 2 independent experiments are indicated in blue. The genomewide P values for positive SNPs are shown as follows: *P < .05; **P < .01; ***P < .001. The intronic SNP (rs9945924) controlling the alternative splicing of HMSD transcripts and expression of encoded ACC-6 minor H antigen is indicated in red. (B) LD blocks identified by pairwise r2 plot from HapMap JPT data are overlayed with SNPs available from Affymetrix GeneChip SNP-genotyping microarrays (arrows) and exon 1 of the BCL2A1 gene. The only SNP showing a high association with CTL-1B9 immunophenotypes (rs1879894) is shown as an open arrow. The nonsynonymous SNP (rs1138357) controlling the expression of the minor H antigen recognized by CTL-1B9 is indicated by a red arrowhead. ***SNP with genomewide P < .001. The 2 SNPs adjacent to the 26 kb LD block (rs2562756 and rs6495463) never gave a significant genomewide P value.
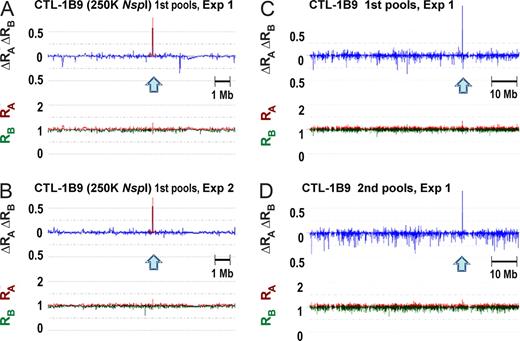
On the other hand, positive association of the target minor H antigen with CTL-1B9 was detected in 2 independent loci: SNP rs1879894 at 15q25.1 in 250 K NspI (Table 1, Figure 4A-B, and Figure S5) and SNP rs1842353 at 8q12.3 in 50 K HindIII (Table 1 and Figure S3A). We eventually focused on rs1879894, as it showed a much more significant genome-wide P value than SNP rs1842353 (Table 1). In contrast to the CTL-2A12 case, where many mutually correlated SNPs around the most significant one created a broad peak in the statistic plots (Figure 2 arrows and Figure S3), the adjacent SNPs (rs6495463 and rs2562756; Figure 3B solid arrows) around rs187894 (Figure 3B open arrow) did not show large test statistic values, reflecting the fact that no marker SNPs on 100 K and 500 K arrays exist in high LD (Figure 3B dashed red lines encompassing 26 kb) with this SNP according to the HapMap data. To further confirm the association, we generated additional B-LCL pools consisting of 75 CTX+ and 34 CTX− B-LCLs from another set of 128 B-LCLs, and performed a WGAS. As expected, the WGAS of the second pools also identified the identical SNP with the highest test statistic value in duplicate experiments, unequivocally indicating that this SNP is truly associated with the minor H locus of interest (Figure 4C,D and Table S3). The association was also detected when the references in the first and second pools were swapped (data not shown).
Reproducible detection of association with the immunophenotypes determined by CTL-1B9 at the BCL2A1 locus. The maximum test statistic value was observed at a single SNP (rs1879894) within 15q25.1 in duplicate experiments for the first pools consisting of 57 CTX+ and 38 CTX− B-LCLs (A-C). The peak association at the same SNP was reproduced in the experiments with the second pools consisting of 75 CTX+ and 34 CTX− LCLs (D). Test statistic values (ΔRAΔRB) are plotted by blue lines together with their RA (red) and RB (green) values. The expected ΔRAΔRB values multiplied by r2 correlation coefficients for the adjacent SNPs within 500 kb from the SNP rs1879894 are overlaid by red lines (A,B).
Reproducible detection of association with the immunophenotypes determined by CTL-1B9 at the BCL2A1 locus. The maximum test statistic value was observed at a single SNP (rs1879894) within 15q25.1 in duplicate experiments for the first pools consisting of 57 CTX+ and 38 CTX− B-LCLs (A-C). The peak association at the same SNP was reproduced in the experiments with the second pools consisting of 75 CTX+ and 34 CTX− LCLs (D). Test statistic values (ΔRAΔRB) are plotted by blue lines together with their RA (red) and RB (green) values. The expected ΔRAΔRB values multiplied by r2 correlation coefficients for the adjacent SNPs within 500 kb from the SNP rs1879894 are overlaid by red lines (A,B).
Identification of the minor H epitope recognized by CTL-1B9
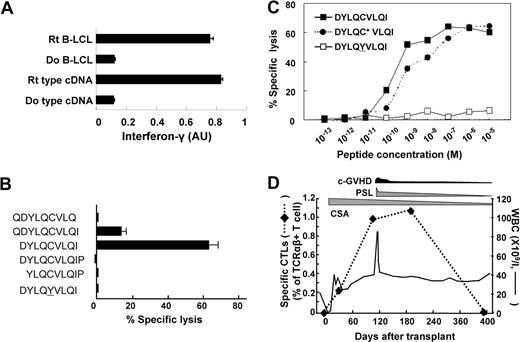
The LD block containing SNP rs1879894 that was singled out from more than 500 000 SNP markers with 2 sets of DNA pools only encodes exon 1 of BCL2A1 (Figure 3B). To our surprise, this was the region to which we had previously mapped an HLA-A24–restricted minor H antigen, ACC-1Y.7 We first confirmed that full-length BCL2A1 cDNA cloned only from the recipient but not his donor could stimulate interferon-γ secretion from CTL-1B9 when transduced into donor B-LCL (Figure 5A), indicating that BCL2A1 is a bona fide gene encoding minor H antigen recognized by CTL-1B9. We next genotyped 3 nonsynonymous SNPs in the BCL2A1 exon 1 sequence (Figure 3B) and comparison was made between the genotypes and the susceptibility to CTL-1B9 of 9 HLA-A*2402+ B-LCLs, including ones generated from the recipient (from whom CTL-1B9 was established) and his donor. Susceptibility to CTL-1B9 correlated completely with the presence of guanine at SNP rs1138357 (nucleotide position 238, according to the mRNA sequence for NM_004049.2) and thymine at SNP rs1138358 (nucleotide position 299) (Table 2), suggesting that the expression of the minor H epitope recognized by CTL-1B9 is controlled by either of these SNPs. We searched for nonameric amino acid sequences spanning the 2 SNPs using BIMAS software,22 since most reported HLA-A*2402 binding peptides contain 9 amino acid residues.23 Among these, a nonameric peptide, DYLQCVLQI (the polymorphic residue being underlined), has a predicted binding score of 75 and was considered as a candidate minor H epitope. As shown in Figure 5B, the DYLQCVLQI was strongly recognized by CTL-1B9, whereas its allelic counterpart, DYLQYVLQI, was not. Decameric peptide, QDYLQCVLQI, on the other hand, appeared to be weakly recognized; however, it is likely that the nonameric form was actually being presented after N-terminal glutamine cleavage by aminopeptidase in the culture medium. Because it was possible that the cystine might be cysteinylated, recognition of synthetic peptides DYLQCVLQI and cysteinylated DYLQC*VLQI were assayed using CTL-1B9. Half-maximal lysis for the former was obtained at a concentration of 200 pM, whereas recognition of the latter was several-fold weaker (Figure 5C). Thus, we concluded that DYLQCVLQI defines the cognate HLA-A*2402–restricted CTL-1B9 epitope, now designated ACC-1C. This incidentally provides a second example of products from both dichotomous SNP alleles being recognized as HLA-A*2402–restricted minor H antigens, the first example being the HB-1 minor H antigen.24 Finally, real-time quantitative PCR revealed that T cells carrying the complementarity-determining region 3 sequence identical to CTL-1B9 became detectable in the patient's blood at the frequencies of 0.22%, 0.91%, 1.07% and 0.01% among TCRαβ+ T cells at days 30, 102, 196, and 395 after transplantation, respectively, suggesting that ACC-1C minor H antigen is indeed immunogenic (Figure 5D).
Identification of the CTL-1B9 minimal minor H epitope. (A) Interferon-γ production from CTL-1B9 against HLA-A*2402–transduced 293T cells transfected with plasmid encoding full-length BCL2A1 cDNA cloned from either the recipient (Rt) from whom CTL-1B9 was isolated or his donor (Do). Rt B-LCL and Do B-LCL were used as positive and negative controls, respectively. Secreted interferon-γ was measured by ELISA and is expressed in arbitrary units (AUs) corresponding to optical density at 630 nm. Results are typical of 2 experiments and data are the mean plus or minus SD of triplicates. (B) A peptide reconstitution assay was conducted to determine the minimal epitope for CTL-1B9. Nonameric peptide (DYLQCVLQI), 2 nonameric peptides shifted by one amino acid to N- or C-terminus, N- and C-terminal extended decameric peptides, and its allelic counterpart (DYLQYVLQI) were synthesized and tested by adding to antigen-negative donor B-LCL at 10 nM in a standard 51Cr release assay. Results are typical of 2 experiments and data are the mean plus or minus SD of triplicates. (C) Titration of the candidate minor H peptide by epitope reconstitution assay. Chromium-labeled donor B-LCLs were distributed to wells of 96-well round-bottomed plates, pulsed with serial dilutions of the indicated peptides for 30 minutes at room temperature, and then used as targets for CTL-1B9 in a standard 51Cr release assay. A cysteinylated peptide (indicated by an asterisk) was included as an alternative form of the potential epitope. Results are typical of 2 experiments. (D) Tracking of ACC-1C–specific T cells in the recipient's peripheral blood. In order to longitudinally analyze the kinetics of the ACC-1C–specific CTLs in peripheral blood from the patient from whom CTL-1B9 was established, a real-time quantitative PCR was conducted. Complementary DNAs of peripheral blood mononuclear cells from the donor and patient before and after HSCT were prepared from the patient. Real-time PCR analysis was performed using a TaqMan assay as described previously.9 The primers and fluorogenic probe sequences spanning the CTL-1B9 complementarity-determining region 3 (CDR3) were used to detect T cells carrying the CDR3 sequences identical to that of CTL-1B9. The primers and fluorogenic probe sequences spanning constant region of TCR beta chain (TCRBC) mRNA were used as internal control. Samples were quantified with the comparative CT method. The delta CT value was determined by subtracting the average CT value for TCRBC from the average CTL-1B9 CDR3 CT value. The standard curve for the proportion of CTL-1B9 among TCRαβ+ T cells was composed by plotting mean delta CT values for each ratio, and the percentages of T cells carrying the CDR3 sequence identical to CTL-1B9 were calculated by using this standard curve. During this period, quiescent chronic GVHD, which required steroid treatment, developed; however, involvement of immune reaction to ACC-1C minor H antigen was unlikely since its frequency increased even after resolution of most chronic GVHD symptoms. c-GVHD, chronic GVHD; CSA, cyclosporine A; PSL, prednisolone; WBC, white blood cell count.
Identification of the CTL-1B9 minimal minor H epitope. (A) Interferon-γ production from CTL-1B9 against HLA-A*2402–transduced 293T cells transfected with plasmid encoding full-length BCL2A1 cDNA cloned from either the recipient (Rt) from whom CTL-1B9 was isolated or his donor (Do). Rt B-LCL and Do B-LCL were used as positive and negative controls, respectively. Secreted interferon-γ was measured by ELISA and is expressed in arbitrary units (AUs) corresponding to optical density at 630 nm. Results are typical of 2 experiments and data are the mean plus or minus SD of triplicates. (B) A peptide reconstitution assay was conducted to determine the minimal epitope for CTL-1B9. Nonameric peptide (DYLQCVLQI), 2 nonameric peptides shifted by one amino acid to N- or C-terminus, N- and C-terminal extended decameric peptides, and its allelic counterpart (DYLQYVLQI) were synthesized and tested by adding to antigen-negative donor B-LCL at 10 nM in a standard 51Cr release assay. Results are typical of 2 experiments and data are the mean plus or minus SD of triplicates. (C) Titration of the candidate minor H peptide by epitope reconstitution assay. Chromium-labeled donor B-LCLs were distributed to wells of 96-well round-bottomed plates, pulsed with serial dilutions of the indicated peptides for 30 minutes at room temperature, and then used as targets for CTL-1B9 in a standard 51Cr release assay. A cysteinylated peptide (indicated by an asterisk) was included as an alternative form of the potential epitope. Results are typical of 2 experiments. (D) Tracking of ACC-1C–specific T cells in the recipient's peripheral blood. In order to longitudinally analyze the kinetics of the ACC-1C–specific CTLs in peripheral blood from the patient from whom CTL-1B9 was established, a real-time quantitative PCR was conducted. Complementary DNAs of peripheral blood mononuclear cells from the donor and patient before and after HSCT were prepared from the patient. Real-time PCR analysis was performed using a TaqMan assay as described previously.9 The primers and fluorogenic probe sequences spanning the CTL-1B9 complementarity-determining region 3 (CDR3) were used to detect T cells carrying the CDR3 sequences identical to that of CTL-1B9. The primers and fluorogenic probe sequences spanning constant region of TCR beta chain (TCRBC) mRNA were used as internal control. Samples were quantified with the comparative CT method. The delta CT value was determined by subtracting the average CT value for TCRBC from the average CTL-1B9 CDR3 CT value. The standard curve for the proportion of CTL-1B9 among TCRαβ+ T cells was composed by plotting mean delta CT values for each ratio, and the percentages of T cells carrying the CDR3 sequence identical to CTL-1B9 were calculated by using this standard curve. During this period, quiescent chronic GVHD, which required steroid treatment, developed; however, involvement of immune reaction to ACC-1C minor H antigen was unlikely since its frequency increased even after resolution of most chronic GVHD symptoms. c-GVHD, chronic GVHD; CSA, cyclosporine A; PSL, prednisolone; WBC, white blood cell count.
Correlation of BCL2A1 sequence polymorphisms with susceptibility to CTL-1B9
| . | HLA-A*2402–positive B-LCLs . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rt . | Do . | UR1 . | UR2 . | UR3 . | UR4 . | UR5 . | UR6 . | UR7 . | |
| Cytolysis by CTL-1B9 | + | − | + | + | + | + | + | − | − |
| Detected SNP, position* | |||||||||
| rs1138357, 238 | G/A | A | G | G | G/A | G/A | G/A | A | A |
| rs1138358, 299 | T/G | G | T | T | T/G | T/G | T/G | G | G |
| rs3826007, 427 | G | G/A | G | G | G | G | G/A | G/A | G |
| . | HLA-A*2402–positive B-LCLs . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rt . | Do . | UR1 . | UR2 . | UR3 . | UR4 . | UR5 . | UR6 . | UR7 . | |
| Cytolysis by CTL-1B9 | + | − | + | + | + | + | + | − | − |
| Detected SNP, position* | |||||||||
| rs1138357, 238 | G/A | A | G | G | G/A | G/A | G/A | A | A |
| rs1138358, 299 | T/G | G | T | T | T/G | T/G | T/G | G | G |
| rs3826007, 427 | G | G/A | G | G | G | G | G/A | G/A | G |
Rt indicates recipient; Do, donor; UR, unrelated; +, yes; and −, no.
Nucleotide positions are shown according to the NM_004092.2 mRNA sequence, available at http://www.ncbi.nlm.nih.gov/ as GEO accession GSE10044.
Discussion
Recent reports have unequivocally demonstrated that WGASs can be successfully used to identify common variants involved in a wide variety of human diseases.25-27 Our report represents a novel application of WGASs to transplantation immunology, which provides a simple but robust method to fine-map the genetic loci of minor H antigens whose expression is readily determined by standard immunophenotyping with CTL clones established from patients who have undergone transplantation.
The current WGA/CTL method has several desirable features that should contribute to the acceleration of minor H locus mapping. In comparing the method to those of linkage analysis and other nongenetic approaches, including direct peptide sequencing of chemically purified minor H antigens5,6,10,13 and conventional expression cloning,8,9,11 there are differences in terms of power, sensitivity, and specificity. Direct sequencing of minor H antigen peptide guarantees that the purified peptide is surely present on the cell surface as antigen, but it requires highly specialized equipment and personnel. Expression screening of cDNA libraries is also widely used and has become feasible with commercially available systems. However, it depends highly on the quality of the cDNA library and expression levels of the target genes. In addition, it often suffers from false-positive results due to the forced expression of cDNA clones under a strong promoter. The current method of WGA/CTL genetically determines the relevant minor H antigen locus, not relying on highly technical protein chemistry using specialized equipment, or repetitive cell cloning procedures. It is also not affected by the expression levels of the target antigens.
As a genetic approach, the current method based on genetic association has several advantages over conventional linkage analysis: the mapping resolution has been greatly improved from several Mb in the conventional linkage analysis to the average haplotype block size of less than 100 kb,17,25-27 usually containing a handful of candidate genes, compared with the dozens as typically found in linkage analysis. This means that the effort needed for the subsequent epitope mapping will be substantially reduced. In fact, the 115 kb region identified for CTL-2A12 contains 4 genes compared with 38 genes as revealed by the previous linkage study (data not shown), and the candidate gene was uniquely identified within the 26 kb region for CTL-1B9, for which linkage analysis had failed due to very rare segregating pedigrees among the CEPH panels with this trait (now ACC-1C; data not shown).15,16 In addition, before moving on to epitope mapping, it would be possible to evaluate the clinical relevance of the minor H antigens by examining the tissue distribution of their expression, based on widely available gene expression databases such as Genomic Institute of the Novartis Research Foundation (GNF, http://symatlas.gnf.org/SymAtlas/).28
Second, the required sample size is generally small, and should be typically no more than 100 B-LCLs for common minor H alleles. This is in marked contrast to the association studies for common diseases, in which frequently thousands of samples are required.17,25-27 In the current approach, sufficiently high test statistic values could be obtained for the relevant loci with a relatively small sample size, since the minor H allele is correctly segregated between the CTX+ and CTX− pools by the highly specific immunologic assay. Combined with high accuracy in allelic measurements, this feature allows for the use of pooled DNAs in WGAS, which substantially saves cost and time, compared with the genotyping of individual samples. Unexpectedly, our method allows for a considerable degree of error in the immunophenotyping, indicating the robustness of the current method; in fact, the minor H locus for CTL-2A12 was successfully identified in spite of the presence of 8 (∼10%) immunophenotyping errors. When the minor H allele has an extreme allele frequency (eg, < 5% or > 95%), which could be predicted by preliminary immunophenotyping, WGAS/CTL may not be an efficient method of mapping, due to the impractically large numbers of B-LCLs that would need to be screened to obtain enough CTX+ or CTX− B-LCLs. However, such minor H antigens would likely have limited clinical impact or applicability.
Sensitivity of the microarray analysis seems to be very high when the target SNP has good proxy SNPs on the array, because we were able to correctly identify the single SNP correlated with the target of CTL-1B9 from more than 500 000 SNP markers. On the other hand, genome coverage of the microarray is definitely important. In our experiments on CTL-2A12, the association was successfully identified by the marker SNPs showing r2 values of approximately 0.74 with the target locus of ACC-6. Since the GeneChip 500 K array set captures approximately 65% of all the HapMap phase II SNPs with more than 0.74 of r2;29 and higher coverage will be obtained with the SNP 6.0 arrays having more than 1 000 K SNP markers, these arrays can be satisfactorily used as platforms for the WGA/CTL method.
As shown in the current study, the intrinsic sensitivity and specificity of the WGA/CTL method in detecting associated SNPs were excellent. In other words, as long as target SNPs are captured in high r2 values with one or more marker SNPs within the Affymetrix 500 K SNP set, there is a high likelihood of capturing the SNP with the current approach. To evaluate the probability of a given minor H antigen being captured in high r2 with marker SNPs, we checked the maximum r2 values of known minor H antigen SNPs with the Affymetrix 500 K SNPs, according to empirical data from the HapMap project (www.hapmap.org). Among 13 known minor H antigens, 7 have their entries (designated minor H SNP) in the HapMap phase II SNP set (HA-3,30 HA-8,31 HB-1,11 ACC-1 and ACC-2,7 LB-ADIR-1F,10 and 7A7-PANE113 ), and were used for this purpose (note that absence of their entries in the HapMap data set does not necessarily mean that they could not be captured by a particular marker SNP set). As shown in Table S4, all 7 minor H SNPs are captured by at least one flanking SNP that is included in the Affymetrix 500 K SNP set with r2 values of more than 0.74 in at least one HapMap panel. The situation should be more favorable in the recently available SNP 6.0 array set with 1 000 K SNPs, indicating the genome coverage with currently available SNP arrays would be sufficient to capture typical minor H antigens with our approach.
Most patients who have received allo-HSCT could be a source of minor H antigen-specific CTL clones to be used for this assay, since the donor T cells are in vivo primed and many CTL clones could be established using currently available methods. In fact, substantial numbers of CTL clones have been established worldwide and could serve as the probes to identify novel minor H antigens.32,33 Once constructed, a panel of B-LCLs, including those transduced with HLA cDNAs, could be commonly applied to immunophenotyping with different CTL clones, especially when CTLs are obtained from the same ethnic group. In addition, by adopting other immunophenotyping readouts such as production of IL-2 from CD4+ T cells, this method could be applied to identification of MHC class II–restricted minor H antigens which have crucial roles in controlling CTL functions upstream. This may open a new field in the study of allo-HSCT since MHC class II–restricted mHags have been technically difficult to identify by conventional methods.
Finally, the discovery of ACC-1C as a novel minor H antigen indicates that all the mismatched transplants at this locus could be eligible for allo-immune therapies, since we have previously demonstrated that the counter allele also encodes a minor H antigen, ACC-1Y, which is preferentially expressed and presented on blood components including leukemic cells and may serve as a target of allo-immunity.7,34 Indeed, CTLs specific for ACC-2, an HLA-B44–restricted minor H antigen restricted by the third exonic SNP on BCL2A1,7 was independently isolated from the peripheral blood of a patient with recurrent leukemia re-entering complete remission after donor lymphocyte infusion.32 The number of eligible allo-HSCT recipients has now been effectively doubled, accounting for 50% of transplants with HLA-A24 or 20% of all transplantations performed in the Asian population. In conclusion, we have described a simple but powerful method for minor H mapping to efficiently accelerate the discovery of novel minor H antigens that will be needed to contribute to our understanding of the molecular mechanism of human allo-immunity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr W. Ho for critically reading the manuscript; Dr Keitaro Matsuo, Dr Hiroo Saji, Dr Etsuko Maruya, Dr Mamoru Ito, Ms Keiko Nishida, Dr Ayako Demachi-Okamura, Ms Yasuko Ogino, Ms Hiromi Tamaki, and the staff members of the transplant center, donor centers, and the Japan Marrow Donor Program office for their generous cooperation and expert technical assistance.
This study was supported in part by Scientific Research on Priority Areas (B01; no.17016089) from the Ministry of Education, Culture, Science, Sports, and Technology, Japan; Research on Human Genome, Tissue Engineering Food Biotechnology and the Second and Third Team Comprehensive 10-year Strategy for Cancer Control (no. 26), from the Ministry of Health, Labor, and Welfare, Japan; and a Grant-in-Aid from Core Research for Evolutional Science and Technology (CREST) of Japan.
Authorship
Contribution: T.K. performed most immunologic experiments and preparation of pooled DNA and quantative PCR, analyzed data, and wrote the manuscript; Y.N. performed the majority of genetic analyses and analyzed the data; H.T. performed T-cell receptor analysis and designed q-PCR primers and probes; G.Y. contributed to the organization of software for linkage analysis and simulation; S.M. prepared the pooled DNA; M.O., K.M., Y.K, and Y.M. collected clinical data and specimens; T.T. and K.K. contributed to data analysis and interpretation, and to the writing of the article; S.O. and Y.A. supervised the entire project, designed and coordinated most of the experiments in this study, contributed to manuscript preparation, and are senior coauthors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seishi Ogawa, Department of Hematology and Oncology, Department of Regeneration Medicine for Hematopoiesis, The 21st Century COE Program, Graduate School of Medicine, University of Tokyo, 7-3-1, Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; e-mail: sogawa-tky@umin.ac.jp; or Yoshiki Akatsuka, Division of Immunology, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan; e-mail: yakatsuk@aichi-cc.jp.
References
Author notes
T.K. and Y.N. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal