Anthracycline action has been thought to involve the neosynthesis of proapoptotic gene products and to therefore depend on protein synthesis for optimal effect. We found that inhibition of general, but not rapamycin-sensitive (cap-dependent), protein synthesis in the preapoptotic period enhanced anthracycline-induced acute myelogenous leukemia (AML) cell death, both in vitro and in several animal AML models. Pre-apoptotic anthracycline-exposed AML cells had altered translational specificity, with enhanced synthesis of a subset of proteins, including endoplasmatic reticulum chaperones. The altered translational specificity could be explained by perturbation (protein degradation, truncation, or dephosphorylation) of the cap-dependent translation initiation machinery and of proteins control-ing translation of specific mRNAs. We propose that judiciously timed inhibition of cap-independent translation is considered for combination therapy with anthracyclines in AML.
Introduction
Despite recent achievements,1,2 progress in therapy is slow for most malignancies, including acute myelogenous leukemia (AML).3 A more thorough understanding of how the most successful current drugs, such as the anthracyclines, induce cancer cell death can reveal novel therapeutically valuable death mechanisms, which can be targeted more efficiently by specifically tailored compounds than by current drugs. It can also reveal that present drugs activate cancer cell survival pathways in ways that can be overcome by, for example, coadministering complementary agents.
The aim of the present study was to undertake a proteomics-based survey4 of protein expression and modification in anthracycline-treated AML cells to explain in molecular terms previous observations that inhibitors of translation could protect completely5 or partially6,,–9 against anthracycline-induced cell death. Anthracycline treatment up-regulates the activity of signaling pathways stimulating the transcription factor nuclear factor-κB,10 and we have found that drug-resistant AML cells have up-regulated signal transducers and activators of transcriptions 3 and 5.11 Because these factors induce genes coding mainly, although not exclusively, for presumed survival promoting proteins,12,,–15 the reported dependence on protein synthesis for death implies either that the proapoptotic genes are induced in excess, that anthracycline treatment tips the balance between translation of mRNA coding for proapoptotic and prosurvival proteins in favor of the former, or can convert short-lived prosurvival proteins to prodeath proteins by, for example, dephosphorylation16 or cleavage.17,–19
We found that the first-line AML anthracycline drugs, such as daunorubicin (DNR), modified or altered the expression of several proteins known to alter the translation of specific mRNAs or direct translation from cap dependence to internal ribosome entry site dependence. This might explain that the DNR-treated AML cells in the late preapoptotic phase had enhanced synthesis of a subset of proteins, including ER chaperones. The preapoptotic protein synthesis served, contrary to expectations based on previous reports, a prosurvival function, because anthracycline-induced AML cell death was enhanced by protein synthesis inhibitor in vitro as well as in 3 separate intact animal (rodent) models for AML. Judiciously timed inhibition of the stress-induced cap-independent translation might therefore be considered in combination with anthracyclines for AML therapy.
Methods
Reagents and cells
Cycloheximide (CHX), rapamycin, puromycin, and emetine were from Sigma-Aldrich (St Louis, MO). Calyculin A was from Calbiochem (San Diego, CA). DNR was from Sigma-Aldrich (cell experiments) or sanofi-aventis (Bridgewater, NJ; animal experiments). Idarubicin (IDA) was from Sigma-Aldrich (cell experiments) or Pfizer (New York, NY; animal experiments).
HL60 cells were from GCMCC (Braunschweig, Germany; http://www.dsmz.de), and NB4 cells were from Dr M. Lanotte (Hôpital St Louis, Paris, France). They were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS) and l-glutamine (2 mmol/L), and kept in logarithmic growth until studied at approximately 0.5 × 106 cells/mL. The IPC-81 rat promyelocytic leukemia cell line was cultured as described previously.20 BN rat acute myelocytic leukemia (BNML) cells were a gift from Dr P. O. Iversen (University of Oslo, Oslo, Norway) and serially passaged in vivo. Blasts from AML patients were isolated and prepared as described previously.21 Detailed information about patient material is given in Table S2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Assessment of apoptosis
Cells were fixed and screened for apoptosis using differential interference contrast microscopy to visualize surface budding, and fluorescence microscopy for chromatin condensation.18 The chromatin pattern in cells incubated with the autofluorescent anthracyclines was identical to that observed with the DNA-specific dyes bisbenzimide (Hoechst 33342) and 4,6-diamidino-2-phenylindole. Cell viability was also scored by the WST1 assay (Millipore Bioscience Research Reagents, Temecula, CA), and flow cytometry. For examination of ultrastructure, HL60 cells were fixed and further processed as described previously.22
Metabolic labeling of cells
Metabolic labeling of phosphoproteins with 32Pi was performed as described previously.23
For protein labeling, the cells were transferred to methionine-free medium supplemented with dialyzed FBS and [35S]methionine (0.02 mCi/mL; SJ1515; GE Healthcare, Chalfont St Giles, United Kingdom). For pulse-chase, labeled cells were washed in medium with unlabeled methionine, left for 0.5 hours, and then exposed to experimental agents. For determination of protein synthesis, cells labeled with [35S]methionine were washed and precipitated in 7% trichloroacetic acid (TCA). The pellets were washed in 5% TCA, and then with water-saturated ether before being dissolved in 5 mmol/L Tris buffer, pH 8, containing 8.4 M urea, 2% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS), and 100 mmol/L dithioerythritol. Aliquots were diluted in sodium dodecyl sulfate (SDS) loading buffer for SDS-polyacrylamide gel electrophoresis (PAGE) analysis,22 or the incorporated radioactivity determined by liquid scintillation counting.24
Isolation of RNA and its use as template for in vitro protein synthesis
HL60 cells (107 cells) were pelleted, the pellet was lysed in 1 mL of TRIzol (Invitrogen), and RNA was isolated.25 In vitro protein synthesis was performed in rabbit reticulocyte lysate (Promega, Madison, WI) as described previously26 with 0.8 mCi/mL [35S]methionine and 0.2 mg/mL RNA. Aliquots were precipitated with 7% TCA and processed for 2-dimensional gel analysis.18
Gel electrophoresis, immunoblotting, evaluation of spot intensity, and picking for MS analysis
One-dimensional SDS-PAGE, two-dimensional PAGE (2DE), and immunoblotting were performed essentially as described previously.18 Antibody against pICln was from Dr K. Strange (Vanderbilt University, Nashville, TN), caspase 3 and actin were from Santa Cruz Biotechnology (Santa Cruz, CA), and p70S6K, phospho-p70S6K, 4E-BP1, phospho-4E-BP1, eEF2K, eEF2, mTOR, and phospho-mTOR were from Cell Signaling Technology (Danvers, MA). Some gels were silver-stained, dried, and exposed to autoradiography films. For mass spectrometry (MS) analysis, gels were stained with Sypro Ruby (Bio-Rad Laboratories, Hercules, CA), and spots were picked manually under illumination in a Fuji LAS3000 (FujiFilm, Tokyo, Japan). [35S]Methionine-labeled spots were excised from dried, unfixed gels using filmless autoradiographic analysis printout as overlay.
Gel spots were quantified based on isotope content using a Fuji Bas5000 PhosphoImager (GE Healthcare) or by densitometry (silver-stained gels), using mainly MultiGauge v2.3 (FujiFilm). To obtain the relative intensity, the intensity of each spot was divided by the sum of the total spot intensity in the gel. The theoretical molecular weight and isoelectric point (pI) of identified proteins was calculated using the ProtParam tool (http://au.expasy.org/tools/protparam.html).
COFRADIC, liquid chromatography-tandem mass spectrometry, and MALDI-TOF-MS analysis
For each analysis, 100 mL of NB4 cells (0.4 × 106 cells/mL) were washed and lysed in 1.5 mL of 20 mmol/L potassium phosphate, pH 6.8, with 1 mmol/L EDTA, 10 mmol/L CHAPS, 50 mmol/L NaF, 0.3 mmol/L NaVO3, and Complete mini protease inhibitor (Roche Molecular Biochemicals, Indianapolis, IN). Extract was centrifuged (10 000g, 20 minutes, 4°C) and kept at −80°C until analysis.
The combined fractional diagonal chromatography (COFRADIC)–based isolation and identification of the protein N-terminal peptides was performed as described previously.4,27 In brief, the technique allows the specific sorting of the trypsin-cleaved N-terminal parts of the proteins present in the complete lysate. The relative intensities of 16O- and 18O-tagged peptides allows relative changes of protein expression between 2 different states to be measured. When a new protein N terminus is generated, as the result of cleavage, this site is easily isolated and identified. Thus, information of the exact cleavage site is obtained. Matrix-assisted laser desorption ionization/time-of-flight (MALDI-TOF)-MS analysis, including the acquirement of postsource decay (PSD) spectra, was performed as described previously.18,28,29
AML animal models and estimation of the antileukemic efficacy of anthracyclines and CHX in vivo
NOD/SCID/B2mnull mice and Rowett (rnu/rnu) rats were bred and maintained under defined flora conditions in a high-efficiency particulate arrester-filtered atmosphere. Male BN/Rij rats were from TNO (Rijswijk, the Netherlands). All experiments were approved by the Norwegian Animal Research Authority and conducted according to the European Convention for the Protection of Vertebrates Used for Scientific Purposes.
Male NOD/SCID/B2mnull mice 6 to 8 weeks old were irradiated (BCC Dynaray CH4.4 megavolt irradiation source) with 2.5 Gy (100 cGy/min) 24 hours before transplantation of 10 × 106 NB4 cells (day 0) via the tail vein. rnu/rnu rats (60-80 g) were similarly injected with 20 × 106 IPC-81 cells, whereas BN/Rij rats (170-230 g) received 5 × 106 BNML cells from the spleen of a terminal stage leukemic animal.
IDA (1 mg/mL in water) was given by mouth to male rnu/rnu rats once daily (3 mg/kg) on days 14 to 16 after transplantation, whereas BN/Rij rats received 1.5 mg/kg on days 3 to 5. DNR (1 mg/mL in saline for rat and 0.1 mg/mL for mouse) was injected intravenously into BN/Rij rats (1.5 mg/kg per day) on days 3 to 5 and to NOD/SCID/B2mnull mice (0.5 mg/kg) on days 10 to 12. CHX (1 mg/mL in saline for rat and 5 mg/mL for mice) was injected intraperitoneally 1 hour after the administration of anthracycline, at 1.5, 0.8, and 5 mg/kg for rnu/rnu rats, BN/Rij rats, and NOD/SCID mice, respectively. Control animals received relevant vehicles. Condition and weight was monitored daily, and animals were killed when moribund.
The survival data were presented according to Kaplan and Meier and survival distributions analyzed by the Mantel-Haenszel log rank statistics, using Prism software (version 3.0; GraphPad Software, San Diego, CA).
Results
Inhibition of protein synthesis enhanced anthracycline-induced AML cell death in vitro and in animal models of transplanted AML
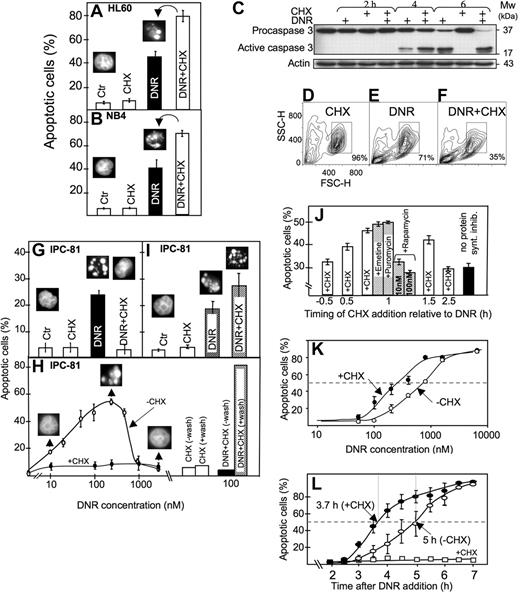
Anthracyclines such as DNR are first-line drugs in the treatment of AML and are believed to depend on protein synthesis to be maximally effective.5,,,–9 Contrary to expectations, we observed more apoptosis in HL60 and NB4 AML cells exposed to DNR and the protein synthesis inhibitor CHX than to DNR alone (Figure 1A,B). CHX intensified the DNR-induced conversion of procaspase3 to caspase3 (Figure 1C), the decrease of mitochondrial dehydrogenase activity (data not shown), and the ultrastructural (Figure 2A-E) and flow cytometric (Figure 1D-F) features of apoptosis.
Protein synthesis inhibitor enhanced anthracycline-induced AML cell death, although suppressing death phenotype features in some cells. (A) HL60 cells were treated for 6 hours with DNR (1.6 μmol/L) and CHX (3.6 μmol/L), alone or in combination, and scored for apoptosis (insets; see “Assessment of apoptosis”). (B) NB4 cells were treated for 6 hours with DNR (5 μmol/L) and CHX (3.6 μmol/L), alone or in combination, and scored for apoptosis. (C) Extracts from HL60 cells treated with DNR (8 μmol/L) for 2, 4, or 6 hours in the absence or presence of CHX (3.6 μmol/L, added 1 hour after DNR) were immunoblotted for caspase 3. (D-F) Flow cytometric analysis (representative of 4 separate experiments) of HL60 cells treated for 4 hours with vehicle (D), 8 μmol/L DNR (E) or DNR + 3.6 μmol/L CHX (F). Note that DNR + CHX converted twice as many cells as DNR alone to a compartment with decreased forward (FSC-H) and side (SSC-H) scatter. (G) Scoring of morphologically evident apoptosis in IPC-81 cells treated for 5 hours with DNR (0.1 μmol/L) alone or in combination CHX (3.6 μM). (H) IPC-81 cells were treated for 6 hours with various concentrations of DNR alone or with CHX (3.6 μmol/L) and scored for apoptotic morphology. Note the lack of cell budding, nuclear fragmentation, and chromatin condensation with DNR + CHX and with DNR more than ∼1 μmol/L. The bars show apoptosis in cells incubated for 6 hours with DNR + CHX or CHX alone, washed in excess medium, and incubated without DNR and CHX for another 12 hours before assessment of apoptosis. Note the high proportion of apoptotic cells after washing. (I) As for panel G, except that cells were treated for 3 hours with DNR (0.1 μmol/L) and CHX (3.6 μmol/L), washed and further incubated for 9 hours (as explained in panel H) before scoring of apoptosis. (J) HL60 cells received the protein synthesis inhibitors emetine (10 nmol/L), puromycin (180 nmol/L), rapamycin (10 or 100 nmol/L), or CHX (3.6 μmol/L) at various time points relative to DNR (1.6 μmol/L), and apoptosis scored 4.5 hours after DNR addition. (K) HL60 cells were treated for 6.5 hours with various concentrations of DNR (50-8000 nmol/L) in the absence (○) or presence (●) of CHX (3.6 μmol/L) and scored for apoptosis. (L) The accumulation of apoptotic HL60 cells as a function of time after addition of DNR (8 μmol/L) alone (○), CHX (3.6 μmol/L) alone (□), or a combination of the 2 (●). CHX was added 1 hour after DNR. Apoptosis scores represent the means (± SEM) of 3 to 6 experiments.
Protein synthesis inhibitor enhanced anthracycline-induced AML cell death, although suppressing death phenotype features in some cells. (A) HL60 cells were treated for 6 hours with DNR (1.6 μmol/L) and CHX (3.6 μmol/L), alone or in combination, and scored for apoptosis (insets; see “Assessment of apoptosis”). (B) NB4 cells were treated for 6 hours with DNR (5 μmol/L) and CHX (3.6 μmol/L), alone or in combination, and scored for apoptosis. (C) Extracts from HL60 cells treated with DNR (8 μmol/L) for 2, 4, or 6 hours in the absence or presence of CHX (3.6 μmol/L, added 1 hour after DNR) were immunoblotted for caspase 3. (D-F) Flow cytometric analysis (representative of 4 separate experiments) of HL60 cells treated for 4 hours with vehicle (D), 8 μmol/L DNR (E) or DNR + 3.6 μmol/L CHX (F). Note that DNR + CHX converted twice as many cells as DNR alone to a compartment with decreased forward (FSC-H) and side (SSC-H) scatter. (G) Scoring of morphologically evident apoptosis in IPC-81 cells treated for 5 hours with DNR (0.1 μmol/L) alone or in combination CHX (3.6 μM). (H) IPC-81 cells were treated for 6 hours with various concentrations of DNR alone or with CHX (3.6 μmol/L) and scored for apoptotic morphology. Note the lack of cell budding, nuclear fragmentation, and chromatin condensation with DNR + CHX and with DNR more than ∼1 μmol/L. The bars show apoptosis in cells incubated for 6 hours with DNR + CHX or CHX alone, washed in excess medium, and incubated without DNR and CHX for another 12 hours before assessment of apoptosis. Note the high proportion of apoptotic cells after washing. (I) As for panel G, except that cells were treated for 3 hours with DNR (0.1 μmol/L) and CHX (3.6 μmol/L), washed and further incubated for 9 hours (as explained in panel H) before scoring of apoptosis. (J) HL60 cells received the protein synthesis inhibitors emetine (10 nmol/L), puromycin (180 nmol/L), rapamycin (10 or 100 nmol/L), or CHX (3.6 μmol/L) at various time points relative to DNR (1.6 μmol/L), and apoptosis scored 4.5 hours after DNR addition. (K) HL60 cells were treated for 6.5 hours with various concentrations of DNR (50-8000 nmol/L) in the absence (○) or presence (●) of CHX (3.6 μmol/L) and scored for apoptosis. (L) The accumulation of apoptotic HL60 cells as a function of time after addition of DNR (8 μmol/L) alone (○), CHX (3.6 μmol/L) alone (□), or a combination of the 2 (●). CHX was added 1 hour after DNR. Apoptosis scores represent the means (± SEM) of 3 to 6 experiments.
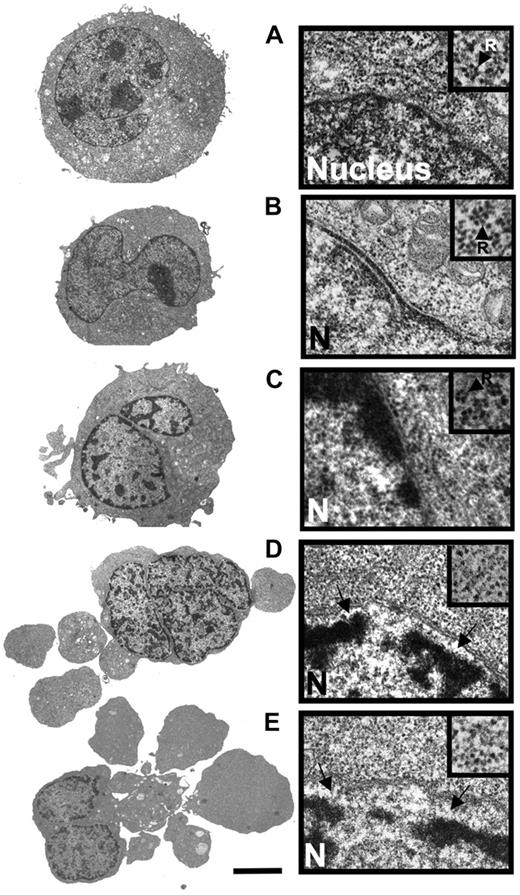
Comparison of the ultrastructural properties of HL60 cells treated with DNR alone and in combination with CHX. (A) Transmission electron microscopic overview (left) and detail showing part of nucleus and perinuclear region of control HL60 cell (incubated with vehicle for 5 hours). The inset (arrow; R) shows a polysomal rosette. (B) Cell treated with CHX (3.6 μmol/L) for 4 hours. (C) Cell treated with DNR (8 μmol/L) for 2.5 hours. Note beginning thickening of microvilli, perinuclear accumulation of condensed chromatin and polysomes (inset). (D) Cell treated for 2.5 hours with DNR (8 μmol/L) + CHX (3.6 μmol/L, added 1 hour after DNR). Note budding, separation of nuclear lamina from chromatin (arrows), and dispersal of polysomes (inset). (E) Cell treated with DNR (8 μmol/L) for 5 hours. Note similarity of features with cell in panel D. Panels A-E: scale bar: 5 μm for left subpanels, 0.3 μm for right subpanels, and 0.18 μm for right subpanel insets. The cells in panels A, B, and E are typical for the average cell in the population. The cells in panels C and D are representative of the 10% of cell population with most advanced apoptosis.
Comparison of the ultrastructural properties of HL60 cells treated with DNR alone and in combination with CHX. (A) Transmission electron microscopic overview (left) and detail showing part of nucleus and perinuclear region of control HL60 cell (incubated with vehicle for 5 hours). The inset (arrow; R) shows a polysomal rosette. (B) Cell treated with CHX (3.6 μmol/L) for 4 hours. (C) Cell treated with DNR (8 μmol/L) for 2.5 hours. Note beginning thickening of microvilli, perinuclear accumulation of condensed chromatin and polysomes (inset). (D) Cell treated for 2.5 hours with DNR (8 μmol/L) + CHX (3.6 μmol/L, added 1 hour after DNR). Note budding, separation of nuclear lamina from chromatin (arrows), and dispersal of polysomes (inset). (E) Cell treated with DNR (8 μmol/L) for 5 hours. Note similarity of features with cell in panel D. Panels A-E: scale bar: 5 μm for left subpanels, 0.3 μm for right subpanels, and 0.18 μm for right subpanel insets. The cells in panels A, B, and E are typical for the average cell in the population. The cells in panels C and D are representative of the 10% of cell population with most advanced apoptosis.
The results obtained with the IPC-81 cells may explain the discrepancy with at least some previous studies. In these cells, CHX blocked signs of visible apoptosis induced by DNR (Figure 1G,H). This was due not to protection against death but rather to the induction of a “frozen” death type, because the cells underwent massive conventional apoptosis after removal of DNR/CHX (Figure 1H). In fact, a higher percentage of apoptosis was noted after washing of cells treated with DNR + CHX than DNR alone (Figure 1I). We noted that the cells could undergo “frozen” death also when exposed to concentrations of DNR above 1 μmol/L (Figure 1H), indicating that CHX aggravated an inherent tendency of these cells to become blocked in certain aspects of apoptosis execution when exposed to an overwhelming anthracycline challenge, as noted previously in other cell systems.30,31
That CHX acted via inhibition of protein synthesis was supported by the similar efficiency of other general translation inhibitors like emetine and puromycin. The inhibitors were most efficient when given in the early preapoptotic phase after DNR addition (Figure 1J). They were much less efficient when given before DNR (Figure 1J; and data not shown). Rapamycin, which counteracts mTOR action to decrease cap-dependent protein synthesis while enhancing internal ribosomal entry site (IRES)–dependent translation,32 was inefficient (Figure 1J).
The presence of CHX from 1 hour after DNR addition served to both sensitize the HL60 cells to respond to lower concentrations of DNR and accelerate the death in response to a maximally effective (8 μmol/L) DNR concentration (Figure 1K,L). The synergistic effects noted at 0.1 μmol/L DNR (Figure 1I,K) indicated that this mechanism might be operating in clinical settings, because peak plasma concentrations of DNR in patients with AML range from 0.3 to 5 μmol/L.33
By transmission electron microscopy, the DNR-induced and DNR/CHX-induced HL60 cell death had similar morphology when comparing similar stages of death (Figure 2A-E). The death type was apoptotic with early margination of chromatin, separation of the nuclear lamina and matrix, and swelling of microvilli, followed by cell budding and hypercondensation of chromatin. Disaggregation of polysomal rosettes (Figure 2D,E right panels, including insets) occurred after separation of the nuclear lamina and commencement of surface budding. In some cells, the endoplasmic reticulum could still have some attached polysomes even if the soluble polysomes had disaggregated (data not shown). Disaggregated ribosomes remained numerous. We conclude that because the protein synthesis machinery was morphologically intact even in cells with clear signs of early apoptosis, protein synthesis might occur in the late preapoptotic and early apoptotic period of cell death.
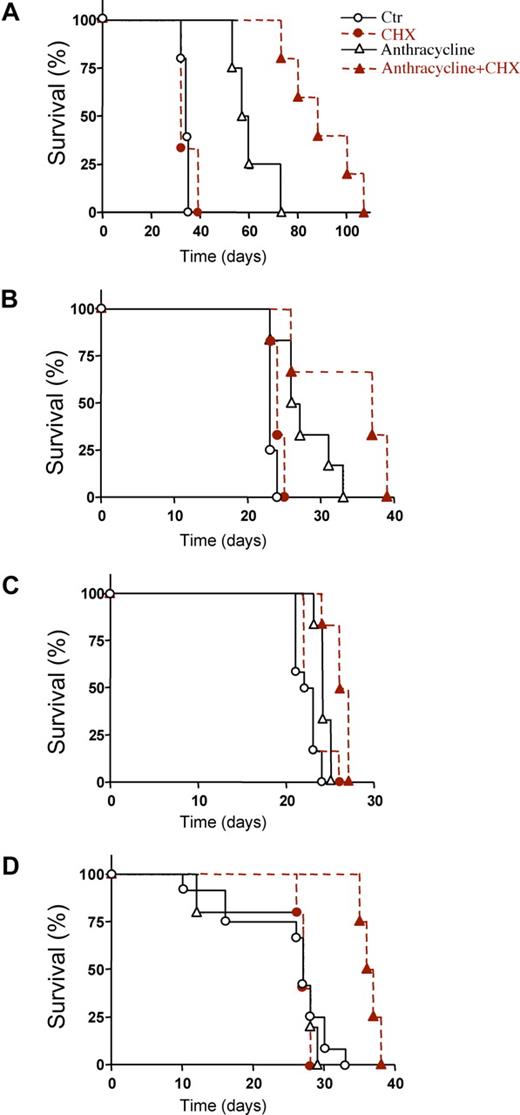
To know whether CHX could enhance anthracycline-induced AML cell death in vivo, we first studied irradiated rnu/rnu rats transplanted with the IPC-81 cells. The combined treatment with CHX and idarubicin (IDA) significantly prolonged survival compared with IDA alone (Figure 3A). The transplantable syngenic BNML model of leukemia, from which the IPC-81 cell line is derived, has been extensively validated as a reliable predictor for drug efficiency against AML in humans.34 In addition, in this AML model, survival was enhanced by CHX, whether combined with IDA (Figure 3B) or with DNR (Figure 3C). Finally, NOD/SCID/B2mnull mice transplanted with human NB4 cells were studied. Again, a significantly prolonged survival was observed when CHX was combined with DNR (Figure 3D). The prolonged survival was due to decreased tumor burden, because autopsied animals given either anthracycline alone or CHX alone, had more AML infiltrate in spleen, bone marrow, and lungs than animals given anthracycline + CHX (not shown), even if the latter animals on the average had lived longer after AML cell inoculation. In conclusion, CHX enhanced the death-inducing effect of anthracyclines on AML cells in animals with deficient as well as with intact (BNML-transplanted BN/Rij rats) immune system.
Prolonged survival of AML transplanted animals treated with protein synthesis inhibitor in addition to anthracycline. (A) Survival of IPC-81 transplanted rnu/rnu rats treated with vehicle (○; n = 5), idarubicin (▵; IDA, 3 mg/kg; n = 4), cycloheximide (●; CHX, 1.5 mg/kg; n = 3), or IDA + CHX (▴; n = 5). The IDA + CHX group had median survival time of 88 days compared with 58.5 days for the IDA mono-therapy group (P = .0072). (B) Survival of syngenic BNML-transplanted BN/Rij rats treated with vehicle (○; n = 12), IDA (▵; 1.5 mg/kg; n = 6), CHX (●; 0.8 mg/kg; n = 6), or IDA + CHX (▴; n = 6). The combination of IDA + CHX (mean survival time, 37 days) significantly increased (P = .03) survival over IDA alone (mean survival time, 26.5 days). (C) Survival of BNML rats treated with vehicle (○; n = 12), DNR (▵; 1.5 mg/kg; n = 6), CHX (●; 0.8 mg/kg; n = 6), or DNR + CHX (▴; n = 6). The combination of CHX with DNR increased the median survival from 22.5 to 26 days (P = .0065). (D) Survival of NOD/SCID/B2mnull mice transplanted with NB4 cells and treated with vehicle (○; n = 7), DNR (▵; 0.5 mg/kg; n = 5), CHX (●; 5 mg/kg; n = 5), or DNR + CHX (▴; n = 4). The combination of CHX with DNR increased the median survival from 27 to 36.5 days (P = .0046). For panels A-D: animal models, type and number of cells injected, and schedules are defined under “AML animal models and estimation of the antileukemic efficacy of anthracyclines and CHX in vivo.”
Prolonged survival of AML transplanted animals treated with protein synthesis inhibitor in addition to anthracycline. (A) Survival of IPC-81 transplanted rnu/rnu rats treated with vehicle (○; n = 5), idarubicin (▵; IDA, 3 mg/kg; n = 4), cycloheximide (●; CHX, 1.5 mg/kg; n = 3), or IDA + CHX (▴; n = 5). The IDA + CHX group had median survival time of 88 days compared with 58.5 days for the IDA mono-therapy group (P = .0072). (B) Survival of syngenic BNML-transplanted BN/Rij rats treated with vehicle (○; n = 12), IDA (▵; 1.5 mg/kg; n = 6), CHX (●; 0.8 mg/kg; n = 6), or IDA + CHX (▴; n = 6). The combination of IDA + CHX (mean survival time, 37 days) significantly increased (P = .03) survival over IDA alone (mean survival time, 26.5 days). (C) Survival of BNML rats treated with vehicle (○; n = 12), DNR (▵; 1.5 mg/kg; n = 6), CHX (●; 0.8 mg/kg; n = 6), or DNR + CHX (▴; n = 6). The combination of CHX with DNR increased the median survival from 22.5 to 26 days (P = .0065). (D) Survival of NOD/SCID/B2mnull mice transplanted with NB4 cells and treated with vehicle (○; n = 7), DNR (▵; 0.5 mg/kg; n = 5), CHX (●; 5 mg/kg; n = 5), or DNR + CHX (▴; n = 4). The combination of CHX with DNR increased the median survival from 27 to 36.5 days (P = .0046). For panels A-D: animal models, type and number of cells injected, and schedules are defined under “AML animal models and estimation of the antileukemic efficacy of anthracyclines and CHX in vivo.”
Anthracycline-treated preapoptotic AML cells had enhanced relative synthesis of a subset of survival-associated proteins
To estimate the effect of DNR and established protein synthesis inhibitors on protein synthesis, HL60 cells were pulse-labeled with [35S]methionine and the synthesis rate estimated as the ratio between autoradiographic intensity and protein staining intensity of SDS-gels of protein extracts from the cells (Figure 4A,B). The percentage synthesis declined as a function of time after addition of DNR (Figure 4A) but remained slightly higher than the percentage of nonapoptotic cells under similar conditions (Figure 1L). This suggested that translation was slightly enhanced in preapoptotic DNR-treated cells, not completely compromised in apoptotic cells, or both. Although CHX at 3.6 μmol/L led to strong inhibition of protein synthesis, rapamycin, as expected from previous studies,35 inhibited the total synthesis by only 10% to 15% (Figure 4B; data not shown).
DNR altered the control of AML cell protein translation. (A) Total protein synthesis rate of HL60 cells as a function of incubation with DNR (8 μmol/L) or CHX (3.6 μmol/L) relative to vehicle-treated cells. Cells were pulse-labeled with [35S]methionine during the last 15 minutes of the incubation. Synthesis is the ratio between the labeling, determined by autoradiography of SDS-PAGE gels of extracted proteins, and the protein staining of the same gels (see right inset). The data represent the means (± SEM) of 4 separate experiments. (B) HL60 cells were treated for 2 hours with CHX (3.6 or 36 μmol/L) or rapamycin (100 nmol/L), the last 0.5 hours of which was in the presence of [35S]methionine. Autoradiograms of SDS-gels (right panel) show that CHX was a far more efficient protein synthesis inhibitor than rapamycin in these cells. The left panel shows protein staining of the same gels. (C) Autoradiogram of 2DE gels (pI 4-5) of extract from HL60 cells treated with vehicle or 8 μmol/L DNR for 5 hours. A pulse of [35S]methionine was given during the last 30 minutes of the incubation. (D) Autoradiograms of proteins translated in vitro in the presence [35S]methionine with RNA isolated from HL60 cells incubated with vehicle (left subpanel) or 8 μmol/L DNR for 4.5 hours (right subpanel) as template. (E) Autoradiograms of proteins from HL60 cells prelabeled with [35S]methionine for 90 minutes and chased with unlabeled methionine for 6 hours in the absence and presence of 8 μmol/L DNR. (F) HL60 cells were treated with DNR (1.6 or 8 μmol/L) or rapamycin (100 nmol/L) for the periods indicated; cell extracts were immunoblotted and probed for P-Ser371-p70S6K, p70S6K, P-Thr37/46-4E-BP1, 4E-BP1, as well as P-Ser2448-mTOR, mTOR, and β-actin. (G) IPC-81 cells were treated for 6.5 hours with various concentrations of DNR (10-300 nmol/L), and extracts were immunoblotted and probed as for (F). For panels C-E: The circled spots correspond to proteins in Table 1. These proteins were subjected to comparative quantitative analysis. Spots found in all gels are circled with closed lines; spots found only in panel B and E are circled with dashed lines. The gels shown are representative of 3 to 5 experiments.
DNR altered the control of AML cell protein translation. (A) Total protein synthesis rate of HL60 cells as a function of incubation with DNR (8 μmol/L) or CHX (3.6 μmol/L) relative to vehicle-treated cells. Cells were pulse-labeled with [35S]methionine during the last 15 minutes of the incubation. Synthesis is the ratio between the labeling, determined by autoradiography of SDS-PAGE gels of extracted proteins, and the protein staining of the same gels (see right inset). The data represent the means (± SEM) of 4 separate experiments. (B) HL60 cells were treated for 2 hours with CHX (3.6 or 36 μmol/L) or rapamycin (100 nmol/L), the last 0.5 hours of which was in the presence of [35S]methionine. Autoradiograms of SDS-gels (right panel) show that CHX was a far more efficient protein synthesis inhibitor than rapamycin in these cells. The left panel shows protein staining of the same gels. (C) Autoradiogram of 2DE gels (pI 4-5) of extract from HL60 cells treated with vehicle or 8 μmol/L DNR for 5 hours. A pulse of [35S]methionine was given during the last 30 minutes of the incubation. (D) Autoradiograms of proteins translated in vitro in the presence [35S]methionine with RNA isolated from HL60 cells incubated with vehicle (left subpanel) or 8 μmol/L DNR for 4.5 hours (right subpanel) as template. (E) Autoradiograms of proteins from HL60 cells prelabeled with [35S]methionine for 90 minutes and chased with unlabeled methionine for 6 hours in the absence and presence of 8 μmol/L DNR. (F) HL60 cells were treated with DNR (1.6 or 8 μmol/L) or rapamycin (100 nmol/L) for the periods indicated; cell extracts were immunoblotted and probed for P-Ser371-p70S6K, p70S6K, P-Thr37/46-4E-BP1, 4E-BP1, as well as P-Ser2448-mTOR, mTOR, and β-actin. (G) IPC-81 cells were treated for 6.5 hours with various concentrations of DNR (10-300 nmol/L), and extracts were immunoblotted and probed as for (F). For panels C-E: The circled spots correspond to proteins in Table 1. These proteins were subjected to comparative quantitative analysis. Spots found in all gels are circled with closed lines; spots found only in panel B and E are circled with dashed lines. The gels shown are representative of 3 to 5 experiments.
To search for altered translation of specific transcripts, vehicle- and DNR-treated HL60 cells were pulse-labeled with [35S]methionine, and individual protein spot intensity compared in 2DE gel autoradiographs of the pI 4-5 sub-proteome. The DNR-treated cells had more than 2-fold increased relative synthesis of ribosomal protein P2 (rpP2, 3 spots labeled 1 in Figure 4C), protein disulfide isomerase (PDI; spot 2), proliferating cell nuclear antigen (PCNA; spot 3), and divalent cation tolerance homolog isoform 2 (CutA; spot 4). Three protein spots (11, 12, and 13) decreased more than 2-fold in [35S]methionine labeling intensity.
The effects of DNR were probably translational rather than transcriptional, because protein spots labeled during in vitro translation had similar relative intensity whether the template was RNA isolated from cells treated for 4.5 hours with vehicle or with DNR (Figure 4D). Comparison of individual spot intensity in cells and after in vitro translation revealed that the synthesis of PCNA, PDI, rpP2, and, to a lesser extent, calreticulin may be translationally turned on even in nonstressed HL60 cells and that their synthesis was additionally increased in DNR-stressed cells (Table 1). Conversely, proteins like MLC had lower synthesis rate in intact cells than expected from their in vitro synthesis rate (Figure 4C,D; Table 1).
Proteins affected by treatment with DNR
| Spotnumber . | Proteinabbreviation . | pI . | Molecular weight . | Relative spot intensity . | |||
|---|---|---|---|---|---|---|---|
| Observed . | Predicted . | Observed . | Predicted . | Control, in cellulo/in vitro . | DNR, in cellulo/in vitro . | ||
| 1 | rpP2 | 4.48/4.39/4.3 | 4.42/4.37/4.31 | 13 | 12 | 5.0 | 26 |
| 2 | PDI | 4.75 | 4.76 | 58 | 57 | >5.0 | >20 |
| 3 | PCNA | 4.57 | 4.57 | 31 | 29 | 4.4 | 12 |
| 4 | cutA | 4.45 | 5.15 | 15 | 17 | n.d. | n.d. |
| 5 | CALR | 4.31 | 4.29 | 46 | 48 | 1.5 | 2.0 |
| 6 | EF1Ba | 4.50 | 4.50 | 25 | 25 | 0.89 | 1.8* |
| 7 | PSMA5 | 4.63 | 4.74 | 24 | 26 | 0.21 | 0.62 |
| 8 | CTM | 4.65 | 4.80 | 33 | 28 | 0.98 | 0.36 |
| 9 | MLC | 4.32 | 4.46 | 16 | 17 | 0.46 | 0.25 |
| 10 | n.i. | 4.62 | n.d. | 20 | n.d. | 0.12 | 0.11 |
| 11 | n.i. | 4.73 | n.d. | 15 | n.d. | 0.09 | 0.03 |
| Spotnumber . | Proteinabbreviation . | pI . | Molecular weight . | Relative spot intensity . | |||
|---|---|---|---|---|---|---|---|
| Observed . | Predicted . | Observed . | Predicted . | Control, in cellulo/in vitro . | DNR, in cellulo/in vitro . | ||
| 1 | rpP2 | 4.48/4.39/4.3 | 4.42/4.37/4.31 | 13 | 12 | 5.0 | 26 |
| 2 | PDI | 4.75 | 4.76 | 58 | 57 | >5.0 | >20 |
| 3 | PCNA | 4.57 | 4.57 | 31 | 29 | 4.4 | 12 |
| 4 | cutA | 4.45 | 5.15 | 15 | 17 | n.d. | n.d. |
| 5 | CALR | 4.31 | 4.29 | 46 | 48 | 1.5 | 2.0 |
| 6 | EF1Ba | 4.50 | 4.50 | 25 | 25 | 0.89 | 1.8* |
| 7 | PSMA5 | 4.63 | 4.74 | 24 | 26 | 0.21 | 0.62 |
| 8 | CTM | 4.65 | 4.80 | 33 | 28 | 0.98 | 0.36 |
| 9 | MLC | 4.32 | 4.46 | 16 | 17 | 0.46 | 0.25 |
| 10 | n.i. | 4.62 | n.d. | 20 | n.d. | 0.12 | 0.11 |
| 11 | n.i. | 4.73 | n.d. | 15 | n.d. | 0.09 | 0.03 |
Cells were treated with vehicle or 8 μmol/L DNR for 4.5 hours, pulse-labeled for the last 30 minutes with [35S]methionine, and protein spots 1 to 11 in 2DE gels (see Figure 4 for details) analyzed for relative labeling intensity. Proteins translated from mRNA isolated from vehicle-treated (Ctr) or DNR-treated cells were similarly separated and analyzed. The ratio between relative spot intensity in cells (in cellulo) and after in vitro translation (in vitro) is listed in the two rightmost columns. The data are averages from 3 separate experiments. Protein abbreviations refer to the following proteins (Swissprot accession numbers in parentheses). rpP2 indicates 60S acidic ribosomal protein P2 (P05387); PDI, protein disulfide isomerase/prolyl 4-hydroxylase β subunit (NP_000909); PCNA, proliferating cell nuclear antigen (NP_002583); cutA, divalent cation tolerance homolog isoform 2(NP_057005); CALR, calreticulin precursor (NP_004334); eEF1Ba, elongation factor 1Ba (P24534); PSMA5, proteasome endopeptidase complex, ζ chain (S17521); CTM, cytoskeletal tropomyosin (CAA28258); MLC, myosin light polypeptide 6 (P60662); n.d., not determined; and n.i., not identified.
The value for EF1Ba is slightly underestimated due to cleavage of this protein (see Figure 6).
The preferential synthesis of presumed survival proteins like PCNA and PDI may explain why inhibition of protein synthesis enhanced DNR-induced death.
Proteins involved in translational control are major targets in anthracycline-treated cells
That anthracyclines affect the translation of specific transcripts is a novel finding. Switching from the usual cap-dependent to IRES-dependent initiation of translation is seen in cells exposed to the mTOR activity perturbing drug rapamycin, acting via decreased phosphorylation of p70S6 kinase (p70S6K) and 4E-BP1.35,37 Phosphorylation of rpS6 via p70S6K is considered necessary for efficient cap-dependent translational initiation of ribosomal and other proteins with 5′-untranslated region (UTR) polypyrimidine tracts,38 whereas phospho-4E-BP1 relieves the tonic inhibition of general cap-dependent translation.39
We found that, like rapamycin, DNR not only decreased phospho-p70S6K (P-p70S6K) but also led to degradation of p70S6K, suggesting that DNR, unlike rapamycin, would lead to irreversible loss of p70S6K activity. Likewise, DNR treatment led to more complete conversion of the highly (γ,β) to the less (α) phosphorylated form of P-4E-BP1 (Figure 4F) than rapamycin treatment (data not shown). The effect of DNR on P-p70S6K and P-4E-BP1 was not confined to HL60 cells and high DNR concentrations, because significant effects were observed already at 100 nmol/L DNR in IPC cells (Figure 4G).
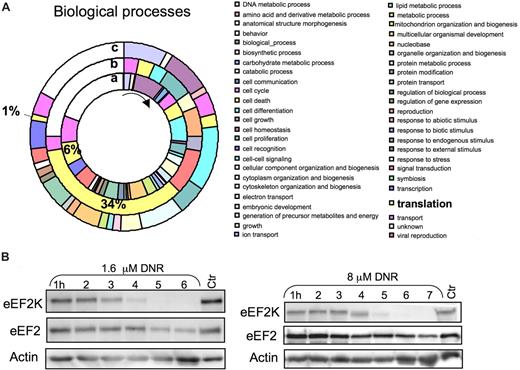
To gain further insight into the effects of anthracyclines on the protein synthesis machinery and other targets, protein extracts from control and DNR/CHX-treated NB4 cells were subjected to differential 16O/18O global peptide labeling, COFRADIC, and selective sorting of protein amino-terminal peptides.4,27 The method, unlike metabolic labeling, avoids exposing the cells to media with decreased amino acid content and dialyzed serum, thus eliminating 2 factors that can influence the protein synthesis machinery. All proteins identified in the NB4 cells (Table S1) were sorted ontologically into 49 biologic processes, and 6% classified under translation. In the subgroup of proteins whose N-terminal peptides were decreased after DNR/CHX treatment, 34% had an assigned role in translation (Figure 5A). This suggested that proteins involved in mRNA translation were preferentially targeted by DNR.
COFRADIC N-terminal analysis revealed preferential down-regulation of proteins related to protein translation/RNA-binding in DNR/CHX-treated NB4 cells. (A) Gene Ontology-based analysis of the COFRADIC protein data according to biologic function. The inner ring (a) represents the biologic process relative values of all the proteins identified in the analysis. The middle ring (b) represents the N-terminal down-regulated proteins, and the outer ring (c) represents the protein substrates producing stable cleavage products. The pie chart visualizes a striking preferential down-regulation of proteins involved in translation (b). The chart was generated using the PIGOK analysis; http://128.40.158.133/pigok.html. (B) HL60 cells were treated with 1.6 or 8 μmol/L DNR for the periods indicated, and extracts were immunoblotted and probed for eEF2K and eEF2.
COFRADIC N-terminal analysis revealed preferential down-regulation of proteins related to protein translation/RNA-binding in DNR/CHX-treated NB4 cells. (A) Gene Ontology-based analysis of the COFRADIC protein data according to biologic function. The inner ring (a) represents the biologic process relative values of all the proteins identified in the analysis. The middle ring (b) represents the N-terminal down-regulated proteins, and the outer ring (c) represents the protein substrates producing stable cleavage products. The pie chart visualizes a striking preferential down-regulation of proteins involved in translation (b). The chart was generated using the PIGOK analysis; http://128.40.158.133/pigok.html. (B) HL60 cells were treated with 1.6 or 8 μmol/L DNR for the periods indicated, and extracts were immunoblotted and probed for eEF2K and eEF2.
The down-regulated proteins included the rapamycin receptor FKBP1240 as well as eIF4H41 and eIF1A42 (Table 2), which support cap-dependent and inhibit IRES-dependent initiation of translation. These findings provide clues about how the action of rapamycin may be blunted (Figure 1J) and suggest, in conjunction with the data of Figure 4F, that the machinery favoring cap-dependent rather than IRES-dependent initiation of translation was severely damaged in DNR-treated cells. Both ribosomal protein L13a43 and hnRNP C1/C244 are involved in the control of specific mRNA translation, and their down-regulation may therefore alter the translational preference, as observed in the DNR-treated cells (Figure 4C, Table 1). The eEF2 kinase (eEF2K) was strongly down-regulated, not only in the DNR/CHX-treated NB4 cells (Table 2) but also in HL60 cells, where it became degraded after 4 to 5 hours' incubation with either 1.6 or 8 μmol/L DNR (Figure 5B). Because eEF2 kinase inhibits elongation factor 2 (eEF2),45 its down-regulation is expected to enhance the efficiency of eEF2. The level of eEF2 itself decreased only moderately (Table 2), mainly after the first 4 to 5 hours of incubation with DNR (Figure 5B).
List of peptides down-regulated more than 2-fold in NB4 cells treated with DNR (1.6 μmol/L) and CHX (3.6 μmol/L) for 8 hours
| Protein description . | Accession no. . | Start . | End . | Identified peptide . | Fold down-regulation . |
|---|---|---|---|---|---|
| EF2 kinase | O00418 | 2 | 9 | Ac-ADEDLIFR | >10 |
| Lysyl-tRNA synthetase | Q15046 | 2 | 25 | Ac-AAVQAAEVKVDGSEPKLSKNELKR | >10 |
| eIF4H, isoform 1 | Q15056 | 2 | 10 | Ac-ADFDTYDDR | >10 |
| 60S ribosomal protein L13 | P40429 | 2 | 12 | Ac-AEVQVLVLDGR | 5.9 |
| Asparaginyl-tRNA synthetase, cytoplasmic | O43776 | 2 | 11 | Ac-VLAELYVSDR | 4.2 |
| hnRNP C1/C2, isoform 1 | P07910 | 2 | 12 | Ac-ASNVTN<Dam>KTDPR | 4.4 |
| Ac-ASNVTNKTDPR | 2.9 | ||||
| 40S ribosomal protein S30 | P62861 | 1 | 8 | Ac-KVHGSLAR | 2.5 |
| 60S ribosomal protein L30 | P62888 | 2 | 17 | Ac-VAAKKTKKSLESINSR | 2.5 |
| LSm3 | P62310 | 2 | 22 | Ac-ADDVDQQQTTNTVEEPLDLIR | 2.5 |
| eEF2 | P13639 | 2 | 10 | Ac-VN<Dam>FTVDQIR | 2.4 |
| eIF1A | O14602 | 2 | 12 | Ac-PKNKGKGGKNR | 2.3 |
| Phenylalanyl-tRNA synthetase alpha chain | Q9Y285 | 2 | 12 | Ac-ADGQVAELLLR | 2.3 |
| 40S ribosomal protein S3a | P61247 | 2 | 8 | Ac-AVGKN<Dam>KR | 2.2 |
| FK506-binding protein 1A/FKBP 12 | P62942 | 2 | 14 | Ac-GVQVETISPGDGR | 2.1 |
| Protein description . | Accession no. . | Start . | End . | Identified peptide . | Fold down-regulation . |
|---|---|---|---|---|---|
| EF2 kinase | O00418 | 2 | 9 | Ac-ADEDLIFR | >10 |
| Lysyl-tRNA synthetase | Q15046 | 2 | 25 | Ac-AAVQAAEVKVDGSEPKLSKNELKR | >10 |
| eIF4H, isoform 1 | Q15056 | 2 | 10 | Ac-ADFDTYDDR | >10 |
| 60S ribosomal protein L13 | P40429 | 2 | 12 | Ac-AEVQVLVLDGR | 5.9 |
| Asparaginyl-tRNA synthetase, cytoplasmic | O43776 | 2 | 11 | Ac-VLAELYVSDR | 4.2 |
| hnRNP C1/C2, isoform 1 | P07910 | 2 | 12 | Ac-ASNVTN<Dam>KTDPR | 4.4 |
| Ac-ASNVTNKTDPR | 2.9 | ||||
| 40S ribosomal protein S30 | P62861 | 1 | 8 | Ac-KVHGSLAR | 2.5 |
| 60S ribosomal protein L30 | P62888 | 2 | 17 | Ac-VAAKKTKKSLESINSR | 2.5 |
| LSm3 | P62310 | 2 | 22 | Ac-ADDVDQQQTTNTVEEPLDLIR | 2.5 |
| eEF2 | P13639 | 2 | 10 | Ac-VN<Dam>FTVDQIR | 2.4 |
| eIF1A | O14602 | 2 | 12 | Ac-PKNKGKGGKNR | 2.3 |
| Phenylalanyl-tRNA synthetase alpha chain | Q9Y285 | 2 | 12 | Ac-ADGQVAELLLR | 2.3 |
| 40S ribosomal protein S3a | P61247 | 2 | 8 | Ac-AVGKN<Dam>KR | 2.2 |
| FK506-binding protein 1A/FKBP 12 | P62942 | 2 | 14 | Ac-GVQVETISPGDGR | 2.1 |
In Tables 2 and 3, only peptides with corresponding parent proteins with functions related to mRNA-binding/processing and/or protein synthesis are shown. The sequence of the identified peptide and its location within the parent protein is indicated Ac- denotes the α-N-acetyl group, and <Dam> indicates deamidation. The parent proteins are referred to by description and UnitProt database accession number, The complete list of peptides is given in Table S1. Peptide identification was done using “Internal Protein Index” databases (http://www.ebi.ac.uk/IPI/IPIhelp.html).
Several of the cleaved RNA-binding proteins (Table 3) may modify the transport of mRNA and thereby its availability for translation, whereas others, like hnRNP A046 and hnRNP U,47 have been incriminated in the control of translation of specific mRNAs. The protein-processing events noted after exposure to DNR could not be explained by generalized proteolysis, because the 2DE protein pattern was quite similar for cells prelabeled with [35S]methionine and chased in the absence or presence of DNR for 6 hours (Figure 4E).
List of internally located α-N-acetylated peptides (formed by cleavage) exclusively found in the same DNR/CHX-treated NB4 cells as in Table 2
| Protein description . | Accession no. . | Start . | End . | Identified peptide . | Site . |
|---|---|---|---|---|---|
| H/ACA ribonucleoprotein complex subunit 4 | O60832 | 419 | 447 | Ac-YSESAKKEVVAEVVKAPQVVAEAAKTAKR | EYVD_ |
| hnRNP A0 | Q13151 | 74 | 81 | Ac-GNTVELKR | HAVD_ |
| hnRNP U, isoform long | Q00839 | 232 | 243 | Ac-GKTEQKGGDKKR | PAGD_ |
| NAC, alpha polypeptide | Q13765 | 43 | 71 | Ac-STQATTQQAQLAAAAEIDEEPVSKAKQSR | EEQD_ |
| Poly(A) binding protein 2 | Q86U42 | 109 | 125 | Ac-PGDGAIEDPELEAIKAR | VEGD_ |
| RNA-binding protein-39 | Q14498 | 332 | 346 | Ac-ASSASSFLDSDELER | ERTD_ |
| Splicing factor U2AF 65 kDa subunit | P26368 | 129 | 146 | Ac-GLAVTPTPVPVVGSQMTR | MTPD_ |
| TAR DNA-binding protein-43 | Q13148 | 90 | 98 | Ac-ASSAVKVKR | DETD_ |
| LSm3 | P62310 | 7 | 22 | Ac-QQQTTNTVEEPLDLIR | DDVD_ |
| Protein description . | Accession no. . | Start . | End . | Identified peptide . | Site . |
|---|---|---|---|---|---|
| H/ACA ribonucleoprotein complex subunit 4 | O60832 | 419 | 447 | Ac-YSESAKKEVVAEVVKAPQVVAEAAKTAKR | EYVD_ |
| hnRNP A0 | Q13151 | 74 | 81 | Ac-GNTVELKR | HAVD_ |
| hnRNP U, isoform long | Q00839 | 232 | 243 | Ac-GKTEQKGGDKKR | PAGD_ |
| NAC, alpha polypeptide | Q13765 | 43 | 71 | Ac-STQATTQQAQLAAAAEIDEEPVSKAKQSR | EEQD_ |
| Poly(A) binding protein 2 | Q86U42 | 109 | 125 | Ac-PGDGAIEDPELEAIKAR | VEGD_ |
| RNA-binding protein-39 | Q14498 | 332 | 346 | Ac-ASSASSFLDSDELER | ERTD_ |
| Splicing factor U2AF 65 kDa subunit | P26368 | 129 | 146 | Ac-GLAVTPTPVPVVGSQMTR | MTPD_ |
| TAR DNA-binding protein-43 | Q13148 | 90 | 98 | Ac-ASSAVKVKR | DETD_ |
| LSm3 | P62310 | 7 | 22 | Ac-QQQTTNTVEEPLDLIR | DDVD_ |
The amino acids preceding the identified peptide are indicated (site). The underlined Asp in the identified peptide for LSm3 indicates the processing site validated by detection of an intermally located α-N-acetylated peptide. In Tables 2 and 3, only peptides with corresponding parent proteins with functions related to mRNA-binding/processing and/or protein synthesis are shown.
In passing, it may be noted that cleavage of lamin B1, B2, the lamin B receptor, and histone 2A (Table S1) could be related to the early loosening of the nuclear lamina and chromatin hypercondensation seen in DNR-treated AML cells (Figure 2D,E).
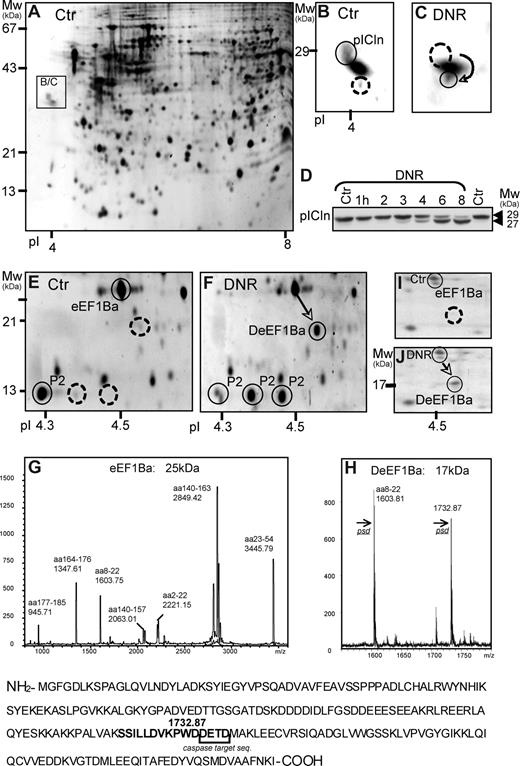
The COFRADIC-based analysis was complemented by a 2DE-based approach, focusing on protein spots altered early in the apoptotic process of DNR-treated HL60 cells. The earliest and most consistent findings with both 1.6 and 8 μmol/L DNR were: changed position of the methylosome subunit pICln as a result of truncation (Figure 6A-D), appearance of 2 rows of spots (47 kDa, pI 6.0/6.3/6.8; 33 kDa, pI 4.8/4.9/5.0/5.1; data not shown), identified as hnRNP K and hnRNP C1/2, respectively, a basic shift of rpP2 (Figure 6E,F), and de novo appearance of a spot (17 kDa, pI 4.53) resulting from cleavage of elongation factor 1Ba (ΔeEF1Ba) at the caspase consensus sequence DETD154 (Figure 6E-H). The DNR-induced appearance of ΔeEF1Ba was noted also in blasts from patients with AML (Figure 6I,J), indicating that it was not restricted to HL60 cells.
Protein truncation and dephosphorylation events detected by 2DE in DNR-treated HL60 cells. (A,B) The boxed area shows the position (A) and detail (B) of the methylosome subunit chloride conductance regulatory protein (pICln) in Sypro Ruby-stained 2DE gels (pI 3-10) with extract of control HL60 cells. (C) Like panel B, except that boxed area is from gel with extract from HL60 cells treated for 6 hours with 8 μmol/L DNR. (D) Immunoblotting of pICLn on extracts of HL60 cells treated with 1.6 μmol/L DNR for the times indicated. Note truncation of pICLn from 29 to 27 kDa, starting after 3 hours of DNR exposure. (E) The spots circled with solid lines show the MS-detected ribosomal protein P2 (rpP2) and eEF1Ba (25 kDa). (F) After 6 hours with 8 μmol/L DNR, a significant proportion of the eEF1Ba appeared as a 17 kDa form (arrow), whereas rpP2 appeared as 3 spots of similar size but with different pI. (G and H) MALDI analysis of the 25-kDa (G) and 17-kDa (H) variants of eEF1Ba revealed a tryptic peptide (1732.87), present only in the 17-kDa ΔeEF1Ba. It was identified by tandem mass spectrometry to have a predicted caspase cleavage site (DETD) at its C terminus. (I-J) The relative distribution of eEF1Ba (circled) and ΔeEF1Ba (arrow) in 2DE gels (pI 4-5) of extract from blasts from patients with AML (M2, patient 5) treated in vitro for 6 hours with vehicle (I) or 8 μmol/L DNR (J). The gels were silver-stained.
Protein truncation and dephosphorylation events detected by 2DE in DNR-treated HL60 cells. (A,B) The boxed area shows the position (A) and detail (B) of the methylosome subunit chloride conductance regulatory protein (pICln) in Sypro Ruby-stained 2DE gels (pI 3-10) with extract of control HL60 cells. (C) Like panel B, except that boxed area is from gel with extract from HL60 cells treated for 6 hours with 8 μmol/L DNR. (D) Immunoblotting of pICLn on extracts of HL60 cells treated with 1.6 μmol/L DNR for the times indicated. Note truncation of pICLn from 29 to 27 kDa, starting after 3 hours of DNR exposure. (E) The spots circled with solid lines show the MS-detected ribosomal protein P2 (rpP2) and eEF1Ba (25 kDa). (F) After 6 hours with 8 μmol/L DNR, a significant proportion of the eEF1Ba appeared as a 17 kDa form (arrow), whereas rpP2 appeared as 3 spots of similar size but with different pI. (G and H) MALDI analysis of the 25-kDa (G) and 17-kDa (H) variants of eEF1Ba revealed a tryptic peptide (1732.87), present only in the 17-kDa ΔeEF1Ba. It was identified by tandem mass spectrometry to have a predicted caspase cleavage site (DETD) at its C terminus. (I-J) The relative distribution of eEF1Ba (circled) and ΔeEF1Ba (arrow) in 2DE gels (pI 4-5) of extract from blasts from patients with AML (M2, patient 5) treated in vitro for 6 hours with vehicle (I) or 8 μmol/L DNR (J). The gels were silver-stained.
The DNR-modified proteins detected by 2DE interact with RNA (hnRNP K,48 C1/C244 ), modify RNA-binding proteins (pICln49 ), or are components of the protein synthesis apparatus (rpP2,50 eEF1Ba45 ), reinforcing the conclusion that protein synthesis is a major target in anthracycline-induced death.
The basic shift of the eEF2 binding rpP2 could be detected already after 2 to 3 hours' incubation with DNR (ie, before the onset of morphologic indices of apoptosis), and dephosphorylation of rpP2 was the most conspicuous effect of DNR on the 2DE phosphoprotein pattern in DNR-treated cells metabolically labeled with [32Pi] (Figure 7A,B; data not shown). Only the 2 acidic rpP2 spots were radioactive, indicating the basic spot as dephospho-rpP2 (Figure 7A,B). Although the phosphorylation rate of newly synthesized rpP2 was slow (Figure 7C) and unaffected by phosphatase inhibitor (data not shown), calyculin A did inhibit the dephosphorylation of already phosphorylated rpP2 (Figure 7D-F). This suggested that phosphatase activation rather than kinase inhibition was responsible for the net dephosphorylation of rpP2, but the enhanced synthesis of rpP2 (Figure 4, Table 1) also contributed to the accumulation of nonphospho-rpP2, because newly synthesized rpP2 stayed unphosphorylated for a considerable period of time (Figure 7C).
The phosphorylation status of ribosomal protein P2 in AML cell lines and patient blasts. The effect of anthracycline in vitro and in vivo. (A,B) IPC-81 cells were treated for 2.5 hours with 0.5 μmol/L DNR, the last 90 minutes of which with 32Pi. Proteins were separated by 2DE (pI 4-7) and silver stained (A) or autoradiographed (B). The spots circled with a solid line were identified by MS analysis as rpP2. Note the lack of labeled phosphate in the most basic spot (right subpanel). (C) HL60 cells were pulse-labeled with [35S]methionine for 5, 10, or 60 minutes and extracts subjected to 2DE (pI 4-5) and autoradiography. The circled spots show rpP2. Note the progressive but slow shift toward the more acidic form as a function of time. (D) HL60 cells were prelabeled with [35S]methionine and chased for 6.5 hours with unlabeled methionine with vehicle (left subpanel) or 8 μmol/L DNR present during the last 6 hours of incubation. The circled spots represent rpP2 from an autoradiogram of 2DE (pI 4-5) gel. (E) Experiment as for panel D, except that gels were silver-stained and incubation time with DNR was 4 hours. (F) As for panel E, except that the phosphatase inhibitor calyculin A (CCA; 0.1 μmol/L) was present during the last 3 hours of incubation. (G) Blasts from patients with AML (M5, patient 11) were treated for 6 hours in vitro with vehicle (left subpanel) or 8 μmol/L DNR (right subpanel) and analyzed by 2DE (pI 4-5). The circled spots represent rpP2. (H) Blasts isolated from 11 patients with various AML classification (M1, M2, M4, M5, M6), one with ALL, and one with LBL were treated in vitro with DNR (8 μmol/L, 6 hours). The percentage increase of nonphosphorylated rpP2 was determined from the relative intensity of PP-rpP2, P-rpP2, and nonphospho-rpP2 in silver-stained gels like the one shown in panel G. (I) AML blasts were isolated from a patient (M4, patient 8) before (left subpanel) and 4 hours after (right subpanel) the onset of induction treatment with IDA and cytarabine and analyzed by 2DE (pI 4-5). Note the significant formation of the monophosphorylated form of rpP2 after induction treatment.
The phosphorylation status of ribosomal protein P2 in AML cell lines and patient blasts. The effect of anthracycline in vitro and in vivo. (A,B) IPC-81 cells were treated for 2.5 hours with 0.5 μmol/L DNR, the last 90 minutes of which with 32Pi. Proteins were separated by 2DE (pI 4-7) and silver stained (A) or autoradiographed (B). The spots circled with a solid line were identified by MS analysis as rpP2. Note the lack of labeled phosphate in the most basic spot (right subpanel). (C) HL60 cells were pulse-labeled with [35S]methionine for 5, 10, or 60 minutes and extracts subjected to 2DE (pI 4-5) and autoradiography. The circled spots show rpP2. Note the progressive but slow shift toward the more acidic form as a function of time. (D) HL60 cells were prelabeled with [35S]methionine and chased for 6.5 hours with unlabeled methionine with vehicle (left subpanel) or 8 μmol/L DNR present during the last 6 hours of incubation. The circled spots represent rpP2 from an autoradiogram of 2DE (pI 4-5) gel. (E) Experiment as for panel D, except that gels were silver-stained and incubation time with DNR was 4 hours. (F) As for panel E, except that the phosphatase inhibitor calyculin A (CCA; 0.1 μmol/L) was present during the last 3 hours of incubation. (G) Blasts from patients with AML (M5, patient 11) were treated for 6 hours in vitro with vehicle (left subpanel) or 8 μmol/L DNR (right subpanel) and analyzed by 2DE (pI 4-5). The circled spots represent rpP2. (H) Blasts isolated from 11 patients with various AML classification (M1, M2, M4, M5, M6), one with ALL, and one with LBL were treated in vitro with DNR (8 μmol/L, 6 hours). The percentage increase of nonphosphorylated rpP2 was determined from the relative intensity of PP-rpP2, P-rpP2, and nonphospho-rpP2 in silver-stained gels like the one shown in panel G. (I) AML blasts were isolated from a patient (M4, patient 8) before (left subpanel) and 4 hours after (right subpanel) the onset of induction treatment with IDA and cytarabine and analyzed by 2DE (pI 4-5). Note the significant formation of the monophosphorylated form of rpP2 after induction treatment.
The dephosphorylation of rpP2 in response to anthracyclines was found also in NB4 cells and mammary carcinoma MCF-7 cells (data not shown). More importantly from the clinical point of view, when blasts from 11 patients with AML, one patient with acute lymphoblastic leukemia (ALL), and one with lymphoblastic leukemia/lymphoma (LBL) were exposed to DNR in vitro, significant DNR-induced dephosphorylation of rpP2 was observed in 10 of the 13 samples (Figure 7G,H). Because dephosphorylation of rpP2 was an early occurrence in DNR-treated AML cells (Figure 7) it might be detected in AML blasts from patients under chemotherapy, even if dying AML cells are expected to be removed from the circulation early in the apoptotic process.51 A basic shift of rpP2 was in fact detected in blasts recovered from a patient with AML 4 hours after commencement of induction chemotherapy (Figure 7I), lending further credibility to the notion that modulation of the translational apparatus could occur early and be of clinical relevance in human AML.
Discussion
We report enhanced synthesis of a subset of proteins, including the ribosomal protein rpP2, cell survival proteins, such as PCNA, and the ER chaperones PDI and calreticulin in AML cells treated with anthracyclines. To our knowledge, specifically altered translational control has not been described in anthracycline-treated cells before.
The effect of anthracyclines on protein synthesis was accompanied by altered phosphorylation, degradation, or limited proteolysis (truncation) of proteins controlling specific mRNA translation, cap-dependent initiation, or polypeptide elongation. We noted massive perturbation of proteins required for cap-dependent translation, which is known to alter the mRNA preference for translation.37 Thus, the down-regulation of eIF1A will relieve its tonic inhibition of IRES-directed initiation52 and disturb the assembly of cap-dependent initiation factors.42 The degradation of the RNA helicase eIF4H will prevent unwinding of secondary structure in the 5′-UTR of mRNA required for cap-dependent initiation.53 The dephosphorylation of 4E-BP1 will inhibit eIF4E and thereby cap-dependent initiation.39 Lack of p70S6K, as observed in the DNR-treated cells, has traditionally been thought to inhibit cap-dependent translation of proteins with polypyrimidine tracts in their 5′-UTR, like the ribosomal proteins.54 A striking effect of DNR was enhanced synthesis of rpP2, whose 5′-UTR has a 17 nucleotide long oligopyrimidine tract55 (REFSEQ: 00100456 ). This supports the notion that phosphorylation of S6 by p70S6K is not required for efficient translation of mRNAs with a oligopyrimidine tract.54
The high rate of protein synthesis in DNR-treated cells was puzzling in view of the many proteomically detected hits on tRNA synthetases and proteins involved in the elongation phase of translation. It should be noted that the tRNA synthetases down-regulated in DNR-treated cells are present in excess relative to the need of the protein translational machinery,57 which may explain why protein translation could persist despite tRNA synthetase down-regulation. The control of elongation (for review, see Browne and Proud45 ) has not been intensively studied in mammalian cells and much must be inferred from yeast studies. The elongation factor eEF1Ba, which became cleaved in DNR-treated AML cell lines and patient blasts, was deleted in yeast without affecting the total protein synthesis.58 This suggests that the cleavage of eEF1Ba can occur and potentially fine-tune elongation without compromising overall elongation efficiency. The second step in elongation depends on eEF2, which is complexed with the ribosomal stalk proteins rpP1 and rpP259 and inhibited by phosphorylation catalyzed by eEF2K.45 The degradation of eEF2K in DNR-treated AML cells will enhance eEF2 activity and thereby elongation. Because dephosphorylation of rpP2 alters translational specificity in yeast,60 it may do so also in AML cells. Dephosphorylation of rpP2 occurred early after onset of DNR treatment in vitro and in blasts from a patient with AML under induction therapy. This supports that the dephosphorylation was preapoptotic because cells are known to be cleared efficiently in vivo before onset of overt apoptosis,61 even sometimes before the commitment to death.62
In addition, several proteins with roles in the control of translation of specific mRNAs were modified in DNR-treated cells. One example is the GAIT complex, which, via the constituent ribosomal protein L13a, is down-regulated in the DNR-treated AML cells, can silence the translation of specific mRNAs by binding to their 3′-UTR.57 Another example is the perturbation of primarily intranuclear proteins, including members of the hnRNP family, which have roles in mRNA transport and control of specific mRNA translation (Figure 5, Table S1). The high synthesis rate of PDI in DNR-stressed cells may be related to modification of such proteins, because its translational induction in hypoxic cells occurs through the action of multiple originally nuclear RNA-binding proteins, including members of the hnRNP family.63
The present study was initiated based on the assumption that the proteins synthesized in anthracycline-treated cells were proapoptotic.6,,–9 The presumed prosurvival actions of the translationally up-regulated PCNA and the ER-associated chaperone PDI64 call this into question. That CHX enhanced rather than blocked DNR-induced AML cell death further challenges this notion. That anthracyclines when combined with CHX could lead to a nonconspicuous form of death in which the cell appeared “frozen,” can explain some of the discrepancy with previous studies.65 Another factor is the timing between the addition of anthracycline and CHX. We found CHX to promote AML cell death most efficiently when given after DNR, whereas most previous studies have administered protein synthesis inhibitors before the anthracyclines.
Calreticulin is a marker for phagocytosis66 with selectively increased surface presentation in anthracycline-treated tumor cells.51 The enhanced relative synthesis of calreticulin in our DNR-treated cells might make more calreticulin available for surface presentation and facilitate AML removal in immune-competent animals.51 Thus questioning the relevance of data obtained in vitro or in immunodeficient animal models of cancer. We found, however, that protein synthesis inhibitor enhanced the anthracycline-induced debulking of AML cells and prolonged survival also in the syngenic rat BNML model of AML.
Although introduced as drugs more than 40 years ago, the mechanisms of action of anthracyclines are still incompletely known and controversial.33,67 Analysis of the proteome changes in DNR-treated cells revealed some actions similar to those of more recently discovered death pathways or drugs. The protein cleavage pattern resembled the one in Jurkat T-cells treated with Fas-activator.27,68,–70 Because the present data were obtained with CHX present, they cannot be explained by anthracycline-induced synthesis of Fas and Fas-ligand,5 suggesting that an effector system similar to that used by Fas was activated without Fas occupancy or that preformed Fas or Fas ligand are externalized.71,72 Another observation was cleavage of fatty acid synthase, which recently has become a promising target for chemotherapy.73 Finally, DNR mimicked known actions of rapamycin, such as dephosphorylation of the major mTOR targets 4E-BP1 and p70S6K, which leads to inhibition of cap-dependent protein synthesis. Rapamycin is under study as an adjunct to conventional chemotherapy in selected cancers.74,,,–78 The mimicry by DNR of important rapamycin actions and the down-regulation of the rapamycin receptor FKBP12 can explain why rapamycin was inefficient as modulator of DNR-induced AML cell death in the present study.
In conclusion, the present study has pointed to several novel actions of anthracyclines in AML cells that help explain the efficacy of this drug, which still is the first choice in several malignancies, including AML. More importantly, we discovered altered translational preference for mRNAs in DNR-treated cells. This effect was found at submicromolar concentrations of DNR, which compares favorably with the peak concentrations (0.3-5 μmol/L; most often 1-2 μmol/L) observed in serum of patients treated with DNR.33 Furthermore, the synergy between CHX and DNR in animal models of AML was observed with clinically relevant concentrations (1-3 mg/kg/day) of DNR. Preapoptotic protein synthesis could enhance AML cell survival, which suggests that it is a partially successful cell reaction to counteract DNR-induced toxicity (ie, an “Achilles heel”) in DNR action. Obviously, general protein synthesis inhibitors such as CHX cannot be used for long-term treatment, although protracted administration to humans has been reported in the past.79 But, by providing an experimental system to test novel drug candidates for the ability to inhibit the non–cap-dependent protein synthesis prevailing in preapoptotic anthracycline-exposed cells, the present study should stimulate the development of agents able to block protein synthesis in the time interval after anthracycline administration when the synthesis of cancer cell survival proteins is critical.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Expert technical assistance was provided by Erna Finsås, Nina Lied-Larsen, Line Wergeland, Odd Harald Oddland, Monica Hals, Kjetil Jacobsen, Kari Espolin Fladmark, Anne Nyhaug, Hans Demol, and Magda Puype.
This work was supported by The Norwegian Cancer Society, The Odd Fellow Medical Research Fund of Norway, the Fund for Scientific Research–Flanders (Belgium), the Concerted Research Actions of the Flemish Community (Belgium), Helse-Vest, and the Norwegian Research Council (The Norwegian Centre for Microarray Technology/The Norwegian Proteomics Center [FUGE/PROBE]).
Authorship
Contribution: All authors contributed intellectually to the work. G.G., B.T.G., and S.D. were involved in most aspects, although E.M. was involved chiefly in animal experiments, P.V.D., K.G., J.V., and R.H. were involved mainly in proteomics and proteomics-related aspects, C.K. in ultrastructural assessment of apopto-sis, and Ø.B. in selecting and characterizing patient AML blasts.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: S.O. Døskeland, Department of Biomedicine, University of Bergen, Jonas Lies vei 91, N-5009 Bergen, Norway; e-mail: stein.doskeland@biomed.uib.no.
References
Author notes
G.G. and B.T.G. contributed equally to this work.

![Figure 4. DNR altered the control of AML cell protein translation. (A) Total protein synthesis rate of HL60 cells as a function of incubation with DNR (8 μmol/L) or CHX (3.6 μmol/L) relative to vehicle-treated cells. Cells were pulse-labeled with [35S]methionine during the last 15 minutes of the incubation. Synthesis is the ratio between the labeling, determined by autoradiography of SDS-PAGE gels of extracted proteins, and the protein staining of the same gels (see right inset). The data represent the means (± SEM) of 4 separate experiments. (B) HL60 cells were treated for 2 hours with CHX (3.6 or 36 μmol/L) or rapamycin (100 nmol/L), the last 0.5 hours of which was in the presence of [35S]methionine. Autoradiograms of SDS-gels (right panel) show that CHX was a far more efficient protein synthesis inhibitor than rapamycin in these cells. The left panel shows protein staining of the same gels. (C) Autoradiogram of 2DE gels (pI 4-5) of extract from HL60 cells treated with vehicle or 8 μmol/L DNR for 5 hours. A pulse of [35S]methionine was given during the last 30 minutes of the incubation. (D) Autoradiograms of proteins translated in vitro in the presence [35S]methionine with RNA isolated from HL60 cells incubated with vehicle (left subpanel) or 8 μmol/L DNR for 4.5 hours (right subpanel) as template. (E) Autoradiograms of proteins from HL60 cells prelabeled with [35S]methionine for 90 minutes and chased with unlabeled methionine for 6 hours in the absence and presence of 8 μmol/L DNR. (F) HL60 cells were treated with DNR (1.6 or 8 μmol/L) or rapamycin (100 nmol/L) for the periods indicated; cell extracts were immunoblotted and probed for P-Ser371-p70S6K, p70S6K, P-Thr37/46-4E-BP1, 4E-BP1, as well as P-Ser2448-mTOR, mTOR, and β-actin. (G) IPC-81 cells were treated for 6.5 hours with various concentrations of DNR (10-300 nmol/L), and extracts were immunoblotted and probed as for (F). For panels C-E: The circled spots correspond to proteins in Table 1. These proteins were subjected to comparative quantitative analysis. Spots found in all gels are circled with closed lines; spots found only in panel B and E are circled with dashed lines. The gels shown are representative of 3 to 5 experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/5/10.1182_blood-2007-07-103242/2/m_zh80060816500004.jpeg?Expires=1769147733&Signature=PyE7e6qNhgi4f8ygJOj~LnNIMwqm4rYO5s-MtNaY2X~kR9UFLjpbaMRHD9LL3hLXJOXQQcGdlEzlIwVF4BnYCUawBGiN~ejV98iz1SKpVWGam5QdK1dCyDA80S-1xYwE7EX7FlS0zgLlWVcftrm8Jo4PbOkCW89avavHX8I32DbbSvrBp13Spd9COtGq4Z81U7XWM-ZEa-kVByEmSQap2sarTs0OUmYONJ2ZDGJWT6NJ33IW7RO50-fx3ZTh3Nfj2lLfnhqjIRNdaQp86Nyw2~gn1MMkdI~xx4gHx~hI1lMs1~lrmiIxQi4ijGCMFfAOFLkdZR0nyKPdDjaCvt-7yA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. The phosphorylation status of ribosomal protein P2 in AML cell lines and patient blasts. The effect of anthracycline in vitro and in vivo. (A,B) IPC-81 cells were treated for 2.5 hours with 0.5 μmol/L DNR, the last 90 minutes of which with 32Pi. Proteins were separated by 2DE (pI 4-7) and silver stained (A) or autoradiographed (B). The spots circled with a solid line were identified by MS analysis as rpP2. Note the lack of labeled phosphate in the most basic spot (right subpanel). (C) HL60 cells were pulse-labeled with [35S]methionine for 5, 10, or 60 minutes and extracts subjected to 2DE (pI 4-5) and autoradiography. The circled spots show rpP2. Note the progressive but slow shift toward the more acidic form as a function of time. (D) HL60 cells were prelabeled with [35S]methionine and chased for 6.5 hours with unlabeled methionine with vehicle (left subpanel) or 8 μmol/L DNR present during the last 6 hours of incubation. The circled spots represent rpP2 from an autoradiogram of 2DE (pI 4-5) gel. (E) Experiment as for panel D, except that gels were silver-stained and incubation time with DNR was 4 hours. (F) As for panel E, except that the phosphatase inhibitor calyculin A (CCA; 0.1 μmol/L) was present during the last 3 hours of incubation. (G) Blasts from patients with AML (M5, patient 11) were treated for 6 hours in vitro with vehicle (left subpanel) or 8 μmol/L DNR (right subpanel) and analyzed by 2DE (pI 4-5). The circled spots represent rpP2. (H) Blasts isolated from 11 patients with various AML classification (M1, M2, M4, M5, M6), one with ALL, and one with LBL were treated in vitro with DNR (8 μmol/L, 6 hours). The percentage increase of nonphosphorylated rpP2 was determined from the relative intensity of PP-rpP2, P-rpP2, and nonphospho-rpP2 in silver-stained gels like the one shown in panel G. (I) AML blasts were isolated from a patient (M4, patient 8) before (left subpanel) and 4 hours after (right subpanel) the onset of induction treatment with IDA and cytarabine and analyzed by 2DE (pI 4-5). Note the significant formation of the monophosphorylated form of rpP2 after induction treatment.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/5/10.1182_blood-2007-07-103242/2/m_zh80060816500007.jpeg?Expires=1769147733&Signature=jJKXZYC4c4aobEOcUgspCXcMueMHOrk8TfaPpLj3KHy8jUiGdMKq0WztEtPbx88zPopTNYHvVy~JNtKUny1pUBpHz71FtZHZn8HLyVpCLCKj8J11pBAkUAnC46wg1n~VkpBU-9FkcHqfneqZ~ElM6sGGV~~bopYgzlj8bOz76iSVD7qnF5R6ATL9zJIHTGj7uj2~MoGG8cuaG7oEbCGX58nZN3lHl1P1h1AvhdXsOo~s8x~Qu8WAi9LkaUTflGobT-d1KWKQm0ElDhidANepoC6OIdvX1mfCRbcok3nh2OyRrqysA8SqBOqAaHVj3cS8~Bs480W1hoR0vFp4v84nMg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)