SPA-1 (signal-induced proliferation associated gene-1) functions as a suppressor of myeloid leukemia by negatively regulating Rap1 signaling in hematopoietic progenitor cells (HPCs). Herein, we showed that transplantation of HPCs expressing farnesylated C3G (C3G-F), a Rap1 guanine nucleotide exchange factor, resulted in a marked expansion of thymocytes bearing unique phenotypes (CD4/CD8 double positive [DP] CD3− TCRβ−) in irradiated recipients. SPA-1−/− HPCs expressing C3G-F caused a more extensive expansion of DP thymocytes, resulting in lethal T-cell acute lymphoblastic leukemia (T-ALL) with massive invasion of clonal T-cell blasts into vital organs. The C3G-F+ blastic thymocytes exhibited constitutive Rap1 activation and markedly enhanced expression of Notch1, 3 as well as the target genes, Hes1, pTα, and c-Myc. All the T-ALL cell lines from C3G-F+ SPA-1−/− HPC recipients expressed high levels of Notch1 with characteristic mutations resulting in the C-terminal truncation. This proliferation was inhibited completely in the presence of a γ-secretase inhibitor. Transplantation of Rag2−/− SPA-1−/− HPCs expressing C3G-F also resulted in a marked expansion and transformation of DP thymocytes. The results suggested that deregulated constitutive Rap1 activation caused abnormal expansion of DP thymocytes, bypassing the pre-T-cell receptor and eventually leading to Notch1 mutations and Notch-dependent T-ALL.
Introduction
Rap1, a Ras-family G protein, mediates diverse functions that are highly dependent on the contexts of the cell types, such as proliferation, differentiation, cell survival, cell adhesion, polarity, and movements.1,2 Rap1 is activated to Rap1GTP by specific guanine nucleotide exchange factors (GEFs) coupled with diverse receptors, whereas specific GTPase-activating proteins (GAPs) hydrolyze Rap1GTP to Rap1GDP.3 We previously reported that mice deficient for SPA-1, a principal Rap1GAP in hematopoietic progenitor cells (HPCs), developed a spectrum of myeloid leukemia or B1-cell leukemia of long latency.4,5 The majority of myeloid leukemia in SPA-1−/− mice resembled human chronic myelogenous leukemia and often was associated with blast crisis of myeloid, erythroid, or B-cell lineage, which strongly suggested a disorder of multipotent HPCs.6 Although the results suggested that SPA-1 functioned as a leukemia suppressor in HPCs, the long latency implied that additional factors might be needed for development of overt leukemia. On the other hand, a Rap1GEF, CalDAG-GEF1, has been reported as a proto-oncogene in BXH-2 myeloid leukemia.7 We also reported that human bcr-abl fusion gene product (Bcr-Abl) was a potent Rap1 activator,8 and that Bcr-Abl+ SPA-1−/− HPCs showed aggravated chronic myelogenous leukemia phenotypes compared with Bcr-Abl+ wild-type HPCs in a mouse model, including prolonged survival of leukemic progenitors and rapid blast crisis in vivo.9 Thus, proto-oncogenes capable of activating Rap1 signal may be potential collaborative factors for leukemia in SPA-1 deficiency.
In the present study, we investigated the effects of C3G expression in normal and SPA-1−/− HPCs in vivo. C3G is a ubiquitous Rap1GEF translocated to the plasma membrane by a wide variety of signals via Crk-family adaptor proteins.10,11 Farnesylated C3G (C3G-F) shows facilitated recruitment to the plasma membrane to activate endogenous Rap1,10 although the activity can be counteracted by SPA-1.12 Results showed that transplantation of normal HPCs expressing C3G-F resulted in a marked expansion of blastic thymocytes with unusual phenotypes, namely, CD4/CD8 double-positive (DP) CD3/TCRβ−, in the irradiated recipients. SPA-1−/− HPCs expressing C3G-F caused a more aggressive thymic DP cell expansion with higher levels of Rap1 activation than C3G-F+ wild-type HPCs, eventually resulting in lethal T-cell acute lymphoblastic leukemia (T-ALL). Leukemic C3G-F+ thymocytes exhibited markedly augmented expression of Notch and the target genes. Furthermore, all the clonal C3G-F+ SPA-1−/− T-ALL cell lines revealed characteristic mutations in Notch1, resulting in C-terminal truncation, which is similar to mutations reported in the majority of T-ALL in humans as well as in genetically predisposed animal models.13,,–16 The proliferation was completely inhibited in the presence of a γ-secretase inhibitor. The current results suggest a functional cross talk of endogenous Rap1 G protein and Notch signaling in thymocyte development and T-ALL genesis.
Methods
Mice
SPA-1−/− mice of C57BL/6 (B6) backgrounds were reported previously.9 B6 and B6 Rag2−/− mice were purchased from CLEA Japan (Tokyo, Japan). All the mice were maintained in a specific pathogen-free condition at the Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University, Kyoto, Japan.
Cells and culture
Plat-E cells, kindly provided by Dr T. Kitamura (University of Tokyo, Tokyo, Japan), were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 1 μg/mL puromycin (Sigma-Aldrich, St Louis, MO), and 10 μg/mL blastocidin (Funakoshi, Tokyo, Japan). T-ALL cell lines were established from the thymi of C3G-F+ SPA-1−/− HPC recipients with overt T-ALL, and maintained in modified Dulbecco modified Eagle medium with 10% fetal calf serum, 10% NCTC109 medium, 10−5 M 2-mercaptoethanol, 100 U/mL insulin, 1 mM sodium pyruvate, 1 mM oxaloacetic acid, 0.1 mM nonessential amino acids, and 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid.
Retroviral infection and transplantation of bone marrow cells
The retroviral plasmid, pMC IRES GFP, was also a gift from Dr T. Kitamura. A full-length C3G cDNA tagged with a CAAX motif of K-Ras at the C terminus (C3G-F) was subcloned into the retroviral vector. Recombinant retrovirus was produced in Plat-E packaging cells by transfecting the plasmids with LipofectAMINE 2000 (Invitrogen, Carlsbad, CA). Bone marrow (BM) cells were harvested from 4- to 8-week-old mice that had been treated with 150 mg/kg 5-fluorouracil (Kyowa Hakko Kogyo, Tokyo, Japan) and further enriched for HPCs by depleting lineage marker (Lin)–positive cells with the use of a cocktail of antibodies (anti-Thy1, anti-B220, anti–Gr-1, anti–Mac-1, and anti-Ter119; BD Biosciences, San Jose, CA) and anti–rat immunoglobulin G–coated magnetic beads (Dynabeads M-450; Dynal, Oslo, Norway), or by fluorescence-activated cell sorting (FACS) with FACSVantage (Becton Dickinson, San Jose, CA). The Lin− HPCs were cultured overnight in complete RPMI 1640 containing 10 ng/mL interleukin-6, 10 ng/mL interleukin-11, 10 ng/mL Flt-3 ligand, and 50 ng/mL stem-cell factor (Genzyme, Minneapolis, MN). The cells were then infected with retroviral supernatants by spinoculation (1800g, 60 minutes, 32°C) followed by culturing in the same medium. The infection procedure was repeated twice. The infected HPCs were injected intravenously into 8.5-Gy γ-ray–irradiated B6 mice (2 × 104 cells per head) together with normal B6 BM cells (2 × 105 cells per head) for rescue.
Immunoblotting, pull-down assay, and Southern blotting
Cells were lysed with lysis buffer (150 mM sodium chloride, 50 mM Tris(hydroxymethyl)aminomethane hydrochloride [pH 7.6], 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, sodium vanadium oxide, 10 mM sodium fluoride, 2 μg/mL leupeptin, and 2 μg/mL aprotinin) and subjected to immunoblotting as described previously.4 Antibodies included anti-Notch1, anti-Rap1, anti–R-Ras (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Ras (Upstate Biotechnology, Lake Placid, NY), anti-Hes1 provided by Dr T. Sudo (Torey Basic Research Institute, Kamakura, Japan), anti–c-Myc (Cell Signaling Technology, Beverly, MA), anti-p27Kip1 (BD Transduction Laboratories, San Diego, CA), anti–SPA-1,4 anti–presenilin-1 (anti–PS-1; Cell Signaling Technology), anti-Flag (Sigma-Aldrich), and anti–glyceraldehyde-3-phosphate dehydrogenase (Chemicon, Temecula, CA). For detection of the NICD1 isoform, an antibody reactive to Val1744-cleaved Notch (Val1744; Cell Signaling Technology) was used. Rap1GTP and RasGTP were determined by a pull-down assay as described previously.12 DNAs of liver and thymocytes were digested with HindIII or PvuII, and Southern blotting was performed with the use of a TCR Cβ or EGFP cDNA probe.
Flow cytometric analysis
Multicolor flow cytometric and DNA content analyses were performed with the use of FACSCalibur (Becton Dickinson) as described previously.4 Antibodies included phycoerythrin-conjugated or allophycocyanin-conjugated anti-CD3, anti-CD4, anti-CD8, anti-CD25, anti-CD44, anti-CD69, biotin anti–mouse TCRβ, and phycoerythrin-conjugated streptavidin (BD Biosciences).
Histohematologic analysis
Peripheral blood leukocyte and red blood cell counts were monitored with the use of an automated cell counter (Nihon Kohden, Tokyo, Japan). Thymocytes were cytospun and stained with May-Giemsa solution (Muto Pure Chemicals, Tokyo, Japan); organs were fixed in 10% formalin and stained with hematoxylin-eosin solution. Thymocytes or T-ALL cells treated with colcemid (Karyo Max; Life Technologies, Grand Island, NY) were subjected to spectral karyotyping analysis using standard procedures.
Real-time PCR
Total RNA was extracted with the use of a NucleoSpin RNA II Kit (Macherey-Nagel, Düren, Germany) and treated with DNase I (Invitrogen) followed by cDNA synthesis with the use of SuperScript III (Invitrogen). Real-time polymerase chain reaction (PCR) was performed with LightCycler 480 SYBR Green I Master Kit (Roche, Basel, Switzerland) on a LightCycler 480 instrument (Roche). PCR primers were as follows: Notch1, 5′-ggacatgcagaacaacaagg-3′ (forward) and 5′-cagtctcatagctgccctca-3′ (reverse); Notch2, 5′-gccgatgtcctcttcacg-3′ (forward) and 5′-acggttgcggatcagaat-3′ (reverse); Notch3, 5′-agctgggtcctgaggtgat-3′ (forward) and 5′-agacagagccggttgtcaat-3′ (reverse); Hes1, 5′-gccagctgatataatggagaaaa-3′ (forward) and 5′-tccatgataggctttgatgactt-3′ (reverse); c-Myc, 5′-cgaaactctggtgcataaactg-3′ (forward) and 5′-gaaccgttctccttagctctca-3′ (reverse); p27Kip1, 5′-tttaattgggtctcaggcaaactct-3′ (forward) and 5′-ccgtctgaaacattttcttctgttc-3′ (reverse); cyclin D1, 5′-gaacaagctcaagtggaacc-3′ (forward) and 5′-cttcaatctgttcctggcag-3′ (reverse); pTα, 5′-ctgctttccggagcctct-3′ (forward) and 5′-gaggagcaggcgcagtag-3′ (reverse); and cyclophilin, 5′-tggagagcaccaagacagaca-3′ (forward) and 5′-tgccggagtcgacaatgat-3′ (reverse). The relative target gene expression levels were normalized to cyclophilin contents.
Cell-proliferation assay
T-ALL cells were cultured at various concentrations in the absence or presence of a γ-secretase inhibitor, N-[N-(3,5-difluorophenacetyl-l-alanyl)]-(S)-phenylglycine t-butyl ester (DAPT; Calbiochem, San Diego, CA) for a various number of days in 96-well plates. The viable cell numbers were assessed with the use of a standard curve by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (ATCC, Manassas, VA).
DNA sequencing
Total RNAs from T-ALL cell lines (Wo1, Wo2, and Wo3) were isolated with the use of ISOGEN RNA extraction reagent (Nippon Gene, Tokyo, Japan), and cDNAs were synthesized as described previously. Notch1 cDNA corresponding to the region surrounding the PEST domain (around 1.7 kb from nt 6212 to nt 7973) was amplified with the use of the following primers: 5′-ggacatgcagaacaacaagg-3′ (6212) and 5′-cttcaccctgaccaggaaaa (7973). PCR products were purified with the use of High Pure PCR Products Purification Kit (Roche) and cloned with the use of pGEM-T Easy Vector (Promega, San Luis Obispo, CA). At least 5 subclones were isolated for each cell line and then sequenced with the use of 4 kinds of primers as follows: 5′-aggcacggaggaaagtcc-3′(6580/Notch1), 5′-agcagcctctccaccaatac-3′ (7104/Notch1), 5′-taatacgactcactataggg-3′ (T7/pGEM), and 5′-cgccaagctatttaggtgac-3′ (RP/pGEM).
Imaging acquisition
The cell and tissue images were visualized using a ZEISS Axiovert200M microscope equipped with an Axiocam MRm camera (Carl Zeiss MicroImaging, Gottingen, Germany). Images were obtained using Plan-APOCHROMAT 40×/0.75 NA and Plan-APOCHROMAT 63×/0.95 NA oil-immersion objective lenses (Carl Zeiss) and processed with Adobe Photoshop CS2 (Adobe Systems, Mountain View, CA).
Results
Enhanced thymic repopulation and expansion of HPCs expressing C3G-F
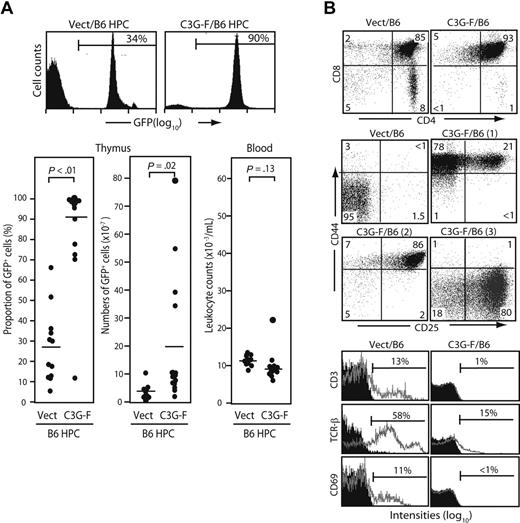
We infected Lin− cells sorted from normal B6 BM with empty or C3G-F–containing retrovirus and transplanted the infected cells together with normal BM cells into lethally irradiated mice. Infection efficiencies of empty and C3G-F retrovirus were comparable (25% on average). The recipients were killed 12 weeks after transplantation. The thymi of C3G-F/B6 HPC recipients showed greater cellularity and much higher proportions of GFP+ thymocytes than those of Vect/B6 HPC recipients (212 ± 56 × 106 and 87 ± 12%, N = 15 vs 106 ± 8 × 106 and 27% ± 8%, N = 12; Figure 1A). Thus, the absolute numbers of GFP+ thymocytes were significantly higher in the recipients of C3G-F/B6 HPCs than in those receiving Vect/B6 HPCs (194 ± 57 × 106 vs 31 ± 8 × 106). Peripheral leukocyte counts were comparable between the 2 groups, with only one recipient of C3G-F/B6 HPCs showing moderate leukocytosis (Figure 1A). The majority of GFP+ thymocytes in C3G-F/B6 HPC recipients were CD4/CD8 DP with marginal double-negative (DN) and single-positive cells (Figure 1B). Although DP cells in the control recipients were CD25− CD44−, C3G-F+ DP cells were CD25+ CD44+ in most recipients, and CD25− CD44+ or CD25+ CD44− in the remaining recipients (Figure 1B). Furthermore, in contrast to control DP cells, C3G-F+ DP cells showed negligible expression of CD3, TCRβ, or CD69 (Figure 1B). T cells with such unusual phenotypes were rarely found in the periphery, including spleen and BM of the C3G-F/B6 HPC recipients (data not shown), and it was unlikely that they immigrated into the thymus from outside.
Enhanced thymic repopulation and expansion of HPCs expressing C3G-F. (A) Lin− BM cells (HPCs) from B6 mice pretreated with 5-fluorouracil were infected with empty (Vect) or C3G-F–containing retrovirus, and transplanted into lethally irradiated B6 mice (2 × 104 cells/head) together with normal rescuing BM cells (2 × 105 cells/head). Twelve weeks later, the proportions and absolute numbers of GFP+ thymocytes as well as blood leukocyte counts were determined. Representative FACS profiles (top panel) and summary of the results (bottom panel) are indicated. Bars represent the mean values, and statistical analysis was done by Student t test. (B) Thymocytes were multicolor analyzed for GFP and indicated antibodies with the use of FACScan; the representative profiles in GFP+ population are shown. CD25/CD44 expression profiles varied in the recipients of C3G-F+ HPCs, and 3 typical profiles (1-3) are indicated. Filled areas in the histograms represent the control staining.
Enhanced thymic repopulation and expansion of HPCs expressing C3G-F. (A) Lin− BM cells (HPCs) from B6 mice pretreated with 5-fluorouracil were infected with empty (Vect) or C3G-F–containing retrovirus, and transplanted into lethally irradiated B6 mice (2 × 104 cells/head) together with normal rescuing BM cells (2 × 105 cells/head). Twelve weeks later, the proportions and absolute numbers of GFP+ thymocytes as well as blood leukocyte counts were determined. Representative FACS profiles (top panel) and summary of the results (bottom panel) are indicated. Bars represent the mean values, and statistical analysis was done by Student t test. (B) Thymocytes were multicolor analyzed for GFP and indicated antibodies with the use of FACScan; the representative profiles in GFP+ population are shown. CD25/CD44 expression profiles varied in the recipients of C3G-F+ HPCs, and 3 typical profiles (1-3) are indicated. Filled areas in the histograms represent the control staining.
Development of frank T-cell leukemia from SPA-1−/− HPCs expressing C3G-F
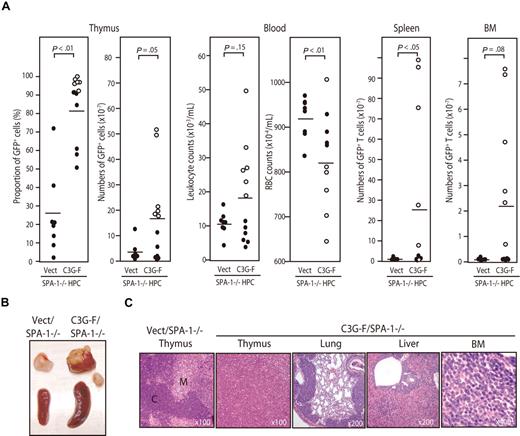
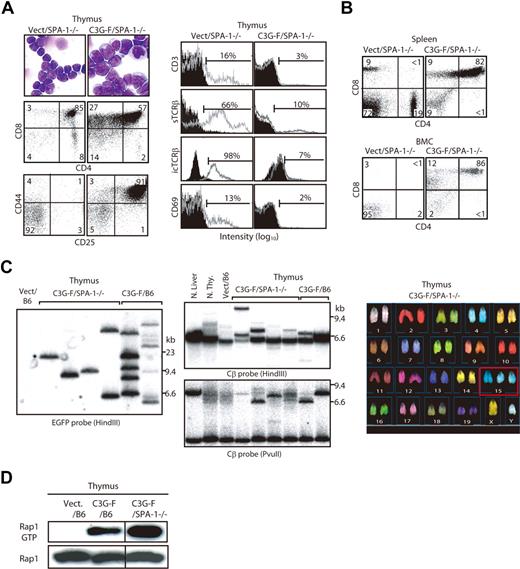
Because it was expected that the Rap1-activating effect of C3G-F might be attenuated by endogenous SPA-1,12 we conducted the same experiments using HPCs from SPA-1−/− mice. C3G-F/SPA-1−/− HPCs also exhibited a markedly enhanced expansion in the thymus, whereas Vect/SPA-1−/− HPCs showed a reconstitution pattern indistinguishable from that of Vect/B6 HPCs (Figure 2A). Furthermore, 6 of 12 recipients of C3G-F/SPA-1−/− HPCs became moribund 9 to 12 weeks after transplantation. These mice showed not only thymic enlargement but also peripheral leukocytosis, anemia, frequent lymphadenopathy, and marked increase in GFP+ T cells in their spleen and BM (Figure 2A,B). Histologically, the thymus lost normal corticomedullary structure; BM and other vital organs were heavily infiltrated with blastic lymphoid cells (Figure 2C). The majority of thymocytes in C3G-F/SPA-1−/− HPC recipients were large blastic cells with similar phenotypes to those in C3G-F/B6 HPC recipients, that is, CD4+ CD8+ CD25+ CD44+ CD3− TCRβ−/low CD69− (Figure 3A). Whereas a minor proportion was CD8+ CD4−/low, the blastic thymocytes coexpressed CD24 (data not shown), corresponding to immature single-positive (ISP) cells. It was confirmed that T cells with the same unusual phenotypes predominated in the spleens and BM (Figure 3B). Analysis of retroviral integration sites with Southern blot analysis using an EGFP cDNA probe revealed that the thymocytes in diseased C3G-F/SPA-1−/− HPC recipients were monoclonal or biclonal at most, whereas those in C3G-F/B6 HPC recipients were multiclonal (Figure 3C). The results were compatible with the diagnosis of T-cell acute lymphoblastic leukemia (T-ALL) andmulticentric thymic lymphoma, respectively. Clonal expansion of C3G-F/SPA-1−/− thymocytes was confirmed by karyotype analysis, in which 8 of 10 mitotic thymocytes in a recipient showed identical chromosomal anomaly (Figure 3C). Nonetheless, the thymocytes showed diverse TCRβ (Cβ) rearrangement bands in most cases, except for the case of a C3G-F/B6 HPC recipient showing a germ-line pattern (Figure 3C). These results suggested that the clonal expansion of C3G-F+ SPA-1−/− progenitors was initiated at an early DN stage prior to Cβ rearrangements. As expected, blastic thymocytes from both C3G-F/B6 and C3G-F/SPA-1−/− HPC recipients showed marked accumulation of Rap1GTP, with the latter exhibiting greater amounts than the former (Figure 3D). The results strongly suggested that deregulated, constitutive activation of endogenous Rap1 caused abnormal proliferation and transformation of DP thymocytes, with the disease severity being correlated with the levels of Rap1 activation.
Development of T-cell leukemia in the recipients of SPA-1−/− HPCs expressing C3G-F. (A) HPCs from SPA-1−/− mice were infected with empty (Vect) or C3G-F–containing retrovirus and transplanted into lethally irradiated B6 mice. Nine to 12 weeks later, the proportions and numbers of GFP+ cells in various tissues as well as the leukocyte and red blood cell counts in blood were determined. Six of 12 recipients of C3G-F/SPA-1−/− HPCs (○) became moribund in 9 to 12 weeks, when they were killed. Bars indicate the mean values; statistical analysis was done by Student t test. (B) Representative thymus and spleen of each group. In C3G-F/SPA-1−/− HPC recipients, lymphoadenopathy was frequently detected, and a swollen abdominal lymph node is also shown. (C) Hematoxylin-eosin–stained photographs of the indicated organs of Vect/SPA-1−/− and C3G-F/SPA-1−/− HPC recipients. Magnifications are indicated. C indicates cortex; M, medulla.
Development of T-cell leukemia in the recipients of SPA-1−/− HPCs expressing C3G-F. (A) HPCs from SPA-1−/− mice were infected with empty (Vect) or C3G-F–containing retrovirus and transplanted into lethally irradiated B6 mice. Nine to 12 weeks later, the proportions and numbers of GFP+ cells in various tissues as well as the leukocyte and red blood cell counts in blood were determined. Six of 12 recipients of C3G-F/SPA-1−/− HPCs (○) became moribund in 9 to 12 weeks, when they were killed. Bars indicate the mean values; statistical analysis was done by Student t test. (B) Representative thymus and spleen of each group. In C3G-F/SPA-1−/− HPC recipients, lymphoadenopathy was frequently detected, and a swollen abdominal lymph node is also shown. (C) Hematoxylin-eosin–stained photographs of the indicated organs of Vect/SPA-1−/− and C3G-F/SPA-1−/− HPC recipients. Magnifications are indicated. C indicates cortex; M, medulla.
Clonal expansion of blastic thymocytes with marked accumulation of Rap1GTP in the recipients of C3G-F/SPA-1−/− HPCs. (A) Cells from the thymi of Vect/SPA-1−/− and C3G-F/SPA-1−/− HPC recipients were Giemsa-stained 12 weeks after transplantation. They were also multicolor analyzed for the expression of indicated antigens with the use of FACScan; the representative profiles in GFP+ fraction are shown. Filled areas in histograms indicate the control staining. (B) Spleen and BM cells (BMCs) of Vect/SPA−1−/− and C3G-F/SPA-1−/− HPC recipients were 3-color analyzed with the use of FACScan; CD4/CD8 expression profiles in GFP+ fractions are shown. (C) DNAs from the thymic cells of Vect/B6, C3G-F/SPA-1−/−, and C3G-F/B6 HPC recipients 12 weeks after transplantation were digested with indicated restriction enzymes and Southern blotted with the use of an EGFP (left panel) or a TCR Cβ (middle panels) cDNA probe. Normal liver and thymocytes served as controls for negative and positive Cβ gene rearrangement, respectively. Thymocytes from a C3G-F/SPA-1−/− HPC recipient were treated with colcemid and subjected to spectral karyotyping analysis (right panel). Eight of 10 mitotic cells showed an identical karyotype with chromosome 15 trisomy (boxed). (D) Cells from the thymi of Vect/B6, C3G-F/B6, and C3G-F/SPA-1−/− HPC recipients 12 weeks after transplantation were lysed; Rap1 GTP was detected by a pull-down assay. Vertical lines have been inserted to indicate a repositioned gel lane.
Clonal expansion of blastic thymocytes with marked accumulation of Rap1GTP in the recipients of C3G-F/SPA-1−/− HPCs. (A) Cells from the thymi of Vect/SPA-1−/− and C3G-F/SPA-1−/− HPC recipients were Giemsa-stained 12 weeks after transplantation. They were also multicolor analyzed for the expression of indicated antigens with the use of FACScan; the representative profiles in GFP+ fraction are shown. Filled areas in histograms indicate the control staining. (B) Spleen and BM cells (BMCs) of Vect/SPA−1−/− and C3G-F/SPA-1−/− HPC recipients were 3-color analyzed with the use of FACScan; CD4/CD8 expression profiles in GFP+ fractions are shown. (C) DNAs from the thymic cells of Vect/B6, C3G-F/SPA-1−/−, and C3G-F/B6 HPC recipients 12 weeks after transplantation were digested with indicated restriction enzymes and Southern blotted with the use of an EGFP (left panel) or a TCR Cβ (middle panels) cDNA probe. Normal liver and thymocytes served as controls for negative and positive Cβ gene rearrangement, respectively. Thymocytes from a C3G-F/SPA-1−/− HPC recipient were treated with colcemid and subjected to spectral karyotyping analysis (right panel). Eight of 10 mitotic cells showed an identical karyotype with chromosome 15 trisomy (boxed). (D) Cells from the thymi of Vect/B6, C3G-F/B6, and C3G-F/SPA-1−/− HPC recipients 12 weeks after transplantation were lysed; Rap1 GTP was detected by a pull-down assay. Vertical lines have been inserted to indicate a repositioned gel lane.
Augmented expression and activation of Notch in leukemic C3G-F+ thymocytes
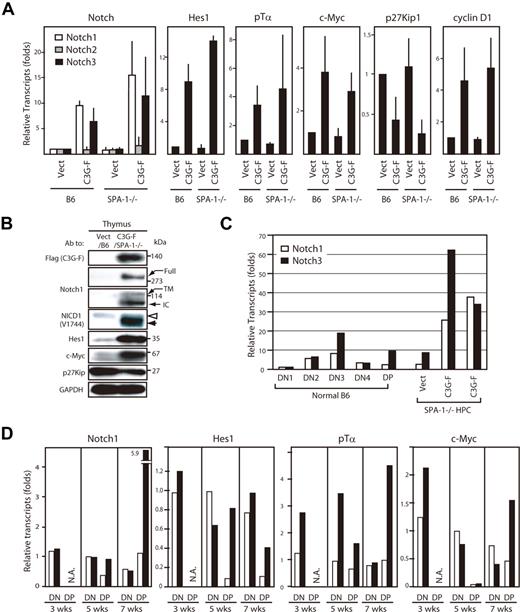
We then analyzed gene expression profiles of the thymocytes using real-time PCR. The thymocytes derived from C3G-F/HPCs 12 weeks after transplantation exhibited markedly augmented expression of Notch1 and 3, but not Notch2, compared with those from Vect/HPCs; the effects tended to be more pronounced in those from C3G-F/SPA-1−/− HPCs (Figure 4A). The blastic thymocytes derived from C3G-F/HPC recipients also showed enhanced expression of putative Notch target genes, Hes1, pTα, c-Myc, and cyclin D1, and reduced expression of p27Kip1, a target of Hes1-mediated gene repression 17 (Figure 4A). The results were confirmed at the protein level by immunoblotting (Figure 4B). Notably, expression of cleaved Notch1 isoforms (transmembrane and intracellular forms) was markedly increased, being consistent with the activation of Notch signaling in vivo. It was noted that NICD1 in leukemic C3G-F+ SPA-1−/− thymocytes tended to be smaller in size than that in control thymocytes (Figure 4B). Expression of Notch is reported to vary in developmental stages of normal thymocytes18,19 ; therefore, we compared Notch expression in leukemic C3G-F/SPA-1−/− thymocytes with that in normal thymocytes at distinct stages. In normal thymocytes, the expression of Notch1 and 3 was the highest at DN3 stage followed by a decline at DN4 and DP stages (Figure 4C). Whereas leukemic C3G-F+ SPA-1−/− thymocytes were mostly DP, they showed much greater amounts of Notch1 and 3 transcripts, even when compared with normal DN3 cells (Figure 4C). Kinetic studies revealed that expression of pTα, c-Myc, and possibly Hes1 tended to be increased in the initial repopulating C3G-F+ DN cells as early as 3 weeks after transplantation, when Notch1 expression was unchanged (Figure 4D). Although the expression of Notch1 and the target genes declined in control DP cells that emerged at 5 weeks, their expression was sustained in C3G-F+ DP cells at high levels, except for c-Myc. At 7 weeks, when the enhanced expansion of C3G-F+ SPA-1−/− DP cells became evident, the expression of Notch1 as well as the target genes in C3G-F+ DP cells, including c-Myc, was markedly augmented compared with control thymocytes (Figure 4D). The results suggested that Notch1 activation was sustained from early DN cells throughout their differentiation to DP cells in C3G-F+ SPA-1−/− thymocytes.
Augmented expression of Notch and the targets in C3G-F+ leukemic thymocytes. (A) RNAs were extracted from sorted GFP+ cells from the thymi of Vect/B6, C3G-F/B6, Vect/SPA-1−/−, and C3G-F/SPA-1−/− HPC recipients 12 weeks after transplantation; transcripts for indicated genes were assessed by real-time PCR. The transcripts of each gene were normalized to cyclophilin; relative amounts of transcripts (folds) to those of Vect/B6 thymocytes are indicated. The means and SE of 3 to 4 mice are shown. (B) Cells from the thymi of Vect/B6 and C3G-F/SPA-1−/− HPC recipients were lysed and immunoblotted with indicated antibodies. Full indicates full-length Notch; TM, transmembrane; and IC, intracellular Notch isoforms. Note different molecular sizes of NICD1 in Vect/B6 and C3G-F/SPA-1−/− HPC recipients (open and solid arrowheads). (C) RNAs were extracted from sorted thymocyte subsets (DN1, DN2, DN3, DN4, and DP) from normal mice as well as total thymocytes of Vect/SPA-1−/− and C3G-F/SPA-1−/− HPC recipients. Notch1 (open columns) and Notch 3 (closed columns) transcripts were assessed by real-time PCR and normalized to cyclophilin; relative amounts of transcripts (folds) to those of normal DN1 cells are indicated. The means of 2 independent analyses are shown. (D) Recipients of Vect/SPA-1−/− HPCs (open columns) and C3G-F/SPA-1−/− HPCs (closed columns) were killed 3, 5, and 7 weeks after transplantation; GFP+ DN and GFP+ DP thymocytes were sorted using FACSVantage. RNAs were extracted and the transcripts of indicated genes were assessed by real-time PCR. Relative amounts of transcripts (folds) to those of control DN cells at 5 weeks are indicated. The means of 2 analyses are shown. NA indicates not applicable because few DP cells were developed yet.
Augmented expression of Notch and the targets in C3G-F+ leukemic thymocytes. (A) RNAs were extracted from sorted GFP+ cells from the thymi of Vect/B6, C3G-F/B6, Vect/SPA-1−/−, and C3G-F/SPA-1−/− HPC recipients 12 weeks after transplantation; transcripts for indicated genes were assessed by real-time PCR. The transcripts of each gene were normalized to cyclophilin; relative amounts of transcripts (folds) to those of Vect/B6 thymocytes are indicated. The means and SE of 3 to 4 mice are shown. (B) Cells from the thymi of Vect/B6 and C3G-F/SPA-1−/− HPC recipients were lysed and immunoblotted with indicated antibodies. Full indicates full-length Notch; TM, transmembrane; and IC, intracellular Notch isoforms. Note different molecular sizes of NICD1 in Vect/B6 and C3G-F/SPA-1−/− HPC recipients (open and solid arrowheads). (C) RNAs were extracted from sorted thymocyte subsets (DN1, DN2, DN3, DN4, and DP) from normal mice as well as total thymocytes of Vect/SPA-1−/− and C3G-F/SPA-1−/− HPC recipients. Notch1 (open columns) and Notch 3 (closed columns) transcripts were assessed by real-time PCR and normalized to cyclophilin; relative amounts of transcripts (folds) to those of normal DN1 cells are indicated. The means of 2 independent analyses are shown. (D) Recipients of Vect/SPA-1−/− HPCs (open columns) and C3G-F/SPA-1−/− HPCs (closed columns) were killed 3, 5, and 7 weeks after transplantation; GFP+ DN and GFP+ DP thymocytes were sorted using FACSVantage. RNAs were extracted and the transcripts of indicated genes were assessed by real-time PCR. Relative amounts of transcripts (folds) to those of control DN cells at 5 weeks are indicated. The means of 2 analyses are shown. NA indicates not applicable because few DP cells were developed yet.
Notch1 mutations and Notch-dependent proliferation of C3G-F+ SPA-1−/− T-ALL cells
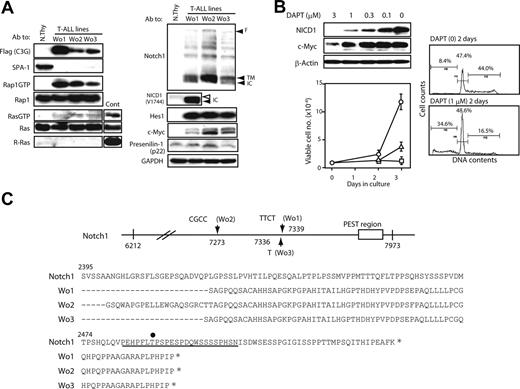
We established continuous cell lines from the thymi of C3G-F/SPA-1−/− HPC recipients bearing frank T-ALL (Wo1, 2, 3). It was confirmed by retroviral integration site analysis that the cell lines originated from clonally expanded thymocytes in the recipients (data not shown). All the independently established T-ALL lines showed similar phenotypes (DP/CD8+ ISP CD3− TCRβ−) and caused rapidly lethal leukemia upon transfer into normal mice (data not shown). They expressed C3G-F and showed constitutive Rap1 activation as expected, whereas activation of classical Ras was marginal (Figure 5A). Although C3G-F was reported to show a weak GEF activity for R-Ras,20 the T-ALL lines expressed no detectable R-Ras (Figure 5A). They also exhibited abundant expression of NICD1 as well as Hes1 and c-Myc proteins, whereas PS-1 expression was largely comparable with that in normal thymocytes (Figure 5A). It was again noted that NICD1 in T-ALL was smaller in size than that in normal thymocytes (Figure 5A). Culture of T-ALL cells in the presence of a γ-secretase inhibitor (DAPT) for 2 days resulted in the reduction of NICD1 and c-Myc expression in a dose-dependent manner (Figure 5B). Concomitantly, the proliferation of T-ALL cells was almost completely inhibited in the presence of DAPT in the comparable range of concentrations (Figure 5B). Essentially identical results were obtained in all 3 T-ALL cell lines (data not shown). FACS analysis revealed that impaired growth was due to reduced S phase entry and apoptotic cell death (Figure 5B). The proliferation of a Moloney leukemia virus–induced T-cell leukemia line was unaffected in the presence of 1 μM DAPT (data not shown). The results strongly suggested that the proliferation of C3G-F+ SPA-1−/−T-ALL cells was dependent on continuous Notch signaling.
Notch1 mutations and Notch-dependent proliferation of C3G-F+ SPA-1−/− T-ALL cell lines. (A) T-ALL cell lines (Wo1, Wo2, and Wo3) established from the thymi of C3G-F/SPA-1−/− HPC recipients as well as normal thymocytes were lysed and immunoblotted with indicated antibodies. Rap1GTP and Ras GTP were detected by a pull-down assay. F indicates full-length Notch; TM; transmembrane form; and NICD, Notch intracellular domain. Note smaller NICD1 size in Wo1 line (solid arrowhead) than in normal thymocytes (open arrowhead). (B) T-ALL cell lines were cultured in the presence of various concentrations of DAPT for 48 hours, and the expression of indicated proteins was detected by immunoblotting (left upper panel). They were also cultured in the absence (circles) or presence of 0.3 μM (triangles) or 1 μM (rectangles) DAPT, and the viable cell numbers were determined by MTT assay at days 2 and 3 (left lower panel). Means and SE of triplicate cultures are indicated. T-ALL cells were cultured in the presence or absence of 1 μM DAPT for 2 days and analyzed for DNA contents (right panels). (C) Notch1 cDNAs of approximately 1.7-kb upstream of PEST region (from nt 6212 to nt 7973) were cloned from 3 independent T-ALL lines, and more than 5 subclones of each were sequenced. Insertions detected in more than 3 subclones were considered to represent genuine mutations and are indicated (top panel). Deduced amino acid sequences are also indicated (bottom panel). An underline indicates the PEST domain, and an asterisk marks the stop codon. T2512 required for the binding of FBW7 ubiquitin ligase is indicated by a dot.
Notch1 mutations and Notch-dependent proliferation of C3G-F+ SPA-1−/− T-ALL cell lines. (A) T-ALL cell lines (Wo1, Wo2, and Wo3) established from the thymi of C3G-F/SPA-1−/− HPC recipients as well as normal thymocytes were lysed and immunoblotted with indicated antibodies. Rap1GTP and Ras GTP were detected by a pull-down assay. F indicates full-length Notch; TM; transmembrane form; and NICD, Notch intracellular domain. Note smaller NICD1 size in Wo1 line (solid arrowhead) than in normal thymocytes (open arrowhead). (B) T-ALL cell lines were cultured in the presence of various concentrations of DAPT for 48 hours, and the expression of indicated proteins was detected by immunoblotting (left upper panel). They were also cultured in the absence (circles) or presence of 0.3 μM (triangles) or 1 μM (rectangles) DAPT, and the viable cell numbers were determined by MTT assay at days 2 and 3 (left lower panel). Means and SE of triplicate cultures are indicated. T-ALL cells were cultured in the presence or absence of 1 μM DAPT for 2 days and analyzed for DNA contents (right panels). (C) Notch1 cDNAs of approximately 1.7-kb upstream of PEST region (from nt 6212 to nt 7973) were cloned from 3 independent T-ALL lines, and more than 5 subclones of each were sequenced. Insertions detected in more than 3 subclones were considered to represent genuine mutations and are indicated (top panel). Deduced amino acid sequences are also indicated (bottom panel). An underline indicates the PEST domain, and an asterisk marks the stop codon. T2512 required for the binding of FBW7 ubiquitin ligase is indicated by a dot.
Recent reports revealed characteristic mutations in Notch1 in the majority of T-ALL cells developed in various genetically prone mouse models13,–15 ; therefore, we examined the relevant region (approximately 1.7-kb upstream of the PEST domain) of Notch1 of Wo cell lines. It was revealed that all 3 T-ALL lines showed variable nucleotide insertions, resulting in frame shifts and premature termination within the PEST domain (Figure 5C). Accordingly, the PEST domain including a crucial residue (T2512) for ubiquitination of Notch1 was abrogated in all cell lines. The results suggested that Notch1 mutations might be related to the continuous Notch-dependent proliferation of C3G-F+ SPA-1−/− T-ALL clones.
C3G-F–induced, aberrant thymocyte expansion is independent of pre-TCR/TCR
We finally examined the requirement of pre-TCR/TCR for abnormal expansion of C3G-F+ thymocytes. HPCs from B6 Rag2−/− or Rag2−/− SPA-1−/− mice were infected with empty or C3G-F–containing retrovirus and transplanted into irradiated B6 mice together with normal BM cells. All the mice were killed 12 weeks after transplantation. As expected, Vect/Rag2−/− HPCs hardly repopulated the thymus (GFP+ cell proportions and numbers: 0.12% ± 0.07% and 0.13 ± 0.07 × 106, N = 5; Figure 6A). In contrast, the recipients of C3G-F/Rag2−/− HPCs (3 mice) and C3G-F/Rag2−/− SPA-1−/− HPCs (2 mice) exhibited significant thymic repopulation (GFP+ cell proportions and numbers: 84% ± 12% and 249 ± 111 × 106, N = 5), and the majority of thymocytes were DP with some CD8+ ISP cells (Figure 6A,B). The extent of DP cell expansion again tended to be more striking in the recipients of C3G-F/Rag2−/− SPA-1−/− HPCs, one of which developed massive thymic lymphoma 12 weeks after transplantation (Figure 6A). The results demonstrated that aberrant expansion and transformation of C3G-F+ DP cells were independent of pre-TCR/TCR expression.
Expansion of C3G-F+ DN cells is independent of pre-TCR. (A,B) Rag2−/− (•) and Rag2−/− SPA-1−/− (○) HPCs were infected with empty or C3G-F–containing retrovirus followed by transplantation into irradiated B6 mice. Twelve weeks later, mice were killed, and the proportions and numbers of GFP+ thymocytes were determined. The thymocytes were 3-color analyzed by FACSCalibur, and CD4/CD8 expression profiles in GFP+ population are indicated.
Expansion of C3G-F+ DN cells is independent of pre-TCR. (A,B) Rag2−/− (•) and Rag2−/− SPA-1−/− (○) HPCs were infected with empty or C3G-F–containing retrovirus followed by transplantation into irradiated B6 mice. Twelve weeks later, mice were killed, and the proportions and numbers of GFP+ thymocytes were determined. The thymocytes were 3-color analyzed by FACSCalibur, and CD4/CD8 expression profiles in GFP+ population are indicated.
Discussion
In this study, we showed that C3G-F/B6 HPCs exhibited much greater thymic repopulation in irradiated recipients than control HPCs. C3G-F/B6 HPCs, however, resulted in a marked expansion of oligoclonal thymocytes with unusual phenotypes (CD4+ CD8+, CD44+ and/or CD25+, CD3− TCRβ−) 12 weeks after transplantation, suggesting multicentric thymic lymphoma. Transplantation of C3G-F/SPA-1−/− HPCs exhibited more pronounced phenotypes. In addition to thymic enlargement, half of the recipients showed a systemic spread of blastic C3G-F+ DP T cells, with marked invasion into vital organs, and became moribund in 9 to 12 weeks. These blastic DP T cells were mostly monoclonal, being consistent with the diagnosis of T-ALL. As expected, the amounts of Rap1GTP were higher in leukemic C3G-F+ SPA-1−/− than in C3G-F+ B6 thymocytes; thus, the difference in disease severity was correlated with the level of Rap1 activation. C3G and SPA-1 hardly exhibited GEF and GAP activity for classical Ras, respectively,20,21 and consistently no significant Ras activation was observed in C3G-F+ SPA-1−/− T-ALL cells. In addition, our preliminary results indicated that transplantation of SPA-1−/− HPCs that expressed an active form of Rap1 (Rap1E63) induced abnormal expansion of DP thymocytes, which could transfer the leukemia to secondary recipients, albeit less frequently (Y.N. et al, unpublished observation, December 2007). These results collectively suggested that deregulated, constitutive activation of endogenous Rap1 signaling was primarily responsible for aberrant proliferation and transformation of thymocytes. Although SPA-1−/− HPCs, per se, showed no apparent abnormality in the thymocyte development attributable to the redundant expression of additional Rap1GAPs such as Rap1GAP1 and SPA-Ls,22 the current results suggested that endogenous SPA-1 significantly contributed to determining the threshold levels for Rap1 signaling in thymocytes.
In contrast to normal DP thymocytes, C3G-F+ DP cells 12 weeks after transplantation marginally expressed CD3/TCRβ but continued to express either CD25 or CD44 or both, being reminiscent of DN cells. Curiously, the sorted GFP+ leukemic thymocytes from C3G-F/SPA-1−/− HPC recipients showed rather diverse TCRβ (probably Dβ/Jβ) rearrangements in spite of clonalexpansion as judged by both retroviral integration sites and karyotypes. The results suggested that aberrant expansion of thymocytes was actually initiated prior to, not following, TCRβ rearrangements. Deregulated Rap1 activation thus apparently induced abnormal proliferation of DN thymocytes without interfering with subsequent TCRβ rearrangements and differentiation to DP cells. Notch receptors play a crucial role in the initial expansion of early T-cell precursors.23,,–26 Present results revealed that the initial repopulating C3G-F+ DN cells tended to express higher levels of Notch1 target genes than control DN cells as early as 3 weeks after transplantation, including Hes1, c-Myc, and pTα, which played crucial roles in promoting DN cell proliferation and pre-TCR expression,27,–29 although Notch1 expression was comparable. Furthermore, whereas Notch1 expression in normal DN cells was down-regulated as they developed into DP stage, C3G-F+ SPA-1−/− DP cells showed a progressive increase in Notch1 expression. Notch expression is autonomously induced by Notch signaling, per se30 ; thus, this finding might reflect the positive feedback mechanism of sustained Notch activation. Alternatively, thymocyte clones with higher Notch expression might have been selected because of the proliferation advantage. In either case, it was suggested that deregulated activation of endogenous Notch signaling underlay the aberrant expansion and transformation of C3G-F+ thymocytes.
Notch signaling is also essential in pre-TCR–mediated expansion of DN cells and their differentiation to αβTCR+ DP cells, called β-selection.31,32 We recently found that mice conditionally expressing SPA-1 transgene under lck promoter showed a severe defect in DP cell development attributable to apoptotic death of DN4 cells, albeit expression of pre-TCR was normal (Kometani et al, manuscript in preparation). We also found that expression of a dominant-negative Rap1 in normal DN thymocytes interfered with the differentiation to DP cells in the presence of stroma cells expressing delta-like 1 (DLL1) in vitro. This finding strongly suggests that Rap1 mediated pre-TCR signaling, at least in part (Kometani et al, manuscript in preparation). As a probable reflection of the synergism of pre-TCR and Notch signaling in normal β selection, the development of T-cell leukemia by NICD-expressing HPCs was reported to depend on pre-TCR.33,34 A more recent report indicated that Rag2−/− HPCs expressing high levels of NICD did develop T-cell leukemia, albeit the latency was much longer than that of normal HPCs expressing NICD. The leukemia development, however, was hastened by anti-CD3 monoclonal antibody, confirming an assisting role of pre-TCR signaling in NICD-induced leukemia.35 Whereas NICD-induced T-ALLs were DP CD3/TCRβ+ of mostly extrathymic origin33 attributable to premature T-cell fate commitment in BM,23,24 C3G-F+ SPA-1−/− T-ALLs originated exclusively within the thymus and thus were dependent on thymic microenvironments. Furthermore, C3G-F/Rag2−/− SPA-1−/− HPCs also caused a massive DP cell expansion in the thymus. Thus, it was suggested that premature, constitutive Rap1 activation promoted a Notch-dependent expansion and DP conversion of DN CD3/TCRβ− cells, bypassing pre-TCR expression.
Supporting this idea, the proliferation of C3G-F+ SPA-1−/− T-ALL cell lines, which showed constitutive Rap1 activation, was completely inhibited in the presence of a γ-secretase inhibitor (DAPT). It was reported that the proliferation of NICD-induced T-ALL cells was mediated by the induction of c-Myc,36 and c-Myc expression consistently was also suppressed by DAPT. These results confirmed that the proliferation of C3G-F+ SPA-1−/− T-ALL cells was dependent on Notch signaling. A recent study has revealed mutations of Notch1 at a few hot spots including the PEST region in the majority of human T-ALL.16 More recently, Notch1 mutations at the PEST region were detected in high proportions of T-ALL that developed in mice with various genetic predispositions, such as Tal1 (SCL)–transgenic and Ikaros−/− mice.13,–15 Their proliferation was also inhibited in the presence of γ-secretase inhibitors.13 Present results indicated that all the C3G-F+ SPA-1−/− T-ALL cell lines harbored insertions at the similar region of Notch1, resulting in premature termination within the PEST domain and C-terminal truncation. The most recent report indicated that threonine residue in the PEST domain (T2512) was crucial for NICD1 ubiquitination by FBW7 and degradation.37 The mutant Notch1 in C3G-F+ SPA-1−/− T-ALL cells lost T2512, and thus might be expected to show increased protein half-life. The results indicated that deregulated activation of endogenous Rap1 signaling also increased the chance for activating Notch1 mutations, and supported a view that Notch1 mutations represented a common secondary event for progression into frank T-ALL.
Possible mechanisms for the synergy of Rap1 and Notch signaling remain to be investigated. It was noted that C3G-F+ SPA-1−/− T-ALL cells grew in a dense aggregate form mediated by multiple adhesion molecules that likely are activated by Rap1 signaling. Albeit the cells showed extensive proliferation in bulk culture, they showed poor clonogenic growth or anchorage-independent colony formation in agar, implying the requirement of cell interactions for optimal growth (M.A. et al, unpublished observation). Furthermore, attenuation of Rap1 activation in T-ALL cells resulted in their reduced growth rate (M.A. et al, unpublished observation, May 2007). It may be possible that enhanced cell-to-cell interaction via Rap1 signaling leads to efficient Notch/ligand interaction. Generation of even small amounts of NICD1 from mutant Notch1 by suboptimal ligands may suffice for efficient signaling in T-ALL cells. Alternatively, Rap1 signaling may induce permissive conditions for Notch signaling. For instance, it was reported that oncogenic Ras enhanced the expression of PS-1 via the p38MAPK pathway38 ; however, present results provided no evidence for enhanced PS-1 expression in C3G-F+ SPA-1−/− T-ALL cell lines. Our unpublished results indicate that endogenous Rap1 signaling is crucial for the rescue of normal DN thymocytes from death at the β-selection checkpoint (Kometani et al, manuscript in preparation), and that the effect may contribute to the enhanced Notch signaling. In either case, present results strongly suggest the synergism of Rap1 and Notch signaling in thymocyte development and transformation. Underlying factors leading to Notch1 mutations and Notch-dependent T-ALL in humans remain to be identified, and further analysis may provide a clue to understanding the pathogenesis of human T-ALL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs T. Kitamura, T. Sudo, and Y. Tanaka for providing a retroviral plasmid and an anti-Hes1 antibody, and for proofreading the article.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Culture, Sports, and Technology of Japan to N.M. and M.H., and Takeda Science Foundation to N.M.
Authorship
Contribution: N.M. and M.H. designed the research; S-F.W., M.A., Y.N., Y.S., H.T., M.T., and M.H. performed experiments and analyzed data; and N.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nagahiro Minato, Department of Immunology and Cell Biology, Graduate School of Medicine, Kyoto University, Yoshida-konoe-cho, Sakyo-ku, Kyoto 606-8501, Japan; e-mail: minato@imm.med.kyoto-u.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal