MicroRNAs are regulated by gene alteration, transcription, and processing. Thus far, few studies have simultaneously assessed all 3 levels of regulation. Using real-time quantitative polymerase chain reaction (QPCR)–based arrays, we determined changes in gene copy number, pre-miRNA, and mature miRNA levels for the largest set of primary effusion lymphomas (PELs) to date. We detected PEL-specific miRNA gene amplifications, and concordant changes in pre-miRNA and mature miRNA. We identified 68 PEL-specific miRNAs. This defines the miRNA signature of PEL and shows that transcriptional regulation of pre-miRNA as well as mature miRNA levels contribute nonredundant information that can be used for the classification of human tumors.
Introduction
MicroRNAs (miRNAs) are approximately 22-nt–long noncoding RNAs, which are found in all metazoans. Approximately 500 miRNAs have been identified in humans.1 Many miRNAs are evolutionarily conserved. MiRNA homologs have been identified in Drosophila, humans, and plants. Each miRNA can regulate the expression of multiple target messenger RNAs (mRNAs). MiRNAs regulate many processes including developmental stages, cellular responses such as inflammation, the differentiation of hematopoietic stem cells into lymphocytes, and the transformation of normal cells into cancer cells (reviewed in Calin and Croce2 ). MiRNAs themselves are transcriptionally regulated and often show stringent tissue specificity.
Most miRNA gene loci are interspersed between coding regions or located within introns, though some can be embedded within an open reading frame. Thirty-seven percent of human miRNAs are organized in multi-miRNA clusters,3 many of which can be found around fragile sites.4,5 Clustered miRNAs are regulated by a common promoter and processed from a single primary transcript (pri-miRNA) that may contain several miRNAs, as well as coding exons. Hence, miRNAs are subject to (1) genomic alterations at the DNA level, (2) transcriptional regulation at the pre-miRNA level, and (3) processing control at the mature miRNA level. Thus far, few studies have evaluated these 3 modes of regulation simultaneously.
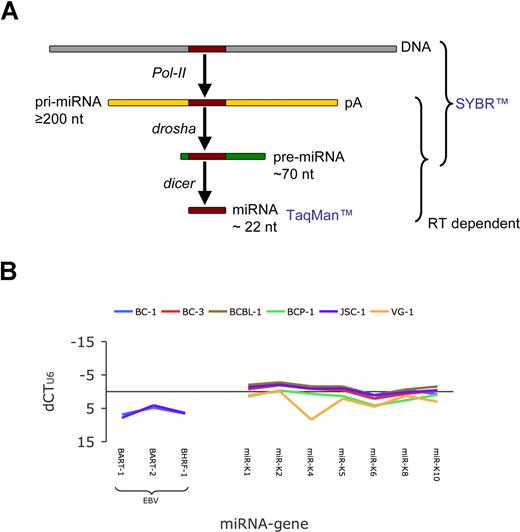
In the nucleus, Drosha initiates miRNA processing (Figure 1A) by cleaving the primary miRNA (pri-miRNA) to release the approximately 60-nt-long precursor miRNAs (pre-miRNAs).6 After export to the cytoplasm, the pre-miRNAs are further processed by Dicer to yield an approximately 22-bp miRNA duplex.7,8 One strand of this duplex is then incorporated into the RNA-induced silencing complex (RISC), where it guides the RISC to mRNAs bearing complementary sequences.9,10 If the mRNA contains a perfectly complementary sequence, the RISC component Ago2 cleaves the target leading to mRNA degradation.11,12 In the case of an imperfect complementary target, RISC binding can induce translational inhibition.12 Translation inhibition is highly cooperative and requires several RISCs, potentially each with a different miRNA.13,14
We hypothesized that cancer-specific miRNA profiles are determined by a combination of changes at the level of the gene locus, that is, mutations, deletions, or amplifications, changes at the level of transcriptional regulation, and changes at the level of miRNA processing. Hence, comprehensive miRNA profiling should query genomic loci, precursor miRNAs, as well as mature miRNAs. We hypothesized further that this information can be used for the differential diagnosis of lymphomas, specifically of primary effusion lymphoma (PEL).
PELs are a unique type of post-germinal center diffuse large B-cell lymphoma (DLBCL).15,16 Clinically PELs are defined by their effusion phenotype. Furthermore, all PEL tumors carry Kaposi sarcoma–associated herpesvirus (KSHV). KSHV also encodes viral miRNAs.17,18 These are highly conserved among KSHV isolates.19 We assembled the largest group of PEL samples to date, which allowed us to generate the first miRNA signature for PEL. Using a novel real-time quantitative polymerase chain reaction (QPCR)–based assay, we identified miRNAs, which are concordantly transcribed in most PELs and are present at the pre-miRNA and mature miRNA level. The more abundant of these miRNAs were previously cloned, which independently confirms our data. However, high-throughput QPCR-based profiling uncovered twice as many PEL-specific miRNAs as found by cloning methods. We found a second group of miRNAs, which are uniformly present in nontransformed tonsil tissue, but down-regulated in PEL as well as a third group of miRNAs that were uniquely up- or down-regulated in individual PEL cell lines. These data can be used to molecularly classify PEL. Based on the known and predicted targets of miRNAs, which are present in PEL, we propose that PEL-specific miRNAs define the biologic phenotype of PEL.
Methods
Cell lines and tissue samples
All cells were cultured in RPMI containing 25 mM HEPES, 10% fetal bovine serum, 0.05 mM 2-mercaptoethanol, 1 mM sodium pyruvate, 2 mM l-glutamine, 0.05 μg penicillin/mL, and 50 U streptomycin/mL at 37°C and in 5% CO2. Deidentified frozen tonsil tissue biopsies were obtained from the cooperative human tissue network (CHTN). Use of human cell lines and tissue was deemed “exempt” by the UNC IRB and does not require IRB approval. The PEL cell lines are described in Table S3 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
DNA and RNA isolation
DNA was isolated using WizardSV kit (Promega, Madison, WI). Total RNA was isolated using Triazol (Sigma-Aldrich, St Louis, MO) as described.20 RNA was quantitated on a Nanodrop, and equal amounts of RNA were subjected to DNase I treatment (Ambion, Austin, TX). RNA was reverse transcribed using cDNA archive kit (Applied Biosystems, Foster City, CA). RNA integrity was evaluated using a 2100 Bioanalyzer Series C (Agilent, Santa Clara, CA). Total RNA was measured using the RNA 6000 Series II Nano Kit, and small RNA was measured using the Small RNA Kit according to the manufacturer's recommendations (Agilent). All chips were analyzed using 2100 Expert software version B.02.04 (Agilent).
Real-time QPCR
We used two 96-well plates containing a total of 372 different primers representing 168 cellular and 12 viral pre-miRNA targets, and 6 cellular and viral control mRNA targets. All primers conform to universal real-time PCR conditions20,–22 with a predicted Tm of 60 (± 1)°C (mean ± standard deviation [SD]) and 100-bp or smaller amplicon length. Real-time QPCR was conducted under universal cycling conditions (40 cycles) with SYBR as the method of detection following our previously validated methods.20 A 36-μL reaction mix was made and distributed in triplicate into a 384-well plate using a CAS-1200 robot that uses filtered carbon-graphite pipette tips (Tecan, Durham, NC) for liquid level sensing, allowing for a pipetting accuracy of 0.1 μL. The final primer concentration was 250 nM in total of 9.0-μL reaction volume. Real-time QPCR was conducted using a Roche LC420 unit (Roche, Indianapolis, IN). The combined pipetting and instrument error was less than 6% (data not shown). All reactions were done in technical as well as experimental triplicates. As pre-miRNA–specific primers also detect the corresponding gene, we used these primers for DNA gene profiling. For DNA QPCR, each reaction contained 1.67 ng DNA/μL. For RNA QPCR, 40 μL of the 100-μL RT reaction was used for each 384-well plate, yielding a final amount of 0.1 μL cDNA per each 9-μL reaction. Real-time QPCR primers against 165 mature miRNAs were used according to the manufacturer's protocol (Applied Biosystems). Of note we were able to quantify miR-K5 from BC-1 cells and miR-K4 from VG-1 cells, even though the corresponding pre-miRNAs contain a single nucleotide polymorphism (SNP)23 (A.J.O. and D.P.D., unpublished observations, July 2007), which is located within the primer binding site. This underscores the robustness of a SYBR-based pre-miRNA QPCR assay, which tolerates up to 2 SNPs in each primer binding site as opposed to TaqMan-based mature miRNA QPCR assays, which do not tolerate SNPs.24
Calculations
CT represents a logarithmic measure of the underlying target concentration. CT values of technical replicates were averaged to yield a single CT for each primer pair. In case of triplicates, if one and only one replicate yielded CT = 40, the average of the remaining 2 CTs was used. Each experiment was repeated an additional 2 times (biologically replicated). The mean CT values were normalized to the CT of U6 as reference to yield dCTU6. For DNA copy number determination and miRNA profiling, dCTU6 were standardized (Z-transformation25 ) to the median of each array. The standard deviation (SD) of U6 was less than 1 CT unit, evidencing that we can discern 2-fold changes in gene copy number (data not shown). Unsupervised clustering was conducted using ArrayMiner (Optimal Design, Brussels, Belgium) and a correlation metric. Clustering using self-organizing maps was conducted using cluster.26 Additional statistical tests were conducted using SPSS v11.0 (SPSS Science, Chicago, IL) and R version 2.5.0 (www.r-project.org/).
Results
Viral miRNA gene profile in PEL
All PELs are virally infected. They contain 2 classes of miRNAs: those encoded by cellular genes and those encoded by either KSHV or Epstein-Barr virus (EBV). Whereas nothing is known about the cellular miRNAs in PEL, the viral miRNAs have been intensely studied.17,18,23,27 This afforded us an internal positive control. First, we determined relative miRNA gene copy numbers (Figure 1B). Total DNA was isolated and subjected to real-time QPCR. All PELs encode the genes for the KSHV miRNAs,17,18 whereas only EBV coinfected PELs contain the genes for the EBV miRNAs (Figure 1B). Except for VG-1 cells, we detected all KSHV miRNA genome loci at equivalent levels in all PELs. The 95% confidence interval (95%CI) was 0.45 to 1.15 dCTU6 units (n = 5). dCTU6 represents the log2 difference between the U6 and the target miRNA gene copy number. One CT unit represents a 2-fold difference. This experiment established the limit of our assay based on combined technical and biologic variation. Using triplicates we were able to distinguish 5-fold, or twice the 95%CI, differences in gene copy number. The reason for the decreased signal for miR-K4 in VG-1 cells is due to a single nucleotide polymorphism in the gene encoding miR-K4 (A.J.O. and D.P.D., unpublished observation, July 2007). The EBV miRNA loci could be detected only in the 2 EBV genome–positive cell lines BC-1 and JSC-1. The relative abundance of the EBV miRNA genes was lower than those compared with the KSHV miRNA genes (Table S1), evidencing that on average, fewer EBV genomes than KSHV genomes are present in PEL.
Genomic profiling of KSHV and EBV miRNAs. (A) Outline of miRNA maturation and the species that can be detected using pre-miRNA–specific primers and SYBR-based real-time QPCR or miRNA-specific primers and TaqMan-based real-time QPCR. (B) Plot of relative copy number of viral miRNA gene loci. Variations in input DNA were adjusted using U6. This yielded dCTU6, which represents a logarithmic measure of relative gene copy number. Primers are indicated on the horizontal axis. Positive dCTU6 indicate lower copy number. Colors indicate different cell lines. The JSC-1 and BC-1 cell lines are EBV positive.
Genomic profiling of KSHV and EBV miRNAs. (A) Outline of miRNA maturation and the species that can be detected using pre-miRNA–specific primers and SYBR-based real-time QPCR or miRNA-specific primers and TaqMan-based real-time QPCR. (B) Plot of relative copy number of viral miRNA gene loci. Variations in input DNA were adjusted using U6. This yielded dCTU6, which represents a logarithmic measure of relative gene copy number. Primers are indicated on the horizontal axis. Positive dCTU6 indicate lower copy number. Colors indicate different cell lines. The JSC-1 and BC-1 cell lines are EBV positive.
Genomic miRNA gene profile in PEL
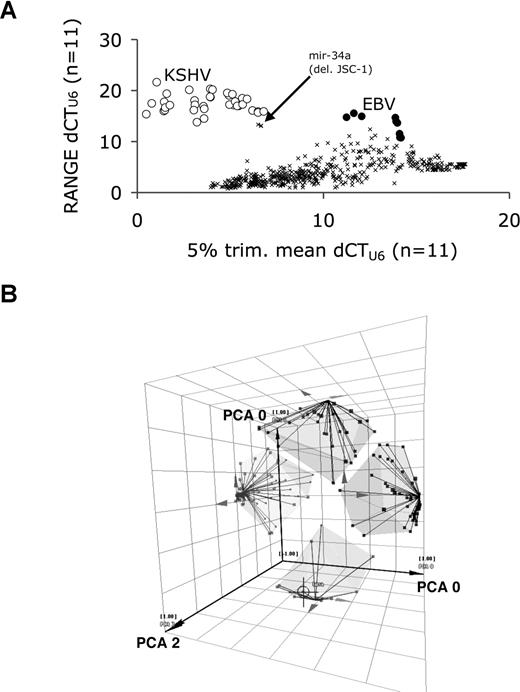
Next, we designed primers for all cellular pre-miRNAs and used these to determine the gene copy number of the cellular miRNA loci. We conducted comparative genomic profiling for 168 miRNA loci. DNA was isolated and adjusted for DNA concentration based on A260 absorption, and real-time QPCR was performed in triplicate (Figure 2A). Most miRNA genes showed little variation across cell lines with dCTU695%CI: 6.15 to 6.80. Genomic profiling of miRNAs in PELs is not trivial, since PELs exhibit severe aneuploidy28,29 and genome coverage based on 168 miRNAs loci was sparse. Nevertheless, we gained important insights. KSHV miRNA loci were easily detected since in PELs the KSHV genome is present in more than 20 copies in all but the virus-negative, non-PEL lymphoma cell lines. Their 5% trimmed mean dCTU6 values were lower that those of most cellular miRNAs (Figure 2A open circles), indicating that on average fewer PCR cycles were needed to yield a significant signal. Lower dCT values correspond to higher copy number. The trimmed mean is computed based on only 100 − 5 = 95% of all observations. It is less susceptible to outliers than a regular arithmetic mean. EBV miRNA gene loci likewise stood out, since they were absent in all but 3 EBV-positive cell lines (Figure 2A closed circles). A large range (dCTU6 > 8 units) in copy number variation represents miRNAs that were deleted in at least one PEL. For instance, miR-34a was deleted in JSC-1 cells (Figure 2A, arrow indicates triplicate measurements). Additional miRNA genes that had sustained deletions or amplifications were identified by pairwise scatter plots (Figure S1): hsa-miR-153-1 (Chr 2), 218-1 (Chr 4), 107 (Chr 10), 188 (Chr X), and 125a (Chr 19) were amplified in more than 50% of PELs. In sum, while miRNA genome loci typing clearly identified miRNA genes that were missing in individual cell lines, no one miRNA locus in our collection was consistently lost or amplified in all PELs. This suggests that the PEL-specific miRNA signature is derived primarily from transcriptional reprogramming rather than a specific mutation in a miRNA gene locus.
Comparative genomic profiling for cellular miRNA loci in PEL. (A) Variation in miRNA gene copy number. Plotted is the range of dCTU6 on the vertical and the 5% trimmed mean dCTU6 on the horizontal axis. The 5% trimmed mean is the arithmetic mean excluding the top and/or bottom 5% of the data. Crosses indicate the values for each cellular miRNA. KSHV miRNAs are shown by open circles; EBV miRNA gene loci, by closed circles; and hsa-miR-34a, by the arrow. Range indicates the difference between the lowest and highest value for each miRNA across all PEL cell lines. If a particular miRNA gene is present in every single cell line at 2 copies per cell, the range is 0 or close to 0, since it reflects only the measurement error. A high range indicates that one or more of the cell lines have more than 2 copies per cell (amplification) or have no copy per cell (deletion). This indicates that one or more cell lines in the sample have sustained amplifications or deletions. These were identified by analyzing the individual scatterplots in Figure S1. (B) Representation of individual clusters of pre-miRNAs as obtained after unsupervised clustering. The axes represent the first 3 principal components of the dataset. The objective of principal component analysis is to test whether all the data are correlated, or if indeed significant patterns or groups of data exist and how many. It serves as a quality control tool for our analysis. As can be seen here, 4, no more and no less, well-separated clusters emerged. These are further analyzed in Figure 3. Individual pre-miRNAs are represented by dots. Cluster membership is indicated by lines and semitransparent hulls.
Comparative genomic profiling for cellular miRNA loci in PEL. (A) Variation in miRNA gene copy number. Plotted is the range of dCTU6 on the vertical and the 5% trimmed mean dCTU6 on the horizontal axis. The 5% trimmed mean is the arithmetic mean excluding the top and/or bottom 5% of the data. Crosses indicate the values for each cellular miRNA. KSHV miRNAs are shown by open circles; EBV miRNA gene loci, by closed circles; and hsa-miR-34a, by the arrow. Range indicates the difference between the lowest and highest value for each miRNA across all PEL cell lines. If a particular miRNA gene is present in every single cell line at 2 copies per cell, the range is 0 or close to 0, since it reflects only the measurement error. A high range indicates that one or more of the cell lines have more than 2 copies per cell (amplification) or have no copy per cell (deletion). This indicates that one or more cell lines in the sample have sustained amplifications or deletions. These were identified by analyzing the individual scatterplots in Figure S1. (B) Representation of individual clusters of pre-miRNAs as obtained after unsupervised clustering. The axes represent the first 3 principal components of the dataset. The objective of principal component analysis is to test whether all the data are correlated, or if indeed significant patterns or groups of data exist and how many. It serves as a quality control tool for our analysis. As can be seen here, 4, no more and no less, well-separated clusters emerged. These are further analyzed in Figure 3. Individual pre-miRNAs are represented by dots. Cluster membership is indicated by lines and semitransparent hulls.
Pre-miRNA profile of PEL
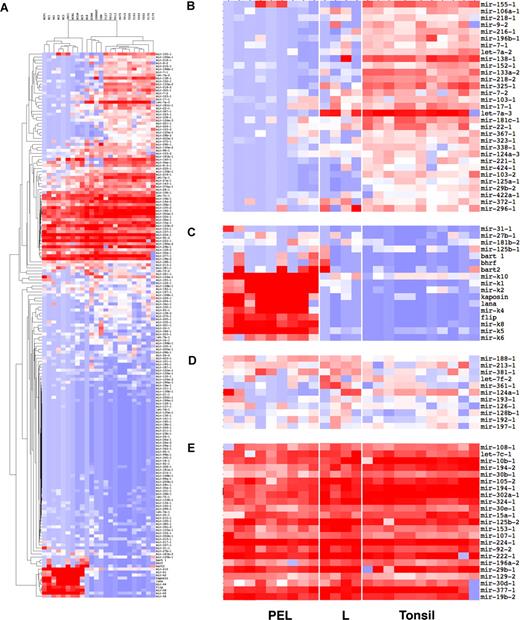
We used our array to identify pre-miRNAs that were transcribed in PEL. Total RNA was isolated, DNAse I–treated, reverse transcribed, and subjected to real-time QPCR (see Figure S2 for detailed experimental data for a representative set of miRNAs). Overall the KSHV miRNAs were present at much higher levels than the cellular miRNAs (see also Figure S4), probably due to the high genome copy number (20–30 copies) of KSHV in PEL.30 In contrast, in EBV+ PEL, the EBV miRNAs were present at approximately equal level of cellular miRNAs. To gain a complete picture, we performed unsupervised clustering. This method of analysis orders miRNAs based solely upon their relative levels of expression without making any a priori assumptions. This is in contrast to classifications schemes that incorporate information upfront, for example, about the tumor cell lineage (PEL or not PEL) and then ask whether a given miRNA is overrepresented in one or the other group. Four distinct clusters of pre-miRNAs were evident (Figure 2B). A detailed picture can be gleaned from the heatmap representation (Figure 3). Here the relative levels of all miRNAs in the array are preserved, such that miRNAs that were not detectable are coded in blue and the most highly abundant miRNAs in the entire array are coded in red. The KSHV miRNAs were abundant in PEL and absent in tonsil or the virus-negative lymphoma lines BJAB, DG75, BL-8, Jurkat (T cell), and CEM (T cell) (Figure 3C).
Pre-miRNA profile of PEL. (A) Unsupervised cluster analysis of standardized dCTU6 for pre-miRNAs using a correlation metric. Standardization was achieved by calculating Z scores across each individual array of dCTU6. This technique eliminates variation in assay performance between individual samples. Red indicates higher abundance; blue, lower than median abundance; white, the median. (B,C) Enlarged view. (B) Cluster of pre-miRNAs with decreased abundance in PEL vis-à-vis tonsil. (C) Cluster of pre-miRNAs with increased abundance in PEL vis-à-vis tonsil. (D) Cluster of pre-miRNAs equal abundance in PEL vis-à-vis tonsil. (E) Cluster of pre-miRNAs with high abundance in PEL as well as in nonvirus-associated lymphoma lines. The cluster of absent pre-miRNAs is not shown separately.
Pre-miRNA profile of PEL. (A) Unsupervised cluster analysis of standardized dCTU6 for pre-miRNAs using a correlation metric. Standardization was achieved by calculating Z scores across each individual array of dCTU6. This technique eliminates variation in assay performance between individual samples. Red indicates higher abundance; blue, lower than median abundance; white, the median. (B,C) Enlarged view. (B) Cluster of pre-miRNAs with decreased abundance in PEL vis-à-vis tonsil. (C) Cluster of pre-miRNAs with increased abundance in PEL vis-à-vis tonsil. (D) Cluster of pre-miRNAs equal abundance in PEL vis-à-vis tonsil. (E) Cluster of pre-miRNAs with high abundance in PEL as well as in nonvirus-associated lymphoma lines. The cluster of absent pre-miRNAs is not shown separately.
Hsa-miR-125b, -181b-2, -27b-1, and -31–1 clustered with the KSHV miRNAs, although their degree of overexpression in PEL was lower than for the KSHV pre-miRNAs (Figure 3C). The mature hsa-miR-125b, -181b, and -27b miRNAs could also be detected by TaqMan QPCR (below). A second cluster of pre-miRNAs was uniformly expressed in B cell–rich tonsil as well as PEL (Figure 3E). For many of those abundant pre-miRNAs, we were also able to detect the corresponding mature miRNAs (below). Many of those were lymphoid lineage–specific miRNAs, suggesting that the PEL tumor miRNA profile reflects the tissue of origin.
A third cluster represents pre-miRNAs that were down-regulated in PEL, as well as the virus-negative B-cell lymphomas compared with tonsil tissue (Figure 3B). This included mir-155, the recently identified cellular ortholog to KSHV mir-K12-11,31 as well as let-7a-2 and let-7a3 among others. Furthermore, we identified pre-miRNAs that were uniquely up-regulated in individual cell lines. As expected, the EBV pre-miRNAs were detected only in the EBV-positive BC1, BC5, and JSC-1 PEL cell lines (Figure 3C).
miRNA profile of PEL
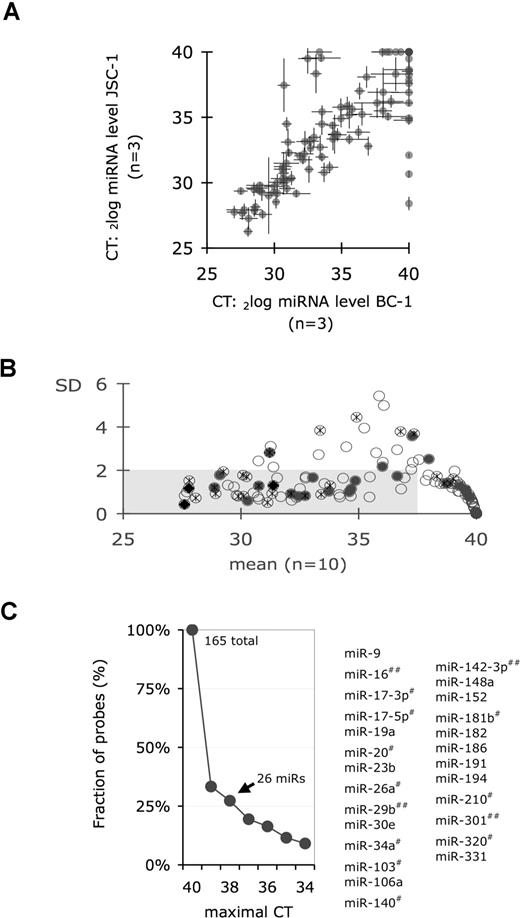
We used a TaqMan-based miRNA array to profile mature miRNAs in PEL. This assay format yielded excellent reproducibility (Figure 4A). We defined the PEL signature as all miRNAs that (a) were detectable in fewer than 38 cycles and (b) exhibited limited variation, as evidenced by a standard deviation (SD) of less than 4-fold, across multiple PEL cell lines (Figure 4B shaded box). This yielded 68 miRNAs, which are listed in Table S2. Alternatively, a stricter definition required that all signature miRNAs were present in every PEL cell line tested. This yielded 26 miRNAs (Figure 4C), namely hsa-miR-9, -16, -17–3p, -17–5p, -19a, -20, -23b, -26a, -29b, -30e, -34a, -103, -106a, -140, -142–3p, -148a, -152, -181b, -182, -186, -191, -194, -210, -301, -320, and -331.
Mature miRNA profiles of PEL. (A) Scatterplot analysis of 2 cell lines JSC-1 (vertical axis) and BC-1 (horizontal axis). Plotted are mean CT values of technical triplicates. Lines indicate the SD. Dots correspond to individual miRNAs. CT = 40 indicates absence of a miRNA signal. (B) Variation of mature miRNA levels in PEL. Plotted is the SD across multiple cell lines on the vertical axis against the mean CT on the horizontal axis (n = 10). Open circles indicate mature miRNAs as detected by TaqMan assay. Dark circles indicate that the corresponding pre-miRNA was also overexpressed. Crossed circles indicate miRNAs that were cloned from PEL (data from Cai et al17 and Samols et al18 ). Shaded area indicates the cutoff used to derive the PEL miRNA signature shown in Table S2. (C) Fraction of total (165) probes on the vertical axis, for which every PEL sample yielded a CT lower than indicated on the horizontal axis. All miRNAs with CT of 38 or less (ie, those that were uniformly detectable in all PEL lines) are tabulated on the right. The # indicates miRNAs that were also cloned once; ##, miRNAs that were cloned independently by 2 groups.17,18
Mature miRNA profiles of PEL. (A) Scatterplot analysis of 2 cell lines JSC-1 (vertical axis) and BC-1 (horizontal axis). Plotted are mean CT values of technical triplicates. Lines indicate the SD. Dots correspond to individual miRNAs. CT = 40 indicates absence of a miRNA signal. (B) Variation of mature miRNA levels in PEL. Plotted is the SD across multiple cell lines on the vertical axis against the mean CT on the horizontal axis (n = 10). Open circles indicate mature miRNAs as detected by TaqMan assay. Dark circles indicate that the corresponding pre-miRNA was also overexpressed. Crossed circles indicate miRNAs that were cloned from PEL (data from Cai et al17 and Samols et al18 ). Shaded area indicates the cutoff used to derive the PEL miRNA signature shown in Table S2. (C) Fraction of total (165) probes on the vertical axis, for which every PEL sample yielded a CT lower than indicated on the horizontal axis. All miRNAs with CT of 38 or less (ie, those that were uniformly detectable in all PEL lines) are tabulated on the right. The # indicates miRNAs that were also cloned once; ##, miRNAs that were cloned independently by 2 groups.17,18
One hundred eight (61%) of the mature miRNA arrays were also represented in our pre-miRNA array. This allowed us to establish a correspondence between pre-miRNA and miRNA levels. Real-time QPCR for mature miRNA detected 11 (10%) miRNAs, which were not detected at the pre-miRNA level, and pre-miRNA profiling detected 45 (42%) of pre-miRNAs for which we could not detect the corresponding miRNA. The majority of miRNAs were detectable as mature as well as pre-miRNAs. Hence, consensus information as well as nonredundant information can be gained from combining mature miRNA and pre-miRNA profiling.
MiRNA cloning previously reported a few cellular miRNAs for the BCBL-1 and BC-1 cell lines.17,18 As these experiments were designed to identify KSHV viral miRNAs, the cloning approach was not saturating. It preferentially identified abundant cellular miRNAs such as hsa-miR-16 (Figure 4C, miRNAs marked with an * or **). Cloning also identified some miRNAs for which we could detect the pre-miRNA, but not the mature miRNA by real-time QPCR (see Table S2 for a summary of all data). Importantly, half of the miRNAs uncovered herein by real-time QPCR were novel and not identified in prior cloning attempts.
Discussion
MiRNA profiling has emerged as a powerful new approach to stratify human cancer and to identify novel pathogenesis determinants (reviewed in Calin and Croce1 ). Some miRNAs are considered bona fide oncogenes based upon their strong transforming properties in animal models.32 To date, cellular miRNAs have not been profiled in PEL. Here we report the miRNA profile of PEL. Multiple layers of regulation control miRNA abundance, and our study for the first time combined querying them all. First, we evaluated miRNA gene locus deletions, amplifications, and mutations using DNA QPCR. This identified cell line–specific deletions of individual miRNA loci; however, no miRNA loci were lost in all PELs. Hsa-miR-218–1, -107, -153–1, -188, and -125a were amplified in multiple PELs and await confirmation by genotyping.
The nascent transcript (pri-miRNA) is regulated at the level of promoter activity. Pri-miRNAs are processed into pre-miRNAs depending on Drosha levels and specificity (Figure 1A). Upon nuclear export, Dicer generates mature miRNAs. We found a high concordance between pre-miRNAs and mature miRNAs. Our results are in agreement with those of Schmittgen and colleagues (Jiang et al33 and Schmittgen et al34 ) who likewise profiled pre-miRNA levels and found them to be good predictors of mature miRNA abundance. This suggests that transcriptional control of miRNAs plays an important role in miRNA regulation.
There exists an additional justification to profile pre-miRNAs, because mature miRNAs of identical sequence can be derived from different pre-miRNAs, each located at a different genomic location and each under control of a different promoter. Thus, profiling pre-miRNA levels provides a stepping stone toward the identification of miRNA regulatory elements.
The real-time QPCR assays for miRNAs and pre-miRNAs revealed a remarkable robustness and sensitivity. On average replicates showed less than 3-fold variation over a linear range of 5 orders of magnitude. This outperformed hybridization or radio-activity-based assays, for which we were not able to establish statistically significant differences of less than 10-fold (data not shown). Equally remarkable was the robustness of the miRNA profile between PEL cell lines. Even though each individual PEL had some pre-miRNAs and miRNAs (≤ 5%) that were unique to a particular cell line, the signature miRNAs were highly up-regulated in most PELs to almost the exact level (SD ≤ 4-fold). This implies that the PEL signature miRNAs are important for and unique to this class of lymphomas.
We feel that integrating multiple approaches of miRNA profiling yielded an accurate picture, as each method is subject to technical limitations and necessarily subjective cutoff limits. Figure 4C shows the minimal set of PEL-specific miRNAs. We could detect each of these miRNAs in every PEL cell line tested. Table S2 shows a slightly larger set allowing for some variation within PELs. As our miRNA profiling included cell lines that were previously analyzed by miRNA cloning approaches, we could establish a concordance between profiling by cloning and profiling by real-time QPCR arrays. Real-time QPCR-based profiling identified many more miRNAs in PEL than cloning, though that may simply be a function of the number of clones sequenced for the particular cell line.35,36
Several miRNAs were identified by multiple methods in PEL cell lines. They fit the biology of PEL. Hsa-miR-181b is B-lymphoid specific and can drive B lymphopoiesis upon ectopic expression in bone marrow progenitor cells.37 Hsa-miR-181 and hsa-miR29b are markers for B-cell chronic lymphocytic leukemia (B-CLL), although they are down-regulated in the most aggressive forms. Recently it was shown that hsa-miR-181 and hsa-miR29b target the mRNA for TCL-1.38 Hsa-miR-15a was easily detectable in PELs, whereas it is commonly down-regulated in B-CLL. In B-CLL, hsa-miR-15a expression was inversely correlated with its target, BCL-2, mRNA levels.4,39 Since KSHV encodes a viral BCL-2 homolog,40 there is no selection pressure to induce human BCL-2. As a result, BCL-2 mRNA is low in PELs by comparison with other DLBCLs (Figure S3). Hsa-miR-34a has been ascribed tumor-suppressor activity in neuroblastomas by targeting E2F3 mRNA,41 but no data have been reported with regard to human lymphoma. Hsa-miR-222 was shown to target c-kit in endothelial cells and other cancers,42,43 and c-kit is induced in KS and KSHV-infected endothelial cells. However, c-kit is neither significantly up- nor down-regulated in PEL,16 suggesting that in lymphoid cancer, hsa-miR-222 has other targets. Interestingly, hsa-miR-222/221 are encoded in close proximity to each other on Xp11.3; but while hsa-mir-222 was easily detectable by pre-miRNA (see Figure S3C for details) and mature miRNA profiling and cloning, hsa-mir-221 was not detected by either of these methods, demonstrating that these 2 adjacent miRNAs are differentially regulated.
Hsa-miR-155 has been studied extensively in the context of human lymphoma, but was underrepresented in PELs. This corroborates the recent observation that Hsa-miR-155 is an ortholog of KSHV miR-K12-1131 and inversely correlated with KSHV miRNA levels (although BC2, BC3, and JSC1 were exceptions). The miR-155 gene locus is embedded within exon 3 of the BIC mRNA, which was initially identified as a viral integration site in avian lymphoma. BIC and miR-155 are highly expressed in HD, DLBCL, and other lymphomas, but interestingly not in BL.44 While BIC mRNA was induced by phorbol ester in EBV-Ramos BL cells, neither the mature hsa-miR-155 miRNA nor the hsa-miR-155 pre-miRNA was detectable.45 This provides another example of discordant regulation between the nascent pri-RNAs comprising both protein and miRNA coding regions, and pre-miRNA/miRNA levels. It supports our contention that combined pre-miRNA and miRNA profiling provides novel information for the classification of human tumors that is independent of messenger RNA profiling.
In summary, PELs express B-cell lineage– and B-cell lymphoma–specific miRNAs, despite the absence of most B-cell CD surface antigens. This study thus contributes to the definition of common and differential miRNA markers for human B-cell lymphoma. We show that pre-miRNA profiling yields corroborative as well as novel, nonredundant information to mature miRNA profiling, which can be used for the stratification of human cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
Gottwein et al46 also recently reported on the inverse correlation between KSHV infection and hsa-mir155 levels and common targets for the cellular and viral orthologs
Acknowledgments
We thank B. Damania for critical reading. We thank especially Drs B. Cullen and R. Renne for communicating unpublished observations and the PEL miRNAs that they identified by cDNA cloning.
This work was supported by National Institues of Health grants CA109232 and DE018304; the AIDS-Associated Malignancies Clinical Trials Consortium (CA121947); the University of North Carolina (UNC) Center for AIDS Research (CfAR); and a translational award from the Leukemia and Lymphoma Society. A.J.O. was supported by NIH training grant T32 AI00741.
National Institutes of Health
Authorship
Contribution: A.J.O. performed research, analyzed data, and contributed to writing the paper; W.V. designed the array; and D.P.D. analyzed data and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dirk P. Dittmer, Department of Microbiology and Immunology, University of North Carolina at Chapel Hill, CB# 7290, 804 Mary Ellen Jones Bldg, Chapel Hill, NC 27599-7290; e-mail: ddittmer@med.unc.edu.