Hodgkin and Reed-Sternberg (HRS) cells in Hodgkin lymphoma (HL) secrete factors that interact with inflammatory background cells and may serve as biomarkers for disease activity. To detect new proteins related to pathogenesis, we analyzed the secretome of HRS cells. Proteins in cell culture supernatant of 4 HL cell lines were identified using 1DGE followed by in-gel trypsin digestion and LC-MS/MS. In total, 1290 proteins, including 368 secreted proteins, were identified. Functional grouping of secreted proteins revealed 37 proteins involved in immune response. Sixteen of the 37 proteins (ie, ALCAM, Cathepsin C, Cathepsin S, CD100, CD150, CD26, CD44, CD63, CD71, Fractal-kine, IL1R2, IL25, IP-10, MIF, RANTES, and TARC) were validated in HL cell lines and patient material using immunohistochemistry and/or ELISA. Expression of all 16 proteins was confirmed in HL cell lines, and 15 were also confirmed in HL tissues. Seven proteins (ALCAM, cathepsin S, CD26, CD44, IL1R2, MIF, and TARC) revealed significantly elevated levels in patient plasma compared with healthy controls. Proteomics analyses of HL cell line supernatant allowed detection of new secreted proteins, which may add to our insights in the interaction between HRS cells and infiltrating lymphocytes and in some instances might serve as biomarkers.
Introduction
Hodgkin lymphoma (HL) is characterized by a minority of neoplastic cells, the Hodgkin and Reed-Sternberg cells (HRS cells), and an extensive inflammatory background. Many studies have documented that HL is associated with disturbed cytokine production.1 Cytokine and chemokine production may not only promote growth of HRS cells and help to evade immune surveillance, but also cause the characteristic histology and the clinical symptoms of HL.2 Cross-talk between HRS cells and surrounding lymphocytes has been studied for many years, and this interaction is regarded to be important for the pathogenesis of HL.
The application of proteomics techniques has been proven to be a powerful tool for the identification of new biomarkers involved in pathogenesis of many diseases (reviewed by Hanash3 ). Some studies on hematologic malignancies using proteomics approaches have been reported,4,5 but application of proteomics technology to HL is still limited. Fujii et al compared HL cell line proteome to anaplastic large cell lymphoma (ALCL) and non-Hodgkin lymphoma (NHL) using a 2-dimensional difference gel electrophoresis approach of total cell lysates.6,7 HL expressed higher levels of pyridoxine-5′-phosphate oxidase, vinculin, dihydropyrimidinase-related protein-2 and NADH-ubiquinone oxidoreductase compared with ALCL. In comparison to NHL cell lines, HL cell lines showed higher expression of pyridoxine-5′-phosphate oxidase, γ-enolase, vinculin, vimentin, galectin-1, annexin A5, and protein kinase C substrate.7 Carvalho et al identified changes in the spectrum using quadrupole–time-of-flight hybrid mass spectrometer in serum of HL patients compared with normal controls, but no further protein identification was performed.8 Overall, only very limited proteomics data are available for HL.
To gain more insight into the proteins secreted by HRS cells that might be involved in the cross-talk with infiltrating lymphocytes or might serve as tumor biomarkers, the secretome of HL cell lines was determined in cell culture supernatant. Protein identification was performed by nanoscale reversed-phase liquid chromatography tandem mass spectrometry (LC-MS/MS) after 1D-SDS-PAGE fractionation. Proteins related to immune response were validated in cell culture supernatant and patient plasma by enzyme-linked immunosorbent assay (ELISA) and in tumor tissues by immunohistochemistry.
Methods
Cell lines and culture
The HL cell lines L1236 and KMH2 were established from HL patients with mixed cellularity subtype, L428 from nodular sclerosis subtype and DEV from nodular lymphocyte predominant HL (NLPHL) subtype. They were cultured in RPMI 1640 medium (Lonza Walkersville, Walkersville, MD) supplemented with fetal calf serum (Lonza Walkersville). For analysis of secreted proteins, to avoid the masking effects of high-abundance serum proteins in the culture medium, the cells were washed 3 times with RPMI 1640 medium to deplete all serum proteins, and cultured in RPMI 1640 medium without serum for 24 hours. Under these conditions, cell viability was high and cells were proliferating. Furthermore, removal of serum did not block the secretion of chemokines CCL17 (TARC) and CCL22 (MDC), known to be produced by HRS cells. Supernatant was harvested by filtration (0.22-μm filter). As a comparison, samples were also collected from the cell lines under normal culture conditions with the same concentration of cells.
Patient and control materials
Plasma samples were obtained from 28 cHL patients (age, 10–64 years; median age, 34 years) before treatment. Plasma of 11 age-matched healthy individuals was taken as a control (age, 23–44 years; median, 28 years). Formalin-fixed, paraffin-embedded or frozen tissue specimens of 11 cases of classic HL (cHL) and 3 cases of NLPHL were retrieved from the tissue bank of the Pathology Department. All diagnoses were established according to the guidelines of the World Health Organisation. The protocol was approved by the medical ethics board of the University Medical Center Groningen. Informed consent was obtained in accordance with the Declaration of Helsinki from all participants.
Protein identification
A total of 30 mL HL cell line culture supernatant of cells cultured in RPMI without serum was concentrated 100-fold by ultrafiltration (Vivaspin 20, 3-kDa cut-off; Vivascience, Stonehouse, United Kingdom) and stored at −80°C until further processing. Protein concentration was measured by Bradford assay (Bio-Rad, Hemel Hempstead, United Kingdom). Proteins in concentrated samples were fractionated by SDS-PAGE on a 4% to 12% Bis-Tris gel with an MES buffer system according to the manufacturer protocol (NuPAGE-Novex, Invitrogen, Carlsbad, CA). Visualization of bands was performed overnight by Coomassie Brilliant Blue G-250 based staining (PageBlue Staining Solution, MBI Fermentas, Glen Burnie, MD). The whole lane was excised and cut into 30 to 35 small pieces. Each piece was washed in ultra pure water and dehydrated in acetonitrile (ACN) to remove residual SDS and Coomassie Blue. In-gel reduction was performed with dithiothreitol (10 mM) for 1 hour at 60°C and carbamidomethylation with iodoacetamide (55 mM) for 45 minutes at room temperature in the dark. Gel pieces were subsequently washed with ultra pure water, 50% ACN, and pure ACN. Next, 0.1 μg of trypsin in 50 mM ammonium bicarbonate was added and gel pieces were allowed to rehydrate on ice for 20 minutes. Protein digestion was carried out overnight at 37°C. Separation of the peptide mixtures was performed by LC-MS/MS as illustrated by Alvarez-Llamas et al.9 ProID 1.1 software10 (Applied Biosystems, Foster City, CA) was used to identify proteins from the mass spectrometric datasets according to Swiss-Prot database.11
Identification of secreted proteins and functional classification
Classification of identified proteins in terms of secretion pathways was performed according to SecretomeP 2.0 Server.12 The proteins with a predicted signal peptide were considered as secreted proteins via a classic pathway (endoplasmic reticulum/Golgi-dependent pathway). If no signal peptide was predicted but the NN-score exceeded a value of 0.5, proteins were considered as secreted via nonclassic pathways. Secreted proteins were classified into groups with similar functions according to biologic function gene ontology13 using Expression Analysis Systematic Explorer.14
Immunohistochemistry
Immunohistochemistry was performed with antibodies against ALCAM (1:5, monoclonal mouse, Vision Biosystems, Wetzlar, Germany), cathepsin C (1:50, polyclonal goat, R&D Systems, Minneapolis, MN), cathepsin S (1:50, polyclonal rabbit, ATLAS Antibodies, Stockholm, Sweden), CD44 (1:20, monoclonal mouse, BD Biosciences PharMingen, San Diego, CA), CD63 (1:20, monoclonal mouse, Chemicon International, Temecula, CA), Fractalkine (1:200, polyclonal rabbit, Proteintech Group, Chicago, IL), IL25 (1:50, polyclonal rabbit, Proteintech Group), IP-10 (1:200, polyclonal rabbit, PeproTech, Rocky Hill, NJ), MIF (1:100, polyclonal rabbit, ATLAS Antibodies), RANTES (1:200, polyclonal goat, Santa Cruz Biotechnologies, Santa Cruz, CA), TARC (1:100, polyclonal goat, R&D Systems) on paraffin-embedded tissues after antigen retrieval. Positive staining was visualized using a HRP-labeled second step and 3,3′-diaminobenzidine (Sigma-Aldrich, St Louis, MO). Staining of CD100 (1:5, monoclonal mouse, Beckman Coulter, Fullerton, CA), IL1R2 (1:20, monoclonal mouse, BioLegend, San Diego, CA), CD150 (1:10, monoclonal mouse, BD Biosciences PharMingen), CD26 (undiluted, monoclonal mouse), and CD71 (1:1000, monoclonal mouse)15 was performed on frozen tissues. Positive staining was visualized using a HRP-labeled second step and 3-amino-9-ethylcarbazole (Sigma-Aldrich). Immunohistochemistry on centrifuge preparations of the 4 cell lines were performed for the 6 proteins (cathepsin C, CD100, CD150, CD63, CD71, and IL25), for which no ELISA kits were available.
ELISA on HL cell line culture supernatant and patient plasma samples
ALCAM, cathepsin S, CD26, CD44 (Bender Medsystems, Vienna, Austria), Fractalkine, IL1R2, IP-10, MIF, RANTES, and TARC protein levels were measured in cell culture supernatant from 4 HL cells lines, plasma of 28 patients and 11 controls by ELISA (R&D Systems).
Statistics
The data were compiled with the software package SPSS, version 12.0.2. Results of the HL patients were compared with the controls to determine a possible significant increase or decrease in plasma protein levels. Differences in plasma protein levels between patients and controls were explored using t test if the raw data or log transformed data (ln) were normally distributed. Otherwise the nonparametric analysis (Mann-Whitney U test) was performed using the raw data (P < .05, statistically significant).
Results
Protein identification
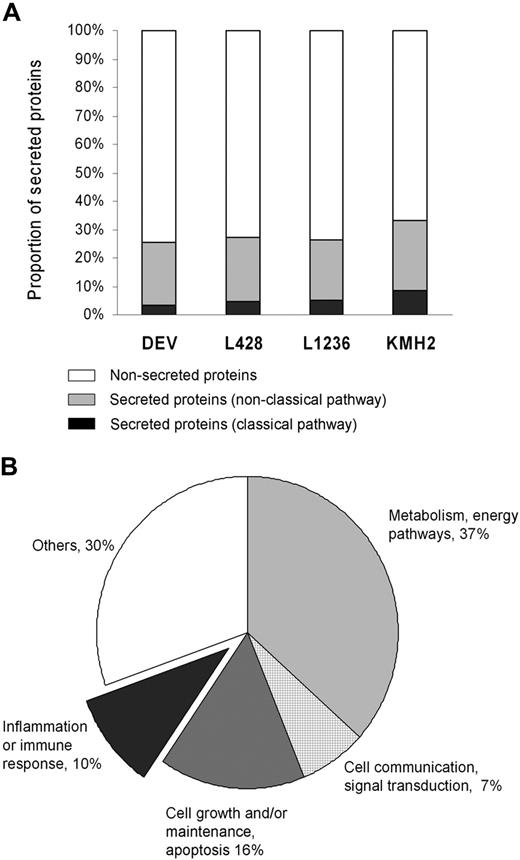
Combination of all data from LC-MS/MS revealed a total of 1290 proteins that were identified with more than or equal to 95% confidence in the 4 HL cell lines. A total of 85 proteins were secreted following a classic pathway based on the presence of a signal peptide as predicted by SecretomeP. Another 283 proteins were indicated to be secreted following nonclassic pathways. A list of all 368 secreted proteins (both classic and nonclassic pathways) can be found in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article), and a list of nonsecreted proteins can be found in Table S2. In general, 26% to 33% of the proteins were predicted to be secreted among 4 cell lines (Figure 1A). The overlap between the secreted proteins in the 3 cHL cell lines (L428, L1236, and KMH2) was 74 proteins and in all 4 cell lines (ie, including the NLPHL cell line DEV) was 50 proteins. Secreted proteins were classified into 5 categories according to function (Figure 1B). The criteria for classification are not always uniform because many proteins have more than one function, which may place them in more than one functional category. To prevent overlap between the function groups, proteins were listed under only one category. A total of 121 proteins (33%) were grouped in one of the 3 main categories that may be involved in the interaction between tumor cells and the microenvironment (ie, inflammation or immune response, cell growth/maintenance or apoptosis, and cell communication/signal transduction).
Protein identification by LC-MS/MS. (A) Proportion of secreted proteins (classic pathway, nonclassic pathway) and nonsecreted proteins in total identified proteins for each Hodgkin lymphoma (HL) cell line. (B) Classification of 368 secreted proteins by biologic function gene ontology13 using the software Expression Analysis Systematic Explorer.14
Protein identification by LC-MS/MS. (A) Proportion of secreted proteins (classic pathway, nonclassic pathway) and nonsecreted proteins in total identified proteins for each Hodgkin lymphoma (HL) cell line. (B) Classification of 368 secreted proteins by biologic function gene ontology13 using the software Expression Analysis Systematic Explorer.14
Protein expression in HL cell lines
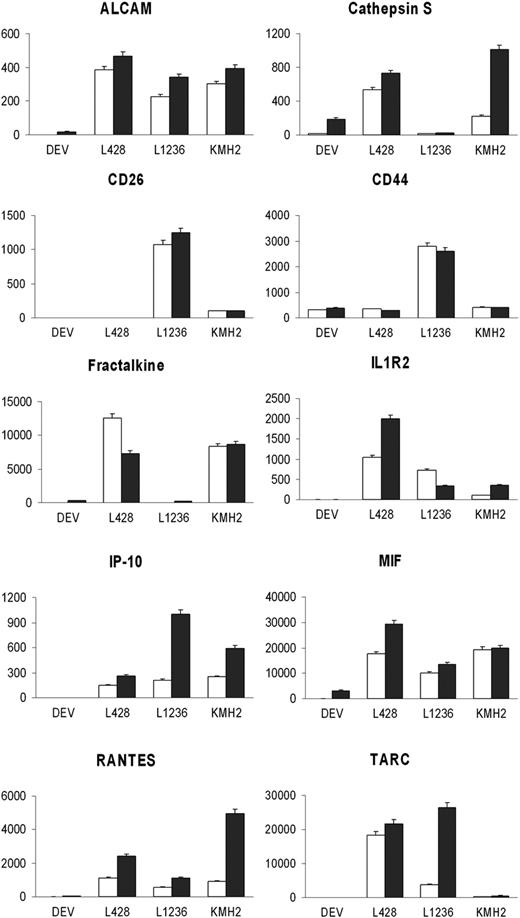
The function group of “Inflammation or immune response” included 37 secreted proteins (Table 1). Based on possible relevance for HL and availability of antibodies, 16 proteins, including all cytokines, CD molecules, and cathepsin family members in this group, were selected for validation in cell lines and HL patient materials. For 10 proteins, ELISA kits were available and secretion could be confirmed in the HL cell line supernatant in which the protein was originally identified and also in supernatant of (part of) the other HL cell lines. Higher protein levels were observed in supernatant of the cells cultured with serum than without serum (Figure 2). CD44 and MIF were present in culture supernatant of all 4 cell lines. ALCAM, IL1R2, IP-10, and RANTES were detected in the supernatant of 3 cHL cell lines (L428, L1236, and KMH2), whereas in DEV these proteins were not present or only at very low levels. Cathepsin S and Fractalkine were present at low levels in both DEV and L1236. CD26 was only present in L1236 and KMH2 supernatants. TARC was secreted at high levels by L428 and L1236, at low level by KMH2, and not by NLPHL cell line DEV.
Secreted proteins in function group of “inflammation or immune response”
| Gene symbol . | Protein name . | Alternative name . | Source† . |
|---|---|---|---|
| ALCAM | Activated leukocyte cell adhesion molecule* | CD166 | 3 |
| ARTS-1 | Adipocyte-derived leucine aminopeptidase precursor* | 3,4 | |
| B2M | Beta-2-microglobulin precursor* | 3,4 | |
| C19ORF10 | Protein C19orf10 precursor (stromal cell-derived growth factor SF20)* | IL25 | 1,2,3,4 |
| C1QBP | Complement component 1 Q subcomponent-binding protein, mitochondrial precursor* | 1,2,4 | |
| C4A | Complement component 4A* | 2 | |
| CCL17 | Small inducible cytokine A17 precursor (thymus and activation-regulated chemokine)* | TARC | 2,3 |
| CCL5 | Small inducible cytokine A5 precursor (T cell-specific RANTES protein)* | RANTES | 2 |
| CD44 | CD44 antigen precursor * | 3 | |
| CD63 | CD63 antigen* | 4 | |
| CORO1A | Coronin, actin binding protein, 1A | 1,2,3 | |
| CST3 | Cystatin C precursor* | 4 | |
| CTSC | Dipeptidyl-peptidase 1 precursor* | DPP-1/Cathepsin C | 2 |
| CTSS | Cathepsin S precursor* | Cathepsin S | 4 |
| CX3CL1 | Fractalkine precursor (CX3C membrane-anchored chemokine)* | Fractalkine | 2 |
| CXCL10 | Small inducible cytokine B10 precursor* | IP-10 | 3 |
| DPP4 | Dipeptidyl peptidase 4 (T-cell activation antigen)* | CD26 | 3 |
| G1P2 | Ubiquitin cross-reactive protein precursor (interferon-induced 17-kDa protein) | 1,3,4 | |
| HLA-B | HLA class I histocompatibility antigen, B-51 alpha chain precursor* | 3,4 | |
| HLA-DRA | HLA class II histocompatibility antigen, DR alpha chain precursor* | 2,3 | |
| IFITM1 | Interferon-induced transmembrane protein 1 | 3 | |
| IL1R2 | Interleukin-1 receptor, type II precursor* | CDw121b/IL-1R-beta | 3 |
| LGALS3 | Galectin-3 | 2,1 | |
| MIF | Macrophage migration inhibitory factor | 1,2,3,4 | |
| NCF4 | Neutrophil cytosol factor 4 | 3 | |
| PRDX5 | Peroxiredoxin 5, mitochondrial precursor | 1,2,3,4 | |
| PRG1 | Secretory granule proteoglycan core protein precursor* | Serglycin | 2,4 |
| PSMB8 | Proteasome subunit beta type 8 precursor | 1,2,3 | |
| PSMB9 | Proteasome subunit beta type 9 precursor | 1,2,3 | |
| RAC2 | Ras-related C3 botulinum toxin substrate 2 precursor | p21-Rac2 | 1,2,3 |
| SCYE1 | Multisynthetase complex auxiliary component p43 | 3 | |
| SH2D1A | Stromal cell-derived factor 2-like protein 1 precursor* | SDF2-like protein 1 | 4 |
| SEMA4D | Semaphorin 4D precursor (leukocyte activation antigen)* | CD100 | 4 |
| SERPINA1 | Alpha-1-antiproteinase precursor* | Alpha-1-antitrypsin | 4 |
| SH2D1A | SH2 domain protein 1A (signaling lymphocyte activation molecule-associated protein) | 3 | |
| SLAMF1 | Signaling lymphocytic activation molecule precursor | CD150 | 4 |
| TFRC | Transferrin receptor protein 1* | CD71 | 1 |
| Gene symbol . | Protein name . | Alternative name . | Source† . |
|---|---|---|---|
| ALCAM | Activated leukocyte cell adhesion molecule* | CD166 | 3 |
| ARTS-1 | Adipocyte-derived leucine aminopeptidase precursor* | 3,4 | |
| B2M | Beta-2-microglobulin precursor* | 3,4 | |
| C19ORF10 | Protein C19orf10 precursor (stromal cell-derived growth factor SF20)* | IL25 | 1,2,3,4 |
| C1QBP | Complement component 1 Q subcomponent-binding protein, mitochondrial precursor* | 1,2,4 | |
| C4A | Complement component 4A* | 2 | |
| CCL17 | Small inducible cytokine A17 precursor (thymus and activation-regulated chemokine)* | TARC | 2,3 |
| CCL5 | Small inducible cytokine A5 precursor (T cell-specific RANTES protein)* | RANTES | 2 |
| CD44 | CD44 antigen precursor * | 3 | |
| CD63 | CD63 antigen* | 4 | |
| CORO1A | Coronin, actin binding protein, 1A | 1,2,3 | |
| CST3 | Cystatin C precursor* | 4 | |
| CTSC | Dipeptidyl-peptidase 1 precursor* | DPP-1/Cathepsin C | 2 |
| CTSS | Cathepsin S precursor* | Cathepsin S | 4 |
| CX3CL1 | Fractalkine precursor (CX3C membrane-anchored chemokine)* | Fractalkine | 2 |
| CXCL10 | Small inducible cytokine B10 precursor* | IP-10 | 3 |
| DPP4 | Dipeptidyl peptidase 4 (T-cell activation antigen)* | CD26 | 3 |
| G1P2 | Ubiquitin cross-reactive protein precursor (interferon-induced 17-kDa protein) | 1,3,4 | |
| HLA-B | HLA class I histocompatibility antigen, B-51 alpha chain precursor* | 3,4 | |
| HLA-DRA | HLA class II histocompatibility antigen, DR alpha chain precursor* | 2,3 | |
| IFITM1 | Interferon-induced transmembrane protein 1 | 3 | |
| IL1R2 | Interleukin-1 receptor, type II precursor* | CDw121b/IL-1R-beta | 3 |
| LGALS3 | Galectin-3 | 2,1 | |
| MIF | Macrophage migration inhibitory factor | 1,2,3,4 | |
| NCF4 | Neutrophil cytosol factor 4 | 3 | |
| PRDX5 | Peroxiredoxin 5, mitochondrial precursor | 1,2,3,4 | |
| PRG1 | Secretory granule proteoglycan core protein precursor* | Serglycin | 2,4 |
| PSMB8 | Proteasome subunit beta type 8 precursor | 1,2,3 | |
| PSMB9 | Proteasome subunit beta type 9 precursor | 1,2,3 | |
| RAC2 | Ras-related C3 botulinum toxin substrate 2 precursor | p21-Rac2 | 1,2,3 |
| SCYE1 | Multisynthetase complex auxiliary component p43 | 3 | |
| SH2D1A | Stromal cell-derived factor 2-like protein 1 precursor* | SDF2-like protein 1 | 4 |
| SEMA4D | Semaphorin 4D precursor (leukocyte activation antigen)* | CD100 | 4 |
| SERPINA1 | Alpha-1-antiproteinase precursor* | Alpha-1-antitrypsin | 4 |
| SH2D1A | SH2 domain protein 1A (signaling lymphocyte activation molecule-associated protein) | 3 | |
| SLAMF1 | Signaling lymphocytic activation molecule precursor | CD150 | 4 |
| TFRC | Transferrin receptor protein 1* | CD71 | 1 |
Secreted proteins that follow a classical pathway predicted by SecretomeP 2.0.
From which cell lines the protein was identified: 1 indicates DEV; 2, L428; 3, L1236; 4, KMH2.
Protein validation in HL cell line supernatant. Protein levels were measured in HL cell line supernatant when cells were cultured for 24 hours in RPMI medium without serum (□) and with serum (■) by ELISA. Protein concentrations are indicated in pg/mL with standard deviation.
Protein validation in HL cell line supernatant. Protein levels were measured in HL cell line supernatant when cells were cultured for 24 hours in RPMI medium without serum (□) and with serum (■) by ELISA. Protein concentrations are indicated in pg/mL with standard deviation.
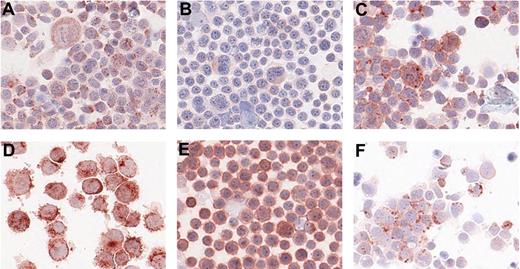
For the other 6 proteins without available ELISA, immunohistochemistry was carried out on centrifuge preparations of the cell lines. The results are summarized in Table 2. Cathepsin C, CD150, CD63, and CD71 were positive in all 4 cell lines; IL25 was positive in L1236, KMH2, and DEV but not in L428; CD100 only showed positive staining in KMH2. Staining examples are given in Figure 3. In all cases, we could confirm protein expression in the cell line from which the protein was originally identified by LC-MS/MS.
Immunohistochemistry on centrifuge preparations of the four HL cell lines
| Protein . | Cell line . | |||
|---|---|---|---|---|
| DEV . | L428 . | L1236 . | KMH2 . | |
| Cathepsin C | + | + | + | + |
| CD100 | − | − | − | + |
| CD150 | + | +* | + | + |
| CD63 | + | + | + | + |
| CD71 | + | + | + | + |
| IL25 | + | − | + | + |
| Protein . | Cell line . | |||
|---|---|---|---|---|
| DEV . | L428 . | L1236 . | KMH2 . | |
| Cathepsin C | + | + | + | + |
| CD100 | − | − | − | + |
| CD150 | + | +* | + | + |
| CD63 | + | + | + | + |
| CD71 | + | + | + | + |
| IL25 | + | − | + | + |
− indicates negative; and +, positive.
Part of the cells were positive.
Immunohistochemistry on centrifuge preparations of HL cell lines. Representative photomicrographs of positive immunohistochemistry staining on centrifuge preparations of HL cell lines for 6 proteins: cathepsin C on L1236 (A), CD100 on KMH2 (B), CD150 on L1236 (C), CD63 on L428 (D), CD71 on KMH2 (E), and IL25 on L1236 (F). Photomicrographs were taken using Aperio ScanScope System and were acquired with Digital Slide Information Management Software (Aperio Technologies, Vista, CA). Image layout was subsequently processed by Microsoft Office XP PowerPoint (Microsoft, Seattle, WA; A-F, original magnifications, ×400).
Immunohistochemistry on centrifuge preparations of HL cell lines. Representative photomicrographs of positive immunohistochemistry staining on centrifuge preparations of HL cell lines for 6 proteins: cathepsin C on L1236 (A), CD100 on KMH2 (B), CD150 on L1236 (C), CD63 on L428 (D), CD71 on KMH2 (E), and IL25 on L1236 (F). Photomicrographs were taken using Aperio ScanScope System and were acquired with Digital Slide Information Management Software (Aperio Technologies, Vista, CA). Image layout was subsequently processed by Microsoft Office XP PowerPoint (Microsoft, Seattle, WA; A-F, original magnifications, ×400).
Protein expression in HL tissues
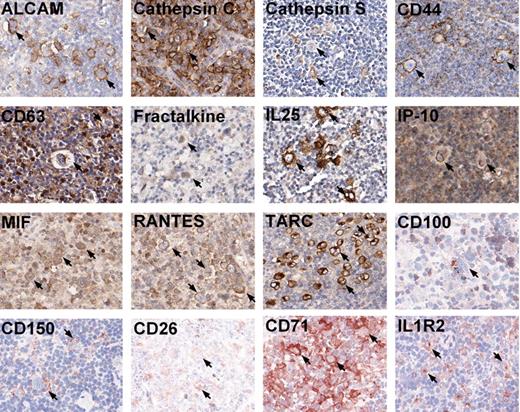
An immunohistochemistry analysis was performed on primary HL tissues for the 16 identified proteins that had been validated on HL cell lines. The scoring analysis was based on the protein expression on the HRS cells. For all 11 cHL cases, CD44, CD71, MIF, RANTES, and TARC expression was found in the majority of HRS cells. The staining for MIF and RANTES was weak in more than half of the cases. IL1R2 was present on most malignant cells in 10 cases. Positive HRS cells for cathepsin C and CD150 were observed in 9 cases. Five cases showed weak staining for cathepsin C. In 7 of 11 cases, ALCAM stained positive on most HRS cells, and in 3 other cases sporadic ALCAM positive HRS cells were observed. Positive HRS cells were observed for IL25 in 8, IP-10 in 5, and fractalkine in 4 cases. In only a small proportion of cases, CD26, cathepsin S, and CD63-positive cells were found in the majority of HRS cells (in 4, 3, and 1 cases, respectively). However, several other cases showed positive staining in part of the HRS cells for all 3 proteins. CD100 was negative for 10 of 11 cases, with only case 7 showing sporadic positive HRS cells. For 3 NLPHL cases, CD44 and CD71 showed positive staining on the majority of L&H cells; cathepsin C, cathepsin S, IL25, and MIF stained (weak) positive in a part of the L&H cells. Only a few L&H cells were weakly stained for ALCAM, CD150, and RANTES in a single case. For CD100, CD26, CD63, Fractalkine, IL1R2, IP-10, and TARC, no positive tumor cells could be observed in all 3 cases. Representative immunohistochemisty examples for these proteins are demonstrated in Figure 4. Detailed results for individual cases can be found in Table S3.
Immunohistochemistry on tissue sections of HL. Representative photomicrographs of positive immunohistochemistry staining on tissue sections for 16 proteins: ALCAM, cathepsin C, cathepsin S, CD44, CD63, fractalkine, IL25, IP-10, MIF, RANTES, and TARC on paraffin-embedded tissues; CD100, CD150, CD26, CD71, and IL1R2 on frozen sections. Photomicrographs were taken using Aperio ScanScope System and were acquired with Digital Slide Information Management Software (Aperio Technologies, Vista, CA). Image layout was subsequently processed by Microsoft Office XP PowerPoint (Microsoft, Seattle, WA; original magnification, ×400). Arrows mark a few of the positive typical HRS cells in all cases.
Immunohistochemistry on tissue sections of HL. Representative photomicrographs of positive immunohistochemistry staining on tissue sections for 16 proteins: ALCAM, cathepsin C, cathepsin S, CD44, CD63, fractalkine, IL25, IP-10, MIF, RANTES, and TARC on paraffin-embedded tissues; CD100, CD150, CD26, CD71, and IL1R2 on frozen sections. Photomicrographs were taken using Aperio ScanScope System and were acquired with Digital Slide Information Management Software (Aperio Technologies, Vista, CA). Image layout was subsequently processed by Microsoft Office XP PowerPoint (Microsoft, Seattle, WA; original magnification, ×400). Arrows mark a few of the positive typical HRS cells in all cases.
Plasma protein levels
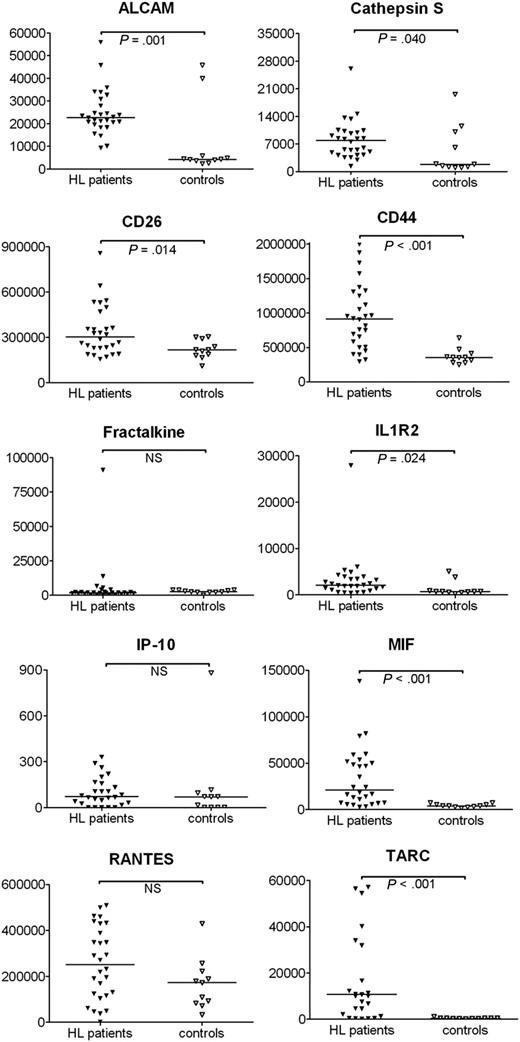
For the 10 proteins with available ELISA kits, we measured the protein levels in plasma of 28 cHL patients before the start of therapy. Plasma ALCAM, cathepsin S, CD26, CD44, IL1R2, MIF, and TARC levels were significantly increased (P < .05) in cHL patients compared with control individuals, whereas fractalkine, IP-10, and RANTES exhibited similar plasma levels between cHL patients and controls (Figure 5).
Protein levels in plasma of cHL patients. Plasma samples from 28 cHL patients and 11 controls were analyzed for protein levels (pg/mL) by ELISA.P values are illustrated to indicate the statistical difference between the groups. NS indicates not significant.
Protein levels in plasma of cHL patients. Plasma samples from 28 cHL patients and 11 controls were analyzed for protein levels (pg/mL) by ELISA.P values are illustrated to indicate the statistical difference between the groups. NS indicates not significant.
Discussion
It is well known that HL is a tumor in which cross-talk between HRS cells and infiltrating cells is of crucial importance for survival and growth of HRS cells.1 Many gene expression profiling studies have been performed using SAGE or microarray approaches and introduced several HL related genes.16,17 It is commonly known that proteins secreted by cancer cells play important roles in cell interaction, adhesion, and invasion.18 To date, no large-scale proteomics studies have been applied to identify secreted factors involved in HL pathogenesis. In the 2 proteomics studies reported by Fujii et al, 2-dimensional difference gel electrophoresis was applied to compare the HL proteome to ALCL and NHL proteome.6,7 Using cell lysate, the whole proteome can be studied, but its high dynamic range makes it difficult to detect the low-abundance, physiologic more interesting secreted proteins. To specifically study the secreted proteins, serum-free culture conditions have been applied successfully in some studies.19,20 This approach has great advantages for the detection and separation of low-abundance proteins secreted by the cells into the culture medium. A disadvantage of culturing cells in medium without serum is the reduced viability and proliferation potential. Careful monitoring of cell viability and secreting potential is thus essential for a reliable secretome analysis. In this study, we checked cell viability and secreting potential, and we decided to culture the cells for 24 hours in medium without serum to achieve a high viability with reasonable production of TARC and MDC. By applying proteomics techniques to analyze HL secretome, we aimed at the identification of putative new players in the interaction between HRS cells and infiltrating cells and proteins that might be used as biomarkers for HL.
Several approaches might be used to study the secretome of HRS cells. Direct analysis of serum or plasma of HL patients contains several putative difficulties because of the presence of high-abundance proteins that mask the potentially more interesting low-abundance proteins. To circumvent this problem, HL cell lines cultured in medium without serum provide a good alternative, despite the putative risk to identify proteins that turn out to be restricted to cell lines or contaminating necrotic cells. Based on confirmation of 15 of 16 selected immune response-related proteins in HRS cells in cHL tissue samples, it is obvious that the use of cell lines is an appropriate method to study the secretome of HL. Furthermore, in NLPHL cases, most proteins detected in the NLPHL cell line DEV were expressed in L&H cells in NLPHL tissues, with the only exception of CD63, which did not show positive staining in L&H cells in 3 cases. Protein identification revealed 368 proteins predicted to be secreted via a classic pathway or nonclassic pathway, and 922 nonsecreted proteins (of intracellular origin). The presence of a relatively large percentage of nonsecreted proteins is most likely related to presence of necrotic cells resulting from the culture conditions without serum, which is necessary for secretome proteomics studies. Remarkably, several of the proteins reported to be specific for HL in other proteomics studies6,7 were also present in our list of identified proteins.
Analysis of protein levels in plasma of 28 cHL patients revealed a significant increase for 7 of 10 proteins compared with controls. These proteins may thus serve as potential biomarkers in HL to evaluate prognosis or disease activity. Feasibility of one of these proteins as potential biomarker has already been demonstrated by Weihrauch et al, who found that significantly increased serum TARC levels in majority of HL patients were related to disease activity.21 In a recent study, Casasnovas et al also demonstrated that the plasma cytokine signature could be used to predict disease-related outcome in cHL.22 For the proteins identified in this study, additional large-scale studies on HL patient plasma or serum need to be performed to elucidate the possible relation of the 7 potential biomarkers to disease activity.
The main goal of this study was to identify new players that might contribute to the pathogenesis of HL. For the chemokines TARC, RANTES, and IP-10, previous studies already demonstrated their putative role and confirmed expression in HRS cells of patient tissues. TARC is a specific chemokine for cHL, which plays an important role in recruiting Th2 and CD4+CD25+ regulatory T cells.23,24 The chemokines IP-10 and RANTES were found to be expressed on HRS cells as well as reactive cells and attract (memory) T cells, NK cells, and eosinophils.25,26 Fractalkine is a chemokine that has not been reported previously in HL and thus provides a potential new player in the pathogenesis of HL. Fractalkine maps to a chemokine gene cluster [CCL22, CX3CL1, CCL17] on 16q13. Immunohistochemistry revealed fractalkine expression in HRS cells only in a few cHL cases, and in most cases it was expressed by the reactive cells. Among all 4 chemokines, only TARC showed elevated plasma levels in cHL patients, which is consistent with the findings by Niens et al.27
A small group of cytokine-related proteins, IL1R2, MIF, CD44, and IL25, was detected by proteomics approach. IL1R2 plays an important physiologic role in the regulation of IL1 action in inflammation sites by binding to IL1α and IL1β with high affinity without inducing signaling pathways.28 In HL, IL1R2 secreted by HRS cells is capable of capturing IL1α or IL1β expressed on HRS cells and reactive cells,29 thus preventing their interaction with the functional receptor IL1R130 and blocking effective IL1 signaling on HRS cells and/or reactive cells. MIF is a pleiotropic cytokine mediator that blocks the cytotoxic T lymphocyte (CTL) response.31 A potential strategy for HRS cells to avoid killing by CTL might thus be the production of MIF. A receptor for MIF is CD74,32 the expression of which was reported on HRS cells.33 CD44, another protein identified in our study, has been reported to be expressed in HL34,35 and is involved in MIF-CD74 receptor complex by providing the required signaling component.36 The MIF-CD74 receptor complex activates mitogen activated protein kinase signaling pathways and results in induction of several target genes important in inflammation and proliferation.37 Thus, in HL microenvironment, MIF expression by the HRS cells and reactive cells may enhance cell proliferation and block a CTL response. IL25 is a recently identified IL17 cytokine family member and has been implicated in the initiation of a Th2 response by driving the expression of Th2 cytokines, such as IL5 and IL13.38,39 The study of Owyang et al suggested the suppression of Th1 or Th17 responses as another role for IL25 in host protection.40 The secretion of IL25 may relate to the IL5 and IL13 expression observed in HL,41,42 and the inhibition of Th1 responses.43 IL25 also induces the expression of CC-chemokines, such as RANTES and Eotaxin (CCL11),44 both of which are found in HL tissue.45,–47
Of the 3 enzymes, CD26, cathepsin S, and cathepsin C, detected in our secretome study, only CD26 was reported to be expressed in HL.48 CD26 is a 110-kDa surface-bound proteolytic enzyme with multiple biologic functions,49 which also exists as a secreted isoform, soluble CD26, that circulates in plasma.50 CD26 is closely related to the functions of chemokines because it is able to truncate and thereby alter the receptor specificities for a number of chemokines, including RANTES, MDC, and IP-10.51,–53 Detection of 2 cystein cathepsins (ie, cathepsin C and S) in HL cells suggests a putative role for these enzymes in HL pathogenesis. This is also supported by expression of a third member of this family of enzymes, cathepsin B, on many HRS cells in HL tissue.54 For both cathepsin B and S, an important role in tumor progression has been reported (reviewed by Gocheva and Joyce55 ). Cathepsin B or S deficiency impaired both angiogenesis and tumor cell proliferation in an animal model.56 Inflammatory cytokines and angiogenic factors, such as vascular endothelial growth factor, basic fibroblast growth factor, and interferon-γ, resulted in increased expression levels of cathepsin S and secretion in human microvascular endothelial cells, supporting its putative role in angiogenesis.57 Cathepsin S secretion by the HRS cells might be closely related to angiogenic activity58 and basic fibroblast growth factor expression59 observed in HL.
The last group of proteins include the CD-markers CD71, CD150, CD63, and ALCAM. CD71 is a T cell activation marker that has been reported to be frequently expressed on HRS cells.60 CD150 is a glycoprotein expressed on activated T and B lymphocytes.61 CD63 is a member of transmembrane 4 superfamily.62 ALCAM is a member of Ig superfamily and is a ligand for the lymphocyte antigen CD6.63 Although a putative involvement for these proteins in HL pathogenesis is still unclear, some studies indicate a role in providing proliferation signals for B cells. Ligation of cell surface CD150 by soluble CD150 (sCD150) enhances the proliferation of T and B cells in vitro.64,65 Expression of SLAMF1 (sCD150) has been found at mRNA level in whole HL tissue samples and cell lines.66 We demonstrated CD150 expression in HRS cells in primary HL cases at protein level. Interaction of sCD150 with cell surface CD150 on HRS cells might provide the tumor cells with proliferation signals. The ALCAM-CD6 interaction is considered as a costimulatory signal involved in lymphocyte activation and proliferation.67 NF-κB, which is constitutively activated in HL,68 could be involved in transcriptional control of the ALCAM gene according to the findings of van Kempen et al.69
Our results demonstrate the power of secretome analysis on HL cell lines to identify relevant proteins. We identified several new putative players that participate in the cross-talk between HRS and infiltrating cells. Fractalkine, a newly identified chemokine, contributes to the recruitment of lymphocytes, monocytes, and NK cells. MIF and CD150 might both enhance proliferation of HRS cells. IL25 is related to HL pathogenesis by inducing a Th2 response, providing host protection signals, and induction of CC-chemokines. Moreover, the newly identified secreted proteins might serve as biomarkers for HL. Further validations with extended clinical studies are necessary to establish clinical relevance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the J.K. de Cock Foundation. Y.M. is a recipient of Bernouilli Bursary.
Authorship
Contribution: L.V., H.R., S.P., R.V., and A.v.d.B. designed the research; Y.M., L.V., and A.v.d.B. wrote the paper; Y.M. and L.V. performed research; Y.M. analyzed data; M.d.V. contributed vital analytical tools (LC-MS/MS); A.D. and S.P. reviewed the histology slides; M.L. and H.V. performed immunohistochemistry and parts of ELISA; G.v.I. and T.v.d.W. managed clinical samples and data; and G.A.-L. helped with preparation of high-quality protein sample for LC-MS/MS.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anke van den Berg, Pathology & Laboratory Medicine, University Medical Center Groningen, PO Box 30.001, Hanzeplein 1, 9700 RB Groningen, The Netherlands; e-mail: a.van.den.berg@path.umcg.nl.