Telomere length is associated with mutation status of the immunoglobulin heavy chain variable (IGHV) gene and clinical course in B-cell chronic lymphocytic leukemia (B-CLL). In a B-CLL cohort of 152 patients, we analyzed telomere length, genomic aberrations, IGHV mutation status, CD38 and ZAP-70 expression to study the prognostic impact and associations among these factors. An inverse correlation existed between telomere length and IGHV homology (P < .001), CD38 (P < .001), and ZAP-70 expression (P = .01). Patients with telomere lengths below median (ie, “short telomeres”) and above median (ie, “long telomeres”) had similar incidences of genomic aberrations (74% vs 68%), 13q− (57% vs 49%), and +12q (5% vs 12%). In contrast, 13q− as a single aberration was more frequent in patients with long telomeres (51% vs 21%; P = .006), whereas 11q− (27% vs 9%; P = .014), 17p− (17% vs 0%; P < .001), and 2 or more genomic aberrations (39% vs 8%; P < .001) were more frequent in patients with short telomeres. Compared with patients with long telomeres, treatment-free survival (TFS) and overall survival (OS) was significantly shorter (P < .001 and P = .015, respectively) in the group with short telomeres, and telomere length was an independent prognostic indicator for TFS. These observations have biological and prognostic implications in B-CLL.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL), the most common leukemia in adults in the Western world, is characterized by a monoclonal expansion of mature B lymphocytes expressing CD19, CD5, and CD23 on the cell surface.1 B-CLL is a clinically heterogeneous disease with survival times ranging from months to normal lifespan.2 As a consequence of the variable clinical course, it has become very crucial to identify reliable prognostic factors useful in planning therapeutic strategies and predicting the outcome. A number of biological features of B-CLL have been described, and several of them can be used to discern different groups of patients with significant differences in clinical course and outcome. The immunoglobulin heavy chain variable (IGHV) gene mutation status analysis reveals 2 subgroups of B-CLL,3,–5 with a more favorable clinical course for patients with mutated IGHV genes.6,,–9
The surface marker CD38 has been proposed as a surrogate marker for IGHV gene mutation status in B-CLL,3 but the association between these markers was shown to be rather weak.7,,,–11 Furthermore, CD38 has been shown to be an independent prognostic marker, although no consensus has been reached concerning cut-off levels.7,8,11,–13 Expression of the protein tyrosine kinase ZAP-70 is strongly associated with IGHV gene mutation status and can be used as an independent prognostic factor.14,–16 Although discordant results have been reported lately, certain IGHV gene usage (eg, IgHV3-21) and presence of high-risk genomic aberrations (eg, 11q− and 17p−) can explain these findings at least in part.17,18
In mature B-cell neoplasms, telomere length correlates with histopathogenesis according to the germinal center.19 This feature includes B-CLL showing a strong correlation between telomere length and IGHV gene mutation status.6,19,,–22 Short telomeres are associated with unmutated IGHV genes and have a significantly shorter median survival time compared with patients with long telomeres.6,20,–22
A hallmark of B-CLL is the occurrence of specific genomic aberrations, and using fluorescence in situ hybridization (FISH), genetic abnormalities have been detected in approximately 80% of patients with B-CLL.23 The most frequent aberration is deletion of 13q14, which observed as a sole abnormality is associated with favorable outcome, whereas patients with deletions of 17p and 11q experience an aggressive disease.24,,,,,,–31 Of interest, the genetic subgroups demonstrate characteristic gene-expression profiles implicating different pathogenetic mechanisms.32
Short telomeres are associated with genetic instability in cell culture, and for solid tumors, telomere dysfunction was found to cause bridge-breakage events leading to a reorganization of the tumor cell genome.33 A similar scenario could prevail in CLL, and in the present study, we investigated the association between telomere length and genetic aberrations, IGHV gene mutation status, and its “surrogate” markers CD38 and ZAP-70. A novel association was identified between telomere length and the type of genomic aberration (high or low risk) as well as karyotype complexity.
Methods
Patients and tumor samples
In this study, we analyzed samples from 152 consenting patients with CLL diagnosed and treated at the University Hospital in Ulm, Germany. Survival data were available for 147 patients. Details on patient characteristics and correlation of clinical as well as biological parameters with telomere length are given in “Results.” Approval was obtained from the University of Ulm institutional review board for these studies. Informed consent was obtained in accordance with the Declaration of Helsinki.
Telomere length determination by quantitative real-time PCR
Telomere length assessment was performed using a quantitative real-time polymerase chain reaction method (Tel-PCR).34 The technique was evaluated in an earlier study on B-CLL from our laboratory,21 showing a significant correlation with Southern blot data. In the present study, modifications to the original protocol included new sets of primers (R. M. Cawthon, written personal communication, October 2004): tel 1b, CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT; tel 2b, GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT; HBG3, TGTGCTGGCCCATCACTTTG; and HBG4, ACCAGCCACCACTTTCTGATAGG.
The primer concentrations were: 100 nM tel 1b primer and 900 nM tel 2b primer for telomere amplification; and 400 nM HBG3 primer and 400 nM HBG4 primer for single copy gene amplification. Genomic Escherichia coli DNA was added to the reaction mix to act as a carrier DNA in order to reduce variation between replicates (R. M. Cawthon, written personal communication, October 2004). All samples were analyzed in triplicates (35 ng DNA/aliquot). The Tel-PCR method provides a relative telomere length value (T/S).
IGHV gene mutation analysis
IGHV gene sequencing was performed as previously described.7 An IGHV gene sequence showing less than 98% homology with the corresponding germ-line gene was considered to be mutated.
Analysis of genomic aberrations
The analysis of genomic aberrations was performed using FISH as previously described.31 The imbalances studied were del(17)(p13), +12q, del(11)(q22-q23), and del(13)(q14).
ZAP-70 expression analysis
Flow cytometry was performed as previously described.17 The antibodies used were: anti–ZAP-70 antibody (clone 2F3.2; Upstate, Hamburg, Germany), goat anti mouse-FITC (Dako, Hamburg, Germany), anti-CD3-PE (clone SK7; BD Bioscience, Heidelberg, Germany), anti-CD56-PE (clone My 31; BD Bioscience), anti-CD19–ECD (clone HD237; Beckman Coulter, Fullerton, CA) and anti-CD5–PC5 (clone BL1a; Beckman Coulter).
CD38 expression analysis
Statistical analyses
The endpoints were overall survival (OS) and treatment-free survival (TFS) from the point of diagnosis. Survival curves were plotted according to the Kaplan-Meier method, and the differences in OS and TFS were analyzed by the log-rank test. Differences between groups were analyzed using the Mann-Whitney U test. For multivariate analysis, the Cox proportional hazards regression model was used.
Results
Patients
Telomere length and IGHV gene mutation status were assessed in all 152 samples. ZAP-70 and CD38 expression data were available for 152 and 144 patients, respectively. Major B-CLL genomic aberrations were analyzed in all but one patient. Full clinical data were available in 147 patients: the median age at diagnosis was 56 years (range, 25–79 years), and the median time from diagnosis to study was 18 months (range, 0–334 months). The male-female ratio was 2.2. Median follow-up time was 52.0 months, median TFS was 41.4 months (95% CI: 31.3–86.1, 81 events), and median OS was 301 months (95% CI: 171 to infinity, 21 events). Treatment prior to study had been administered to 27 (18%) patients with a median of 3 lines of therapy (range, 1–5 lines of therapy). The median time from prior therapy to study was 36.3 months (range, 4–168 months). First-line and subsequent treatments were heterogeneous precluding detailed analyses on the relation between type of prior therapy and telomere length (data not shown). There was no significant correlation between telomere length and number of prior lines of therapy but the group with prior therapy overall showed a trend toward shorter telomeres (P = .055). Data on staging according to Binet were available for 140 patients (96 were stage A and 44 were stage B or C). The incidences of genetic parameters in the different stage groups followed the expected distribution (data not shown).
Telomere length determination
Telomere length was assessed for all 152 patients enrolled in the study, with a median T/S value of 0.3575 (range, 0.0598–1.2305). When converting T/S values to telomere restriction fragment (TRF) length values in kilobase pairs, using the previously published formula,21 the mean telomere length was 4.4 kbp (range, 3.32–7.1 kbp).
IGHV gene mutation status
IGHV gene mutation analysis was successful in all patients in the study. A total of 80 patients presented unmutated IGHV genes (more than 98% homology to germ line), and in 72 patients the IGHV genes were mutated.
Association between telomere length and other clinical parameters
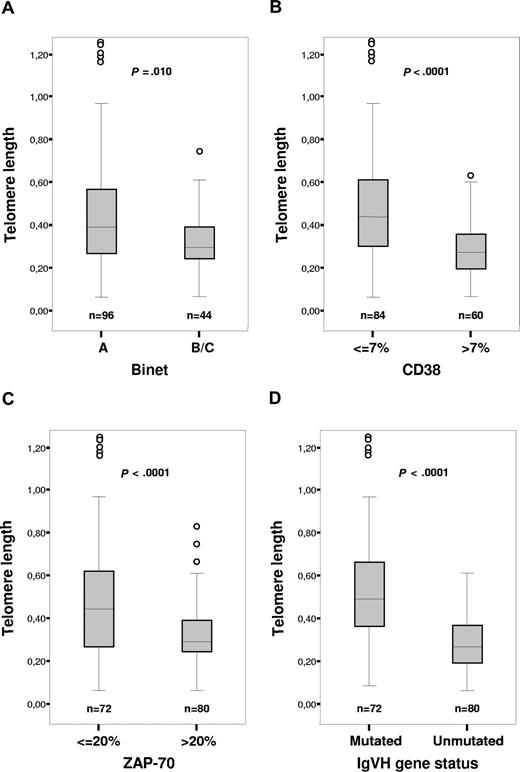
No significant association between telomere length and age or sex was found in the examined cohort (P = .114 and .723, respectively; data not shown). We could show a clear correlation between telomere length and Binet staging (P = .01), CD38 (P < .001), and ZAP-70 expression (P < .001; Figure 1A-C). We could also confirm the difference in telomere length distribution between patients with mutated and unmutated IGHV genes in this independent data set (P < .001), where the mutated cases had longer telomeres (Figure 1D). Several patients showing discordance between telomere length and IGHV mutation status were noted: 21 patients with long telomeres (values above median T/S) and unmutated IGHV genes, and 17 patients presented short telomeres (values below median T/S) and mutated IGHV genes.
Telomere length distribution in relation to established prognostic factors. (A) Telomere length versus stage. (B) Telomere length versus CD38 expression. (C) Telomere length versus ZAP-70 expression. (D) Telomere length versus IGHV mutation status. The boxes shown present median value and interquartile range, and the whiskers minimum and maximal values, except for outliers.
Telomere length distribution in relation to established prognostic factors. (A) Telomere length versus stage. (B) Telomere length versus CD38 expression. (C) Telomere length versus ZAP-70 expression. (D) Telomere length versus IGHV mutation status. The boxes shown present median value and interquartile range, and the whiskers minimum and maximal values, except for outliers.
Association between telomere length and genomic aberrations
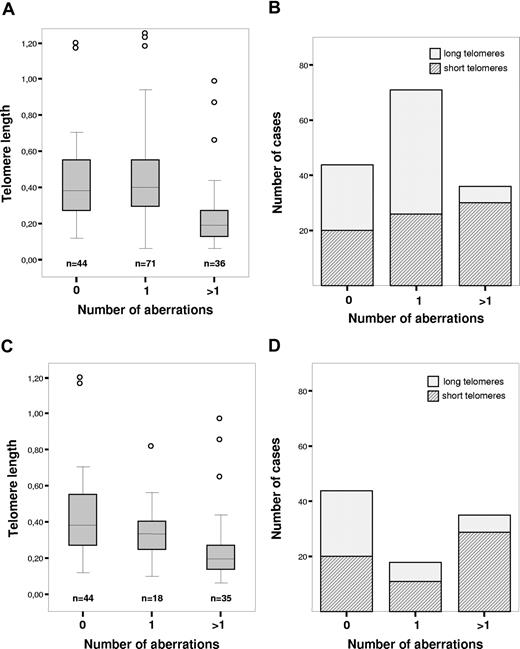
The analysis of genomic aberrations was successful in all patients studied (n = 151). The poor prognostic markers del(17)(p13) and del(11)(q22-q23) were found in 13 and 8 patients, respectively. del(13)(q14) was present in 54 patients and +12 was present in 27 patients; in 44 patients, no abnormality was shown by FISH. We observed a significant difference in telomere length distribution between patients with more than one aberration compared with the patients with normal karyotype or only one abnormality (P < .001; Figure 2A,B). After exclusion of 13q− patients, it was found that the presence of any other cytogenetic abnormality increased the likelihood of shortened telomere length (Figure 2C,D). We could also establish that short telomeres were associated with genetic complexity, indicated by a high number of aberrations and occurrence of unfavorable chromosomal abnormalities such as del(11)(q22-q23) and del(17)(p13) (Table 1). In the subgroup presenting deletion of 17p, all 13 patients demonstrated short telomeres (P < .001; Table 1). Another unfavorable abnormality, del(11q), was significantly more prevalent in patients displaying short telomeres (P = .002; Table 1). No substantial difference in distribution of +12 between groups was discerned (P = .985; Table 1). Deletion 13q, associated with better prognosis, was more frequent among patients with long telomeres (P < .001; Table 1).
Telomere length and genomic aberrations detected by FISH. (A) Telomere length in relation to number of genomic aberrations. (B) Distribution of patients with long and short telomeres in relation to number of genomic aberrations. (C) Telomere length in relation to number of genomic aberrations after exclusion of 13q− patients. (D) Distribution of patients with long and short telomeres in relation to number of genomic aberrations after exclusion of 13q− patients. The boxes shown in panels A and C present medium value and interquartile range, and the whiskers minimum and maximal values, except for outliers.
Telomere length and genomic aberrations detected by FISH. (A) Telomere length in relation to number of genomic aberrations. (B) Distribution of patients with long and short telomeres in relation to number of genomic aberrations. (C) Telomere length in relation to number of genomic aberrations after exclusion of 13q− patients. (D) Distribution of patients with long and short telomeres in relation to number of genomic aberrations after exclusion of 13q− patients. The boxes shown in panels A and C present medium value and interquartile range, and the whiskers minimum and maximal values, except for outliers.
Distribution of FISH-detected genomic aberrations in relation to telomere length and survival
| . | Genomic aberration . | |||
|---|---|---|---|---|
| 17p− | 11q− | +12 | 13q− | |
| Short telomeres, (shorter than median) | 13 | 21 | 4 | 16 |
| Long telomeres, (longer than median) | 0 | 6 | 4 | 38 |
| P | < .001 | .002 | .985 | < .001 |
| Mean survival, mo | 147 | 117 | Not reached | 268 |
| . | Genomic aberration . | |||
|---|---|---|---|---|
| 17p− | 11q− | +12 | 13q− | |
| Short telomeres, (shorter than median) | 13 | 21 | 4 | 16 |
| Long telomeres, (longer than median) | 0 | 6 | 4 | 38 |
| P | < .001 | .002 | .985 | < .001 |
| Mean survival, mo | 147 | 117 | Not reached | 268 |
Prognostic implications of telomere length, CD38 expression, ZAP-70 expression, IGHV mutation status, and high-risk genomic aberrations in univariate and multivariate analyses
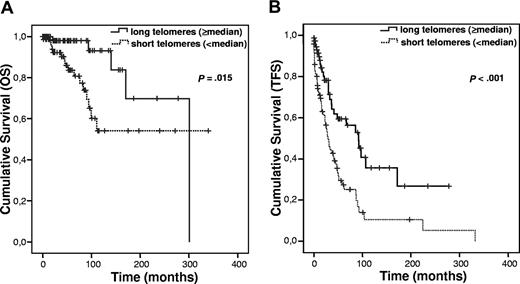
When the material was divided at the median telomere length, a significant difference in OS (P = .015; Figure 3A) and TFS (P < .001; Figure 3B) was observed. Furthermore, the prognostic impact of telomere length on TFS was unchanged after excluding the patients presenting unfavorable genetic abnormalities (P = .006; data not shown). All the other parameters studied (IGHV mutation status, ZAP-70, CD38, and high-risk genomic aberrations) showed a highly significant association to OS and TFS as previously demonstrated (data not shown in figures).
Survival after subdivision of the patients with CLL into 2 groups with a cut-off value at the median telomere length. (A) OS. (B) TFS.
Survival after subdivision of the patients with CLL into 2 groups with a cut-off value at the median telomere length. (A) OS. (B) TFS.
The proportional hazards regression model of Cox was used to investigate the prognostic impact of short telomere length, Binet stage B/C, CD38 and ZAP-70 positivity, unmutated IGHV genes, and presence of high-risk genomic aberrations (17p− or 11q−) on TFS in our cohort of patients with B-CLL. When all factors were incorporated in the model, Binet stage B/C and short telomeres were identified as independent prognostic factors for TFS (Table 2). As advanced stage is clinically the primary marker for initiation of treatment and therefore directly related to TFS, we performed an additional analysis excluding stage. This resulted in the identification of the presence of high-risk genomic aberrations as the sole significant prognostic factor (Table 2). Multivariate analysis was not performed for OS due to too few events in the individual risk groups.
Multivariate analysis of TFS including the high-risk parameters studied
| Parameter . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Stage B/C | 3.27 | 1.85–5.78 | < .001 | — | — | — |
| CD38+ | .94 | .51–1.72 | .845 | 1.31 | .76–2.27 | .330 |
| ZAP-70+ | 1.33 | .78–2.26 | .294 | 1.34 | .79–2.27 | .280 |
| High-risk genomic aberrations | 1.45 | .84–2.51 | .186 | 1.85 | 1.12–3.04 | .016 |
| IGHV unmutated | 1.22 | .65–2.29 | .533 | 1.68 | .91–3.12 | .097 |
| Short telomeres | 1.98 | 1.10–3.56 | .023 | 1.47 | .88–2.45 | .145 |
| Parameter . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Stage B/C | 3.27 | 1.85–5.78 | < .001 | — | — | — |
| CD38+ | .94 | .51–1.72 | .845 | 1.31 | .76–2.27 | .330 |
| ZAP-70+ | 1.33 | .78–2.26 | .294 | 1.34 | .79–2.27 | .280 |
| High-risk genomic aberrations | 1.45 | .84–2.51 | .186 | 1.85 | 1.12–3.04 | .016 |
| IGHV unmutated | 1.22 | .65–2.29 | .533 | 1.68 | .91–3.12 | .097 |
| Short telomeres | 1.98 | 1.10–3.56 | .023 | 1.47 | .88–2.45 | .145 |
Discussion
In the present study, we show that telomere length correlated strongly with established prognostic markers (IGHV mutation status, CD38, and ZAP-70) as well as the clinical course in B-CLL where short telomere length predicted an unfavorable clinical outcome. Interestingly, a novel association was identified between telomere length and the type of genomic aberration (high or low risk) as well as karyotype complexity. However, what could be the biological background for the latter finding?
The original “mortality stages 1 and 2 (M1 and M2) model” states that human cells must overcome 2 mechanisms to become immortalized, namely senescence induction (M1) and cell death due to critically short telomeres (M2).35,36 M1 can be overcome by, for example, p53 and Rb inactivation,36 whereas M2 is characterized by very short telomeres, genetic instability, and high cell death. Immortalization usually occurs by up-regulation of telomerase activity, giving telomere length stabilization. Malignant tumors generally demonstrate active telomerase and shorter telomeres than their normal cellular counterparts.37,,–40 Telomere shortening also plays a role in epithelial carcinogenesis by promoting chromosomal rearrangements in mice,41 a scenario also indicated in human tumorigenesis.33,42
In hematopoietic malignancies, an increased frequency of genetic alterations has been coupled to short telomeres.43,–45 Accordingly, we could demonstrate that short telomeres were associated with a higher number of genomic aberrations in CLL. The collected data are basically in agreement with the M1/M2 model and can be interpreted as an accumulative effect of 2 main events: (1) genetic alterations force cells to bypass senescence (M1), leading to additional telomere attrition; and (2) short telomeres induce genetic instability. Our most interesting observation was the strong association between telomere length and specific cytogenetic abnormalities, since short telomeres and 17p− or 11q− abnormalities were coupled, whereas 13q− single patients were characterized by long telomeres. Thus, bad prognostic cytogenetics was linked to short telomeres and good prognostic cytogenetic features was linked to long telomeres. Ricca et al have reported a nonsignificant trend toward an association between short telomere length and high-risk cytogenetics, partially supporting our findings.46
Normal germinal-center B cells are telomerase activity positive, and significant telomere elongation occurs during the germinal-center reaction concomitant with induction of the IG gene hypermutation machinery.47,48 Thus, B cells with mutated IGHV genes have longer telomeres than B cells with unmutated genes. These characteristics have been argued to form a basis for the strong association demonstrated here and elsewhere between telomere length and IGHV gene mutation status,6,21,22 indicating that the telomere length is “preset” depending on the germinal-center experience of the original B cell giving rise to a CLL clone. It is conceivable that the CLL-initiating B cells differ in telomere length from start.
A kinetic study of CLL demonstrated that the cellular birth rate was considerable in many patients, indicating a dynamic clonal progression, and a correlation between birth rate and disease activity seemed to exist.49 These data suggest that cell kinetic characteristics can contribute to differences in telomere length. Furthermore, cytogenetic subgroups of B-CLL have demonstrated interesting gene expression profiles. Patients with 11q− showed decreased expression of ATM, and p53 expression was low in 17p− patients, indicating a reduced DNA damage response in both cytogenetic groups32 in accordance with a deregulated senescence checkpoint. 17p− was also associated with an up-regulation of c-myc, a putative positive regulator of hTERT, and since p53 is a negative regulator of hTERT, a possible combined effect is telomerase activation.50,–52 This scenario fits with earlier data on high telomerase and short telomeres in poor-prognosis CLL,22 which also strengthens the notion that these patients start with short telomeres.
In the present study, we found a strong correlation between short telomeres and the expression of ZAP-70 and CD38, both established indicators of B-CLL prognosis. CD38 together with ZAP-70 appear to be partners in a pathway that may sustain signals mediated by the B-cell receptor promoting cell proliferation.53 11q− or 17p− aberration in combination with overexpression of ZAP-70 and/or CD38 give cells a survival advantage and facilitates cell-cycle progression, one consequence of which is telomere attrition. Thus, a number of factors characterizing the poor-prognostic group of CLL contribute to the “short telomere phenotype.” Finally, it cannot be fully excluded that short telomeres in patients with 11q− or 17p− may be the result of selection, since it might be a prerequisite with 17p− (p53 low) and/or 11q− (reduced ATM) to survive for cells with very short telomeres.
Using univariate analysis, all parameters investigated in the present CLL cohort (telomere length, IGHV mutation status, ZAP-70, CD38, and cytogenetic group) could subdivide the material into groups with significantly different outcomes with respect to TFS in line with a number of previous studies.3,5,7,10,13,15,–17 In multiparameter analysis, including all negative prognostic indicators studied, only Binet stage B/C and short telomeres were independent indicators of TFS, and when excluding stage, only high-risk genomic aberrations seemed to be an independent parameter. Surprisingly, the IGHV mutation status had no independent prognostic value for TFS in either model. Thus, there are a number of prognostic factors identified in CLL with mutation status as the “role model,” with the other factors often called surrogate markers for mutation status. In the future, it is less likely that one parameter will be used as “the marker” for subdivision of CLL into prognostic subgroups giving guidance for therapy, and a more possible scenario is a prognostic index using a combination of markers. In this context, our present study suggests that telomere length, easily determined by a rapid PCR method, must be taken into serious consideration.
Telomerase is a promising target for cancer therapy.54 Regarding CLL, the “short telomere phenotype” should be of interest, since these patients constitute a bad prognosis group needing better treatment strategies. Increased telomere attrition rate by inhibiting telomerase would theoretically lead to accentuated cell death in this patient cohort.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Silja Groner is thankfully acknowledged for data collection and management.
This study was supported by grants from the Swedish Cancer Society, the Medical Faculty, Umeå University, Lion's Cancer Research Foundation at Umeå University, Krebshilfe (106142), José Carreras Leukämie-Stiftung (R 05/25), DFG STI 296/1-1, and by grant LSHC-CT-2004-502943 Mol Cancer Med from the European Union.
Authorship
Contribution: G.R. and S.S. designed research, analyzed data, and wrote the paper; P.G. performed research, analyzed data, and wrote the paper; A.K., D.K., A.B., and H.D. performed research and analyzed data; and R.R. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Göran Roos, Department of Medical Biosciences, Umeå University, S-90187, Umeå, Sweden; e-mail: goran.roos@medbio.umu.se.
References
Author notes
A.K. and P.G. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal