Fanconi anemia (FA), an inherited syndrome that associates bone marrow failure, cancer predisposition, and genetic instability, is characterized by an overproduction of the myelosuppressive cytokine TNF-α through unknown mechanisms. We demonstrate here that FANC pathway loss-of-function results in the aberrant activation of 2 major stress-signaling pathways: NF-κB and MAPKs. These responses are independent on TNF-α expression. On the contrary, inhibition of the MAPK pathways normalizes TNF-α oversecretion in FA. Moreover, our data show that the overexpression of the matrix metalloproteinase MMP-7 is the key event directly responsible for the high rate of TNF-α shedding and release from the cytoplasmic membrane in FA. TNF-α overproduction is, indeed, normalized by MMP-7 inhibition. Finally, MAPK inhibition impacts on MMP-7 overexpression. Evidence is provided of the existence of a linear pathway in which FANC mutations activate MAPK signaling that induces MMP-7 overexpression leading, in fine, to TNF-α oversecretion. TNF-α may, in turn, sustain or amplify both MAPKs and NF-κB activation. Aberrant expression or activity of NF-κB and/or MAPKs has been already involved in bone marrow failure and leukemia, and their inhibition offered clinical benefit for patients. In conclusion, our data provide a strong rationale for new clinical trials on FA patients.
Introduction
Fanconi anemia (FA) is a rare recessive syndrome featuring progressive bone marrow (BM) failure, multiple developmental abnormalities and cancer predisposition.1,–3 BM failure and its related consequences, such as pancytopenia or acute myeloid leukemia (AML), are the major cause of morbidity and mortality of FA patients.4 The cellular phenotype is characterized by chromosomal instability and hypersensitivity to DNA interstrand cross-link (ICL)–inducing agents such as mitomycin C (MMC), diepoxybutane, and cisplatin.5,,–8 At least 13 complementation groups have been identified (FA-A, B, C, D1, D2, E, F, G, I, J, L, M, and N) and the genes for all of these groups have been cloned.9 One of the major functions of FANC proteins is to deal with DNA damage, thus participating in an as-yet-undefined manner to the repair of DNA lesions induced by cross-linking agents.10,–12 However, the spectrum of clinical and cellular abnormalities of the syndrome suggests that FANC proteins could have other functions or participate in pathways other than DNA repair.13,14 Whether the hematologic problems of the FA patients are a consequence of a defect in DNA repair or in other potential functions of the FANC proteins remains to be determined.
Tumor necrosis factor-α (TNF-α) is a major cytokine involved in hematopoiesis, inflammation, and apoptosis.15,16 TNF-α is synthesized as a membrane-bound precursor of 26 kDa that can be processed to generate a secreted 17-kDa mature TNF-α.17,18 Soluble mature TNF-α is released from the cells by cleavage of the precursor at the Ala76-Val77 bond by the TNF-α converting enzyme (TACE or ADAM17)19,20 or, less efficiently, by the matrix metalloproteinase 7 (MMP-7 or matrilysin).21,22 TNF-α signals through 2 distinct cell-surface receptors, TNFR-1 and TNFR-2.16 The binding of TNF-α to its receptors results, among other events, in the activation of both the mitogen-activated protein kinases (MAPKs) stress signaling cascade16 and the NF-κB transcription factor.23 Activation of the MAPKs and NF-κB plays an important role in the induction of many cytokines including the TNF-α itself.24,25 TNF-α negatively regulates the expansion and self-renewal of pluripotent hematopoietic stem cells (HSCs)26,27 and has inhibitory effects on normal human hematopoietic progenitor cells as well as leukemia progenitor cells.28,–30 Consistently, TNF-α overproduction has been associated with different hematopoietic disorders such as myelodysplastic syndrome (MDS), AML, and aplastic anemia.31,–33 FA syndrome recapitulates all these abnormalities, that is, impaired HSC expansion and development of the myeloid lineages, MDS, aplastic anemia, and AML.2,–4 FA is also characterized by TNF-α overproduction, both in vivo and in vitro. Indeed, it has been reported that TNF-α is (1) overexpressed in BM of FA patients,34,35 (2) increased in the serum of patients,35,36 and (3) overproduced by Epstein-Barr virus (EBV)–transformed FA lymphoblasts.36 Moreover, hematopoietic progenitors from Fancc knockout mice, which fail to spontaneously overproduce the cytokine, show hypersensitivity to the TNF-α myelosuppressive action.37,38 In light of these observations, abnormalities in TNF-α expression could be considered at the origin of the progressive BM failure observed in FA patients. Understanding the origins of TNF-α overproduction could open new therapeutic roads for the BM failure in FA.
In the present work, we investigated the molecular origins of TNF-α overproduction in FA. Our data indicate that the loss-of-function of FANC proteins leads to overactivation of 2 major stress-response pathways: the MAPK and NF-κB networks. As a consequence of MAPKs activation, MMP-7 is expressed over the level normally observed in FANC-proficient cells and it sheds TNF-α from cytoplasmic membrane leading to the extracellular accumulation of the cytokine. Importantly, we show that the inhibition of MAPKs or MMP-7 activity in FA cells leads to a significant decrease in TNF-α overproduction.
Methods
Cell lines and reagents
EBV-transformed lymphoblasts and their growth conditions were previously described.36,39 GM16757 and 293T/NF-κB-Luc cells were purchased from Coriell Cell Repositories (Camden, NJ) and Panomics (Freemont, CA), respectively. HeLa and 293T/NF-κB-Luc cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FCS (Dutcher, Brumath, France) and antibiotics (Gibco-BRL, Gaithersburg, MD). Hygromycin (100 μg/mL; Sigma-Aldrich, St Louis, MO) was added to 293T/NF-κB-Luc cultures.
Dexamethasone, brefeldin A, and curcumin (Sigma) were dissolved in ethanol and MMP inhibitor II, SB203580, PD98059, and SP600125 (Calbiochem, San Diego, CA) in DMSO.
Lentiviral shRNA vector construction, production, and transduction
Lentivirus vectors were obtained from http://tronolab.epfl.ch/. MMP-7 and untargeted shRNA oligonucleotide sequences can be provided on request. Vectors were constructed by cloning the sequences into the Mlu1-Cla1 site of the pLV-TH plasmid and then sequenced. Lentiviruses were produced by transient transfection of 293T cells according to standard protocols.40 For transduction, lentiviruses were added to cells for 4 hours. Cells were sorted 96 hours later for green fluorescence using a FACSDiva flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
RNA extraction and semiquantitative RT-PCR analysis
Total RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA), following the manufacturer's instructions including the optional DNase treatment. Polymerase chain reaction (PCR) reactions were performed as previously described.39 Primers sequences were as follows: TNF-α: forward 5′-CAGAGGGCCTGTACCTCATC-3′, reverse 5′-GGTTGAGGGTGTCTGAAGGA-3′; and actin: forward 5′-AGAGCTACGAGCTGCCTGAC-3′, reverse 5′-AGTACTTGCGCTCAGGAGGA-3′. Reverse-transcription (RT)–PCR quantification was done with Fluor-S-Multilmager (Biorad, Hercules, CA).
Real-time PCR amplification using Taqman assay
Primers were purchased from Applied Biosystems (Foster City, CA). Real-time PCR was performed on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) following the manufacturer's instructions.
Protein extraction and Western blot analysis
Whole-cell extracts were prepared in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40) supplemented with protease inhibitors (Roche Diagnostic, Meylan, France), 10 mM NaF, and 1mM Na3VO4 (Sigma). Supernatants from lymphoblasts cultured in serum-free RPMI for 24 hours were collected by centrifugation, placed in Vivaspin 6 (Sartorius, Goettingen, Germany), and centrifuged to remove proteins with molecular weights of less than 5 kDa. Blots were incubated with primary antibodies: TNF-α (R&D Systems, Minneapolis, MN), TACE, MMP-7 (Oncogene Research, Merck, Darmstadt, Germany), phospho-p38, p38, phospho-ERK, ERK, phospho-JNK, JNK (Cell Signaling, Danvers, MA), FANCA, IκBα, actin (Santa Cruz Biotechnology, Santa Cruz, CA), FANCD2 (Abcam, Cambridge, United Kingdom), and FANCC (FA Research Fund, Eugene, OR) followed by species-specific secondary antibodies coupled to horseradish peroxidase (Santa Cruz Biotechnology). Signals were detected by chemiluminescence using the enhanced chemiluminescence (ECL) kit (Pierce, Rockford, IL).
TNF-α titration
Cell suspensions were collected by centrifugation and supernatants were filtered (0.45 μm, Millipore Filter; Millipore, Paris, France) before storage at −20°C. TNF-α content in supernatants or protein extracts was evaluated by Quantikine enzyme-linked immunosorbent assay (ELISA) as described by the supplier (R&D Systems).
siRNA
Cell transfection and luciferase assay
Lymphoblasts were transfected using the Amaxa nucleofection technology (Amaxa, Cologne, Germany). Briefly, 2 × 106 cells were resuspended in 100 μL Amaxa solution kit V, mixed with 2 μg plasmid (pGL3-hTNF-α,43 pGL3-hMMP-7,44 pNF-κB-Luc [Clontech, Mountain View, CA]) and 10 ng phRL-TK (Promega, Madison, WI). Cells were nucleofected using the program T20 and harvested 48 hours after transfection, and 20 μL cell lysate was assayed for luciferase activity using the dual luciferase assay system (Promega). Firefly luciferase activity was corrected for transfection efficiency using the Renilla luciferase activity.
After 24 hours of siRNA transfection, HeLa cells were transfected with 0.8μg plasmid (pNF-kB-Luc or pGL3-hMMP-7) and 5 ng phRL-TK using FUGENE 6 (Roche Diagnostic) according to the manufacturer's protocol. Cells were always harvested 72 hours after siRNA transfection.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde (10 minutes) and permeabilized with 0.1% SDS (12 minutes) at room temperature. Coverslips were blocked with 2% BSA in PBS and then incubated with primary antibody (p50 and p65; Santa Cruz Biotechnology) for 1 hour. Cells were washed and incubated with secondary antibody (Alexa Fluor; Invitrogen) for 1 hour before mounting with a solution containing DAPI (Vectashield; Vector Laboratories, Peterborough, United Kingdom). Analysis was performed using a laser confocal microscope (LSM 510; Zeiss, Oberkochen, Germany) equipped with an Axiovert 200M microscope and a 63×/1.40 NA oil-immersion objective. Images were acquired with an AxioCam MR camera (Zeiss) using LSM 5 Image Browser (Zeiss). Images were then imported in Adobe Photoshop 8.0 CS (Adobe Systems, San Jose, CA), where adjustments were made for the whole images for brightness and contrast.
Statistical analysis
Results are the mean of at least 3 independent experiments with error bars showing the standard error. The Student t test was used, and P less than .05 was considered statistically significant.
Results
FANCC gene product modulates the spontaneous production of biologically active TNF-α
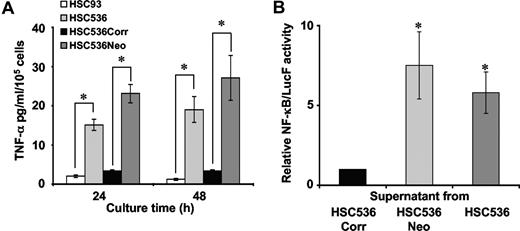
Overproduction of TNF-α in FA syndrome has been previously reported both in vivo and in vitro.35,36 However, previous experimental approaches did not establish whether TNF-α overproduction was directly linked to FA gene mutation. To address this point, we compared TNF-α level and activity in the supernatant of exponentially growing normal (HSC93), FANCC-corrected (HSC536Corr), and FA-C lymphoblasts (HSC536 and HSC536Neo) (Figure 1A, B). FA-C cells accumulated, in a time-dependent manner, 5 to 8 times more TNF-α in their growth medium than lymphoblasts from a healthy donor (Figure 1A). The ectopic expression of FANCC in FA-C cells reduced TNF-α production to the level observed in normal cells (Figure 1A).
Overproduction of biologically active TNF-α by FA-C cells. (A) TNF-α accumulation in the supernatant of normal cells (HSC93), and FA-C (HSC536) and its derived cell lines, HSC536Neo and HSC536Corr. HSC536 cells were transfected with an empty vector (HSC536Neo) or with a vector bearing the wild-type FANCC cDNA (HSC536Corr). Cells were collected by centrifugation, washed, and resuspended in fresh medium (3 × 105 cells/mL/well in 3 mL medium in 6-well tissue culture plates). Supernatants were collected 24 hours and 48 hours after subculturing. The level of TNF-α in supernatants was determined by ELISA. Data presented are the mean (± SD) of at least 5 independent experiments. *P < .01. (B) Luciferase activity in 293T/NF-κB-Luc cells cultured in conditioned medium from lymphoblasts. 293T/NF-κB-Luc cells were plated (106 cells/well in 6-well tissue culture plates) 48 hours before treatment with supernatant collected from 48-hours-old lymphoblasts cultures. One volume of supernatant, equivalent to 106 lymphoblasts, was added per well of 293T/NF-κB-Luc cells. Cells were harvested 24 hours after treatment for luciferase measurement. Luciferase activity of 293T/NF-κB-Luc cells cultured in supernatant from HSC536Corr cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05.
Overproduction of biologically active TNF-α by FA-C cells. (A) TNF-α accumulation in the supernatant of normal cells (HSC93), and FA-C (HSC536) and its derived cell lines, HSC536Neo and HSC536Corr. HSC536 cells were transfected with an empty vector (HSC536Neo) or with a vector bearing the wild-type FANCC cDNA (HSC536Corr). Cells were collected by centrifugation, washed, and resuspended in fresh medium (3 × 105 cells/mL/well in 3 mL medium in 6-well tissue culture plates). Supernatants were collected 24 hours and 48 hours after subculturing. The level of TNF-α in supernatants was determined by ELISA. Data presented are the mean (± SD) of at least 5 independent experiments. *P < .01. (B) Luciferase activity in 293T/NF-κB-Luc cells cultured in conditioned medium from lymphoblasts. 293T/NF-κB-Luc cells were plated (106 cells/well in 6-well tissue culture plates) 48 hours before treatment with supernatant collected from 48-hours-old lymphoblasts cultures. One volume of supernatant, equivalent to 106 lymphoblasts, was added per well of 293T/NF-κB-Luc cells. Cells were harvested 24 hours after treatment for luciferase measurement. Luciferase activity of 293T/NF-κB-Luc cells cultured in supernatant from HSC536Corr cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05.
TNF-α is a potent inducer of NF-κB activity.23 We evaluated the biologic activity of the secreted TNF-α by measuring NF-κB transcriptional activity using a 293T cell line expressing a luciferase reporter construct under the control of NF-κB target sequence. 293T/NF-κB-Luc cells were cultured for 24 hours in 48-hour-old supernatants collected from FA-C and corrected cells. As depicted in Figure 1B, luciferase activity was 6 to 8 times more important when 293T/NF-κB-Luc cells were cultured in supernatant from FA-C cells than in supernatant from FA-C–corrected cells.
All together, these results demonstrate that TNF-α is overproduced in FA-C cells and is biologically active, and that its aberrant expression is directly linked to FANCC loss-of-function.
FA-C cells show a higher TNF-α shedding from cytoplasmic membrane
TNF-α production is controlled at multiple levels from transcription to cytoplasmic membrane shedding. To determine the molecular origin of TNF-α overproduction, we first analyzed, using 2 different approaches, TNF-α mRNA intracellular production and accumulation (Figure 2A-B). First, FA-C cells and their ectopically corrected counterpart were transfected with a reporter plasmid expressing Firefly luciferase under the control of human TNF-α promoter (pGL3-hTNF-α).43 Interestingly, relative TNF-α promoter activity in FA-C cells was 2 to 2.5 times higher compared with corrected FA-C cells (Figure 2A). Then, we performed semiquantitative RT-PCR analysis to assess the steady-state level of TNF-α mRNA in lymphoblasts. Actin mRNA was amplified in the same PCR reactions and used as an internal control. Quantification of RT-PCR products revealed a slight (1.5 times higher) but constant and significant difference in TNF-α mRNA content in FA-C cells compared with corrected cells (Figure 2B).
Analysis of TNF-α mRNA expression and level and protein expression and secretion. (A) TNF-α promoter activity in FA and corrected lymphoblasts. Cells were cotransfected with the reporter pGL3-hTNF-α-LucF and phRL-TK, used as internal control. Luciferase activity in HSC536Corr cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (B) Steady-state level of TNF-α mRNA in FA and corrected lymphoblasts assessed by semiquantitative RT-PCR. The histogram represents the relative level of TNF-α mRNA in FA-C–deficient compared with FANCC-corrected cells. The level in HSC536Corr was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (C) Immunoblot showing the level of TNF-α in total cell extracts prepared from HSC536 cell line either untreated (Unt) or treated for 18 hours with brefeldin A (Bref A) or dexamethasone (Dexa). A cross-reactive band (marked with asterisk) was used as a loading control. (D) Intracellular TNF-α accumulation in FA and corrected lymphoblasts. Cells were treated with dexamethasone (10 μg/mL) for 3 hours. After this incubation, cells were collected by centrifugation, washed, and resuspended in fresh medium in presence of brefeldin A (1 μg/mL) for 24 hours. Proteins were extracted at the indicated time points. TNF-α content was measured by ELISA. Each point represents the mean (± SD) of at least 3 independent experiments. (E) Kinetics of TNF-α accumulation in HSC93, HSC536Corr, HSC536, and HSC536Neo cell lines. Supernatants were collected 3, 6, and 9 hours after cell subculturing in fresh medium and cytokine release was determined by ELISA. Each point represents the mean (± SD) of at least 3 independent experiments. *P < .01.
Analysis of TNF-α mRNA expression and level and protein expression and secretion. (A) TNF-α promoter activity in FA and corrected lymphoblasts. Cells were cotransfected with the reporter pGL3-hTNF-α-LucF and phRL-TK, used as internal control. Luciferase activity in HSC536Corr cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (B) Steady-state level of TNF-α mRNA in FA and corrected lymphoblasts assessed by semiquantitative RT-PCR. The histogram represents the relative level of TNF-α mRNA in FA-C–deficient compared with FANCC-corrected cells. The level in HSC536Corr was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (C) Immunoblot showing the level of TNF-α in total cell extracts prepared from HSC536 cell line either untreated (Unt) or treated for 18 hours with brefeldin A (Bref A) or dexamethasone (Dexa). A cross-reactive band (marked with asterisk) was used as a loading control. (D) Intracellular TNF-α accumulation in FA and corrected lymphoblasts. Cells were treated with dexamethasone (10 μg/mL) for 3 hours. After this incubation, cells were collected by centrifugation, washed, and resuspended in fresh medium in presence of brefeldin A (1 μg/mL) for 24 hours. Proteins were extracted at the indicated time points. TNF-α content was measured by ELISA. Each point represents the mean (± SD) of at least 3 independent experiments. (E) Kinetics of TNF-α accumulation in HSC93, HSC536Corr, HSC536, and HSC536Neo cell lines. Supernatants were collected 3, 6, and 9 hours after cell subculturing in fresh medium and cytokine release was determined by ELISA. Each point represents the mean (± SD) of at least 3 independent experiments. *P < .01.
Having observed a slight increase in both TNF-α mRNA production and accumulation, we then questioned whether this increase in mRNA could be responsible for the accumulation of TNF-α in the supernatant of FA-C cells. To evaluate TNF-α protein synthesis, we first treated the cells with dexamethasone, which inhibits TNF-α mRNA translation,45 to reduce the steady-state intracellular content of TNF-α protein (Figure 2C-D). After 3-hour exposure to dexamethasone (10 μg/mL), intracellular TNF-α content was reduced to approximately half of the starting quantity (Figure 2D). Dexamethasone was then removed and cells were treated with brefeldin A (1 μg/mL). This agent inhibits TNF-α protein transport to the membrane impeding its secretion,46 thereby leading to its intracellular accumulation (Figure 2C). Proteins were extracted 3, 6, 9, and 24 hours following brefeldin A addition, and the TNF-α protein level was assessed by ELISA. As reported in Figure 2D, the kinetics of intracellular accumulation of TNF-α was similar in both FA-C and corrected cells. Importantly, the content of TNF-α in the supernatant of brefeldin A–treated cells was below the detection threshold, showing the ability of the drug to efficiently block TNF-α transport to the cytoplasmic membrane and its consequent release (data not shown). We thus conclude that TNF-α protein accumulated equally in cells independently of the FA gene status.
In view of these results, a difference in the TNF-α shedding may be responsible for the observed overproduction of TNF-α by FA-C cells. To address this point, we determined the kinetics of TNF-α secretion after plating the cells in fresh, TNF-α–free, medium (Figure 2E). As early as 3 hours after cell plating, TNF-α accumulation in the supernatant of FA-C cells was significantly higher (2.5 times more) than proficient FANCC cells. Moreover, TNF-α accumulation in the supernatant of HSC93 and HSC536Corr cells reached a plateau after 6 hours, whereas in FA-C cells this accumulation was constantly increasing even after 9 hours.
All together, these results indicate that an increased release of the cytokine from the cytoplasmic membrane is responsible for TNF-α overproduction in FA-C cells.
MMP-7 overexpression links FANCC to TNF-α oversecretion
To date, only 2 proteases have been described as able to produce a correctly processed and bioactive TNF-α: TACE19,20 and MMP-7.21 Western blot analysis showed that TACE expression was independent of the FANCC status (Figure 3A). On the contrary, MMP-7 was clearly overexpressed in FA-C cell lines as assessed by Western blot analysis on either whole-cell lysates or supernatant (Figure 3B top and bottom panels, respectively). Then, the intracellular production and accumulation of MMP-7 mRNA was analyzed (Figure 3C,D). First, lymphoblasts were transfected with a reporter plasmid expressing Firefly luciferase under the control of human MMP-7 promoter (pGL3-hMMP-7).44 As reported in Figure 3C, relative MMP-7 promoter activity in FA-C lymphoblasts was higher than in corrected and normal cells. The steady-state level of MMP-7 mRNA in lymphoblasts was analyzed by quantitative RT-PCR. 18S rRNA was used as control gene for normalization. As shown in Figure 3D and in agreement with our previous data,39 FA-C cells showed a significant accumulation of MMP-7 mRNA compared with FANCC-proficient cells.
Overexpression of MMP-7 in FA-C cells and its involvement in the aberrant secretion of the TNF-α. (A) Immunoblot showing the level of TACE in total cell extracts. Immature and mature forms of TACE are indicated by i and m, respectively. The doublets observed for each form on Western blot reflect protein glycosylation. Equal loading of the membrane was verified by ponceau red staining and/or evaluated by the intensity of the indicated (*) aspecific band. (B) Immunoblot showing the level of MMP-7 in total cell extracts (top panel) and in supernatant (bottom panel). Equal loading of the membrane was verified by ponceau red staining and/or evaluated by the intensity of the indicated (*) aspecific band. Vertical lines have been inserted to indicate a repositioned gel lane. (C) MMP-7 promoter activity in normal, corrected, and FA cells. Cells were cotransfected with the reporter pGL3-hMMP-7 and phRL-TK, as internal control. Luciferase activity in AHH1 cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (D) MMP-7 mRNA steady-state levels as determined by quantitative RT-PCR on mRNA extracted from exponentially growing lymphoblasts and normalized to 18S rRNA content. To normalize the values among the different experiments, each time the ratio MMP-7/18S was considered equal to 1 in AHH1 cells. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (E) TNF-α release in MMP-7 inhibited FA-C cells. HSC536 and HSC536Neo cells were treated with MMP inhibitor (50 μM) or its solvent (DMSO) or left untreated. Supernatants were collected 24 hours after subculturing and cytokine release was determined by ELISA. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01. (F) TNF-α release in FA-C cells after the down-regulation of MMP-7 expression. Immunoblot shows the level of MMP-7 in supernatant of FANCC siRNA–MMP-7 down-regulated cells. A cross-reactive band (*) was used as a loading control. TNF-α accumulation in the supernatant of infected HSC536Neo cells was evaluated by ELISA. Histogram represents the mean reduction (in percentage) plus or minus SD of TNF-α accumulation observed in at least 3 independent experiments. *P < .01.
Overexpression of MMP-7 in FA-C cells and its involvement in the aberrant secretion of the TNF-α. (A) Immunoblot showing the level of TACE in total cell extracts. Immature and mature forms of TACE are indicated by i and m, respectively. The doublets observed for each form on Western blot reflect protein glycosylation. Equal loading of the membrane was verified by ponceau red staining and/or evaluated by the intensity of the indicated (*) aspecific band. (B) Immunoblot showing the level of MMP-7 in total cell extracts (top panel) and in supernatant (bottom panel). Equal loading of the membrane was verified by ponceau red staining and/or evaluated by the intensity of the indicated (*) aspecific band. Vertical lines have been inserted to indicate a repositioned gel lane. (C) MMP-7 promoter activity in normal, corrected, and FA cells. Cells were cotransfected with the reporter pGL3-hMMP-7 and phRL-TK, as internal control. Luciferase activity in AHH1 cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (D) MMP-7 mRNA steady-state levels as determined by quantitative RT-PCR on mRNA extracted from exponentially growing lymphoblasts and normalized to 18S rRNA content. To normalize the values among the different experiments, each time the ratio MMP-7/18S was considered equal to 1 in AHH1 cells. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (E) TNF-α release in MMP-7 inhibited FA-C cells. HSC536 and HSC536Neo cells were treated with MMP inhibitor (50 μM) or its solvent (DMSO) or left untreated. Supernatants were collected 24 hours after subculturing and cytokine release was determined by ELISA. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01. (F) TNF-α release in FA-C cells after the down-regulation of MMP-7 expression. Immunoblot shows the level of MMP-7 in supernatant of FANCC siRNA–MMP-7 down-regulated cells. A cross-reactive band (*) was used as a loading control. TNF-α accumulation in the supernatant of infected HSC536Neo cells was evaluated by ELISA. Histogram represents the mean reduction (in percentage) plus or minus SD of TNF-α accumulation observed in at least 3 independent experiments. *P < .01.
To validate the role of MMP-7 in TNF-α secretion in FA, we analyzed TNF-α accumulation in supernatant of FA-C cells in which MMP-7 activity has been down-regulated either by pharmacologic inhibition (Figure 3E) or by knocking-down MMP-7 expression by shRNA (Figure 3F). As reported, inhibition of MMP-7 activity or protein down-regulation by shRNA significantly reduced by 2- to 3-fold the level of TNF-α in the supernatant of FA-C cells. All together, these data suggest that MMP-7 overexpression is involved in TNF-α oversecretion in FA-C cells.
Stress-response pathways are aberrantly activated in FA-C cells
We next wanted to determine what could be the link between loss of FANCC function and MMP-7 overexpression/TNF-α oversecretion. Previous studies have implicated 2 masterpieces of the stress responses, the NF-κB and MAPK pathways, in MMP-7 transcriptional induction.47,,–50 TNF-α is able to activate NF-κB and MAPK signaling, which in turn could regulate transcription of genes including TNF-α itself.16,23,–25 Consequently, we questioned the relationship between the stress-response pathway activity and the TNF-α overproduction in FA.
A constitutive NF-κB activity was already found in simian virus 40 (SV40)–transformed FA fibroblasts.51 To determine whether this is also the case in EBV-transformed lymphoblasts, NF-κB transcriptional activity was analyzed by transfecting FA-C cells with a luciferase reporter construct containing 6 NF-κB consensus sequences (pNF-κB-Luc). As reported in Figure 4A, FA-C cells exhibited a higher level of basal NF-κB transcriptional activity compared with FANCC-proficient cells. Then, we attempted to evaluate the consequences of the inhibition of NF-κB activity on MMP-7 promoter activity. Unfortunately, since NF-κB activity is critical for the growth and survival of EBV-transformed B lymphocytes,52 NF-κB inhibition was lethal, impeding the evaluation of the implication of NF-κB in MMP-7 overexpression in FA-C lymphoblasts.
Aberrant activation of stress-response pathways in FA-C cells. (A) NF-κB transcriptional activity in lymphoblasts. NF-κB transcriptional activity was evaluated as increased luciferase activity in cell extracts isolated 48 hours after transfection with the reporter pNF-κB-Luc and phRL-TK. Luciferase activity in AHH1 cells was fixed to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. (B) Immunoblot showing the level of phosphorylation of key players of the MAPK pathways. Proteins were isolated from exponentially growing lymphoblasts. (C) TNF-α level in FA-C cells as function of MAPK activity. HSC536 and HSC536Neo cells were treated with SB203580 (p38 inhibitor, 10 μM), PD98059 (ERK inhibitor, 30 μM), and SP600125 (JNK inhibitor, 25 μM). Supernatants were collected 24 hours after subculturing and cytokine release was determined by ELISA. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01.
Aberrant activation of stress-response pathways in FA-C cells. (A) NF-κB transcriptional activity in lymphoblasts. NF-κB transcriptional activity was evaluated as increased luciferase activity in cell extracts isolated 48 hours after transfection with the reporter pNF-κB-Luc and phRL-TK. Luciferase activity in AHH1 cells was fixed to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. (B) Immunoblot showing the level of phosphorylation of key players of the MAPK pathways. Proteins were isolated from exponentially growing lymphoblasts. (C) TNF-α level in FA-C cells as function of MAPK activity. HSC536 and HSC536Neo cells were treated with SB203580 (p38 inhibitor, 10 μM), PD98059 (ERK inhibitor, 30 μM), and SP600125 (JNK inhibitor, 25 μM). Supernatants were collected 24 hours after subculturing and cytokine release was determined by ELISA. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01.
Successively, we evaluated the basal phosphorylation status of p38, ERK, and JNK, 3 major players of the MAPK pathway, in FANCC-proficient and -deficient lymphoblasts. As reported in Figure 4B, all 3 MAPK pathways appeared to be activated, that is, phosphorylated in absence of exogenous stimuli, in FA-C cells compared with healthy donor-derived or corrected cells. Having determined the spontaneous overactivation of MAPKs in FA-C cells, we sought to determine whether their activation was upstream or a consequence of the TNF-α oversecretion. For this purpose, we inhibited MAPKs activity using SB203580, PD98059, and SP600125 that are, respectively, p38-, ERK-, and JNK-specific inhibitors. As reported in Figure 4C, each of the MAPK inhibitors reduced TNF-α oversecretion in FA-C cells, suggesting that overactivation of MAPK pathways leads to TNF-α oversecretion in FA-C cells.
FANCC down-regulation leads to aberrant activation of stress-response pathways and MMP-7 overexpression
Since the observed anomalies in stress-response pathways could be simply related to the EBV immortalization of B lymphocytes, we decided to validate our data working with another cellular model. The transient inhibition of the expression of a target protein by siRNA approach is a powerful methodology allowing characterization of molecular pathways in heterologous cell models. Down-regulation of FANCC protein in HeLa cells recapitulates the major cellular characteristics of FA, that is, absence of FANCD2 monoubiquitination (Figure 5A) and cellular hypersensitivity to DNA cross-linking agents (data not shown). Knocking down FANCC expression in HeLa cells results in (1) a decrease of the NF-κB inhibitor IκB (Figure 5B), (2) the nuclear translocation of the NF-κB subunits p50 and p65 (Figure 5C), (3) an increased NF-κB transcriptional activity (Figure 5D) and (4) a spontaneous activation of MAPKs as shown by the phosphorylation status of p38, ERK, and JNK (Figure 5E). All these data, combined with the previously reported observations, are consistent with a basal overactivation of NF-κB and MAPK pathways as a consequence of FANCC loss-of-function. Furthermore, a higher production and intracellular accumulation of MMP-7 mRNA were also observed as evaluated by its promoter activity and the consequent mRNA accumulation (Figure 5F,G).
FANCC down-regulation leads to aberrant activation of stress-response pathways and MMP-7 overexpression in HeLa cells. (A) Immunoblot showing monoubiquitination of FANCD2 in FANCC siRNA–transfected cells. Forty-eight hours after siRNA transfection, cells were treated with mitomycin C (MMC) (500 ng/mL) or 8-methoxypsoralen (10−5M) + UVA (10 kJ/m2) (8-MOP) and lysed 24 hours later. FANCC down-regulation impacts on DNA damage–induced FANCD2 monoubiquitination. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Western blot analysis of IκBα in HeLa cells depleted of FANCC for 3 days. Actin was used as a loading control. (C) Detection of p50 and p65 subcellular localization by confocal immunofluorescence microscopy. HeLa cells treated with FANCC siRNA were subjected to immunostaining of p50 (green fluorescence) and p65 (red fluorescence) 72 hours after transfection. (D) NF-κB transcriptional activity in HeLa cells depleted for FANCC. Twenty-four hours after siRNA treatment, cells were cotransfected with the reporter pNF-κB-Luc and phRL-TK for 48 hours before the analysis of the luciferase activity. Luciferase activity in GFP-siRNA–transfected cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (E) Immunoblot showing the level of phosphorylation of MAPK players in cells with a reduced FANCC expression compared with mock-transfected and unperturbated cells. Cells were collected 72 hours after transfection and actin was used as a loading control. Vertical lines have been inserted to indicate a repositioned gel lane. (F) MMP-7 promoter activity in mock- and siRNA FANCC–transfected HeLa cells. Twenty-four hours after siRNA treatment, cells were cotransfected with the reporter pGL3-hMMP-7 and phRL-TK, and induced luciferase activity was measured 24 hours later. Luciferase activity in GFP-siRNA–transfected cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (G) MMP-7 mRNA steady-state level in FANCC-deprived or mock-transfected HeLa cells was determined by quantitative RT-PCR. The relative mRNA level (normalized as function of 18S rRNA content) in GFP-siRNA–transfected cells was fixed to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (H) MMP-7 promoter activity in 50 μM curcumin-treated FANCC-deprived cells assessed by luciferase activity as in panel F. *P < .01. (I) MMP-7 mRNA steady-state level in FANCC-deprived HeLa cells treated with curcumin (50 μM) as assessed by quantitative RT-PCR as in panel G. *P < .01. (J) MMP-7 promoter activity in siRNA FANCC–transfected HeLa cells treated with SB203580 (p38 inhibitor, 10 μM) and PD98059 (ERK inhibitor, 30 μM) for 18 hours. *P < .05 compared with DMSO treatment.
FANCC down-regulation leads to aberrant activation of stress-response pathways and MMP-7 overexpression in HeLa cells. (A) Immunoblot showing monoubiquitination of FANCD2 in FANCC siRNA–transfected cells. Forty-eight hours after siRNA transfection, cells were treated with mitomycin C (MMC) (500 ng/mL) or 8-methoxypsoralen (10−5M) + UVA (10 kJ/m2) (8-MOP) and lysed 24 hours later. FANCC down-regulation impacts on DNA damage–induced FANCD2 monoubiquitination. Vertical lines have been inserted to indicate a repositioned gel lane. (B) Western blot analysis of IκBα in HeLa cells depleted of FANCC for 3 days. Actin was used as a loading control. (C) Detection of p50 and p65 subcellular localization by confocal immunofluorescence microscopy. HeLa cells treated with FANCC siRNA were subjected to immunostaining of p50 (green fluorescence) and p65 (red fluorescence) 72 hours after transfection. (D) NF-κB transcriptional activity in HeLa cells depleted for FANCC. Twenty-four hours after siRNA treatment, cells were cotransfected with the reporter pNF-κB-Luc and phRL-TK for 48 hours before the analysis of the luciferase activity. Luciferase activity in GFP-siRNA–transfected cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .05. (E) Immunoblot showing the level of phosphorylation of MAPK players in cells with a reduced FANCC expression compared with mock-transfected and unperturbated cells. Cells were collected 72 hours after transfection and actin was used as a loading control. Vertical lines have been inserted to indicate a repositioned gel lane. (F) MMP-7 promoter activity in mock- and siRNA FANCC–transfected HeLa cells. Twenty-four hours after siRNA treatment, cells were cotransfected with the reporter pGL3-hMMP-7 and phRL-TK, and induced luciferase activity was measured 24 hours later. Luciferase activity in GFP-siRNA–transfected cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (G) MMP-7 mRNA steady-state level in FANCC-deprived or mock-transfected HeLa cells was determined by quantitative RT-PCR. The relative mRNA level (normalized as function of 18S rRNA content) in GFP-siRNA–transfected cells was fixed to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (H) MMP-7 promoter activity in 50 μM curcumin-treated FANCC-deprived cells assessed by luciferase activity as in panel F. *P < .01. (I) MMP-7 mRNA steady-state level in FANCC-deprived HeLa cells treated with curcumin (50 μM) as assessed by quantitative RT-PCR as in panel G. *P < .01. (J) MMP-7 promoter activity in siRNA FANCC–transfected HeLa cells treated with SB203580 (p38 inhibitor, 10 μM) and PD98059 (ERK inhibitor, 30 μM) for 18 hours. *P < .05 compared with DMSO treatment.
HeLa cells used in this study are able to activate NF-κB transcriptional activity in response to exogenous TNF-α, but were unable to secrete the cytokine after exposure to classical inducers such as phorbol-ester (PMA) or lipopolysaccharide (LPS). This suggests that HeLa cells are intrinsically unable to produce TNF-α, and, consequently, the basal overactivation of NF-κB and MAPK pathways observed following FANCC down-regulation cannot be the consequence of TNF-α production.
To determine whether NF-κB and/or MAPK hyperactivity were potentially involved in FANCC–MMP-7–TNF-α pathway, we measured MMP-7 promoter activity and mRNA cellular content after inhibition of both NF-κB and MAPK pathways in FANCC-depleted HeLa cells. For this purpose, we exposed cells to curcumin, a chemical agent able to inhibit both pathways.53 Firstly, we analyzed MMP-7 promoter activity. Twenty-four hours after FANCC-siRNA transfection, HeLa cells were transfected with the reporter of MMP-7 promoter, cultured for 24 hours, and then incubated with 50 μM curcumin for 24 hours before evaluation of the luciferase activity. As reported in Figure 5H, curcumin reduced MMP-7 promoter activity to basal level. Then, 48 hours after FANCC-siRNA transfection, HeLa cells were incubated for 24 hours with 50 μM curcumin before RNA extraction for quantitative RT-PCR analysis. As shown in Figure 5I, curcumin clearly reduced the amount of MMP-7 mRNA in FANCC-depleted cells. These results suggest that NF-κB, MAPKs or both may be implicated in MMP-7 overexpression in FANCC-depleted HeLa cells.
To decipher which target(s) of curcumin is involved, we transfected a dominant-negative mutant inhibitor of NF-κB activation (pCMV-IκBM) in HeLa cells. Overexpression of IκBM fails to modify MMP-7 promoter activity, whereas NF-κB transcriptional activity was significantly reduced (data not shown). This demonstrates that NF-κB is not responsible for MMP-7 overexpression in FANCC-depleted HeLa cells. Next, MMP-7 promoter activity was measured in these cells exposed to MAPK-specific inhibitors. Treatment with PD98059 (ERK inhibitor) but not SB203580 (p38 inhibitor) decreases MMP-7 promoter activity (Figure 5J). Curiously, treatment with SP600125 (JNK inhibitor) leads to a basal overactivation of MMP-7 promoter activity even in the siGFP-transfected cells, impeding the analysis of JNK involvement in MMP-7 expression in HeLa cells (data not shown). Taken together, these results suggest that MMP-7 overexpression in FANCC-deficient cells is induced mainly via ERK activation.
FA gene mutations lead to MMP-7 overexpression
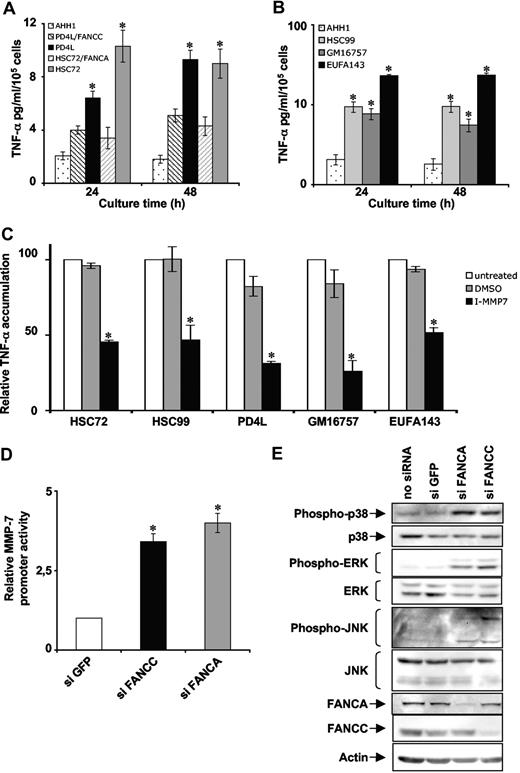
To generalize our findings, we extended our study to other FA complementation groups. As it was observed for HSC536 cells, ectopic expression of FANCC in PD4L cells (another FA-C cell line) or FANCA in HSC72 cells (a FA-A cell line) significantly reduced TNF-α accumulation in the supernatant (Figure 6A). FA-A, FA-B, FA-C, and FA-D1 cells were already shown as overproducers of TNF-α36 (and Figure 1A). We demonstrated here that FA-F and FA-G lymphoblasts display a similar characteristic. Indeed, FA-A (HSC99), FA-F (GM16757), and FA-G (EUFA143) cells accumulated 4 to 10 times more TNF-α in the growth medium than FANC-proficient lymphoblasts (AHH1) (Figure 6B). In addition, MMP-7 inhibition significantly reduced the amount of TNF-α in supernatant of FA-A (HSC72 and HSC99), FA-C (PD4L), FA-F (GM16757), and FA-G (EUFA143) cells as it was observed for the FA-C cell line HSC536 (Figure 6C). So, all the FANCcore complex lymphoblasts examined show the same defect in TNF-α overproduction and MMP-7 overactivation. The consequences of FANCcore complex protein depletion on MMP-7 promoter activity and MAPK network activation were evaluated using HeLa cells subjected to siRNA-mediated FANCA knockdown. To assess MMP-7 mRNA production, FANC-siRNA down-regulated cells were transfected with the reporter of MMP-7 promoter and cultured for 48 hours before the analysis. MMP-7 promoter activity was clearly up-regulated in HeLa cells depleted of FA proteins (Figure 6D). FANCA-depleted HeLa cells also presented a spontaneous overactivation of MAPKs as shown by phosphorylation status of p38, ERK, and JNK (Figure 6E).
FA gene mutation leads to MMP-7 overexpression and MAPK pathway activation. (A) TNF-α accumulation in the supernatant of AHH1 (normal), PD4L (FA-C), PD4L/FANCC (FA-C corrected), HSC72 (FA-A), and HSC72/FANCA (FA-A corrected) lymphoblasts. Supernatants were collected as described in Figure 1A. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01 compared with gene-corrected cells. (B) TNF-α accumulation in the supernatant of AHH1 (normal), HSC99 (FA-A), GM16757 (FA-F), and EUFA143 (FA-G) cell lines. Supernatants were collected as described in Figure 1A. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01. (C) HSC72, HSC99, PD4L, GM16757, and EUFA143 cells were treated with MMP inhibitor as described in Figure 3F and TNF-α content in supernatants was measured by ELISA. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01. (D) MMP-7 promoter activity in siRNA FANC–transfected HeLa cells. Cells were cotransfected with the reporter pGL3-hMMP-7 and phRL-TK, and induced luciferase activity was measured 24 hours later. Luciferase activity in GFP-siRNA–transfected cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (E) Western blot analysis of MAPK phosphorylation in siRNA FANC–transfected HeLa cells. Cells were collected 72 hours after transfection, and actin was used as a loading control.
FA gene mutation leads to MMP-7 overexpression and MAPK pathway activation. (A) TNF-α accumulation in the supernatant of AHH1 (normal), PD4L (FA-C), PD4L/FANCC (FA-C corrected), HSC72 (FA-A), and HSC72/FANCA (FA-A corrected) lymphoblasts. Supernatants were collected as described in Figure 1A. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01 compared with gene-corrected cells. (B) TNF-α accumulation in the supernatant of AHH1 (normal), HSC99 (FA-A), GM16757 (FA-F), and EUFA143 (FA-G) cell lines. Supernatants were collected as described in Figure 1A. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01. (C) HSC72, HSC99, PD4L, GM16757, and EUFA143 cells were treated with MMP inhibitor as described in Figure 3F and TNF-α content in supernatants was measured by ELISA. Data presented are the mean (± SD) of at least 3 independent experiments. *P < .01. (D) MMP-7 promoter activity in siRNA FANC–transfected HeLa cells. Cells were cotransfected with the reporter pGL3-hMMP-7 and phRL-TK, and induced luciferase activity was measured 24 hours later. Luciferase activity in GFP-siRNA–transfected cells was considered equal to 1 in each experiment. Data presented are the mean (± SD) of at least 3 independent experiments done in triplicate. *P < .01. (E) Western blot analysis of MAPK phosphorylation in siRNA FANC–transfected HeLa cells. Cells were collected 72 hours after transfection, and actin was used as a loading control.
Taken together, the present data indicate that TNF-α overproduction, MMP-7 overexpression, and basal MAPK overactivation are general features of FANCcore complex–depleted cells.
Discussion
The aim of this work was to comprehend the origin of the TNF-α overproduction in FA. Combining several approaches, we demonstrated that FANC pathway loss-of-function induces elevated NF-κB transcriptional activity, overactivation of all 3 MAPK pathways, overexpression of MMP-7, and oversecretion of TNF-α (Figure 7A). Using both molecular and pharmacologic inhibitors, we provided evidence that the ERK pathway is the main player responsible for the elevated expression of the MMP-7 mRNA in FA, which is the key event linking FANC mutation to the high TNF-α shedding from the cytoplasmic membrane in FA. Besides this pathway, TNF-α oversecretion is reduced by p38- and JNK-specific inhibitors. This suggests that the aberrant activation of stress-response pathways participates in the clinical and cellular FA phenotype acting on multiple targets and pathways. A speculative working model that recapitulates in a hierarchic order the events induced by FANC loss-of-function is summarized in Figure 7A,B. It is clear that MAPKs and NF-κB stress pathways are involved in the expression of several myelosuppressive cytokines including interferons, TGF-β, MIP-α, and MCP-1, and in this regard the model in Figure 7A is extremely simplified. However, we and others failed to detect the presence of cytokines other than TNF-α in primary as well as in immortalized FA samples34,35 (and data not shown), indicating that overexpression of TNF-α is specifically linked to FANC loss-of-function–mediated stress pathway activation
A model for TNF-α oversecretion in FA. (A) The FANC–TNF-α pathway. (B) The FA network (see “Discussion”).
A model for TNF-α oversecretion in FA. (A) The FANC–TNF-α pathway. (B) The FA network (see “Discussion”).
We used 2 complementary cellular models to perform our analysis: patient-derived lymphoblasts and HeLa cells in which FANC expression was down-regulated using siRNA technology. Comparing FA-C lymphoblasts to their isogenic ectopically corrected counterpart, we showed that wild-type FANCC cDNA is able to normalize TNF-α overproduction in FA-C cells, demonstrating that TNF-α overproduction is directly linked to FANCC mutation. The same results were observed with a FA-A cell line pair, suggesting that at least FANCcore complex loss-of-function leads to TNF-α overproduction. Although a slight increase in TNF-α mRNA transcription/stabilization in FA-C cells was observed, it seems devoid of major biologic significance since we failed to observe any differences in protein synthesis between FANC-mutated and wild-type cells. Instead, we demonstrated that the extracellular accumulation of the cytokine in FA cells is the results of a high rate of cleavage from the cell membrane.
TNF-α shedding is mainly related to TACE activity.19,20 Among other proteases able to cleave membrane-anchored TNF-α,21 MMP-7 is the only one known to correctly cleave TNF-α precursor.22 It has been proposed that TACE and MMP-7 are part of independent mechanisms by which TNF-α could be shed from the cell surface. TACE is involved in inducible release such as in response to PMA or LPS treatment, while MMP-7 is involved in the constitutive release of the cytokine.54,55 In line with these data, MMP-7 is overexpressed at both mRNA and protein level in FA-C cells, and its overexpression was obtained in HeLa cells by siRNA-mediated depletion of FANCC. Furthermore, the pharmacologic inhibition of MMP-7 activity as well as the down-regulation of its expression by specific shRNA significantly reduced the cytokine release in the supernatant of FA-C cells. The key role of MMP-7 in TNF-α overproduction is also observed in FA-A, FA-F, and FA-G cells, suggesting that the aberrant expression of MMP-7 is a general characteristic of FA. These results provide the first evidence of the mechanism involved in the overproduction of TNF-α in FA syndrome.
Exposure of cells to TNF-α activates several signaling pathways, including NF-κB and MAPK networks. Inversely, activation of both NF-κB and MAPKs plays an important role in the induction of many cytokines, including TNF-α itself.16,23,–25 Overactivation of NF-κB activity was previously reported in SV40-transformed FA fibroblasts.51 Here we demonstrated that EBV-transformed FA lymphoblastoid cells are also characterized by an increased NF-κB and MAPK activity, and these phenotypic modifications have been reproduced in FANCC and FANCA siRNA–depleted HeLa cells. Since these cells are intrinsically unable to secrete TNF-α, NF-κB, and MAPKs, activation is not due to TNF-α overproduction but rather linked to FANC depletion. In line with these data, using specific inhibitors of MAPKs in lymphoblasts and in FANC-depleted HeLa cells, evidence is provided that the FANC-dependent TNF-α overproduction and MMP-7 overexpression are dependent on MAPK activity. These data do not exclude that TNF-α may still play a role in maintaining or in exacerbating NF-κB and/or MAPK activity in FA cells.
Observations described in this study corroborate data from other published works and present arguments that propose a functional connection between the TNF-α and the FANC pathways. On one hand, the myelosuppressive cytokine TNF-α is increased in BM and serum of FA patients, as well as in culture medium from EBV-transformed FA lymphoblasts34,–36 as a consequence of MMP-7 induction (this work). Inversely, TNF-α acts on the FANC pathway in normal cells. Indeed, the expression of FANCG and FANCA and the FANCA/FANCG complex in the nucleus increase after TNF-α treatment in an NF-κB–dependent manner.56 In contrast to FANCG, the amount of FANCC associated with FANCA is reduced following treatment with TNF-α.57 Interestingly, in Fancc-null mice, which do not show spontaneously hematologic abnormalities,58,,–61 TNF-α oversecretion was not observed.38 Recently, Pang and collaborators, using different approaches in Fancc−/− mice, including treatment with LPS or TNF-α, reported that the cytokine, besides its negative action on mice BM development, is also implicated in overactivation of p38 and JNK, phosphorylation of p53 and H2AX, induced DNA damage, chromosome aberrations, and apoptosis (Sejas et al62 and Zhang et al63 ). All these characteristics are shared by human FA cells64 (this paper and F.R., unpublished data, January 2007). Moreover, in agreement with a role of TNF-α in FA genetic instability, our previous data have demonstrated that TNF-α inhibition partially rescues FA hypersensitivity to cross-linking agent treatment at both the cellular and chromosomal level.36 All together, these data strongly suggest that TNF-α could play a role in both the BM failure and the cellular and chromosomal hypersensitivity to DNA damage observed in FA.
FANC proteins play a role in defining cellular tolerance to cross-linking agents via their function in DNA repair/cell-cycle checkpoint.65,66 However, they are also functionally or biochemically associated with several other proteins not involved in the enzymology of DNA repair, including several redox regulators: NADPH cytochrome-P450 reductase, glutathione S-transferase P1-1, and the peroxiredoxin-3.67,,–70 The reported interactions with redox regulators are involved in both the pro-oxidant state and the oxygen hypersensitivity that characterize the FA cells.14 Both oxidative stress and impaired DNA repair may contribute to the activation of the ATM pathway recently reported in FA.64 Interestingly, it has been described that both ATM and reactive oxygen species (ROSs) may induce NF-κB and MAPKs.71,72 Consequently, we speculate (Figure 7B) that FA cells suffer permanent stress due to both intracellular ROS increase and accumulation of endogenous DNA damage that leads to MAPK and NF-κB signaling activation. MAPK activation, in turn, contributes, through the alteration of the MMP-7 expression, to the TNF-α overproduction. Since TNF-α is able to activate NF-κB and MAPKs, these factors form an autocrine loop that results in the escalation of their own levels and, consequently, of the severity of the pathogenesis. In accord with this, it has been shown that NF-κB and MAPK overactivation is causative of BM failure, MDS, and leukemia73,–75 and that MMP-7 overexpression promotes leukemia, facilitating migration and invasion of the leukemic cells.76 Importantly, from a clinical point of view, NF-κB and MAPK inhibition was reported as beneficial in BM failure and leukemia.77,–79
Currently bone marrow transplantation is the best treatment available for the hematopoietic manifestations of FA. Our data provide a strong rationale for clinical strategies based on the use of pharmacologic approaches to block stress-response pathways and/or TNF-α activity or shedding in FA patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank J. Grosjean, G. Cherubini, and J. H. Guervilly for helpful discussion and J. A. Dugas Du Villard, Y. Lecluse, S. Queille, and J. Abdelali for excellent technical assistance. We also thank the following for kindly sharing material: Dr G. C. Bagby, Dr J. P. Bidwell, Dr C. Dufour, Dr L. Matrisian, Dr D. Schindler, Dr J. Surrallés, Dr D. Trono, Dr I. Udalova, and the Fanconi Anemia Research Fund.
This work was supported by grant from La Ligue contre le Cancer (Equipe labellisée 2006). D.B. was a fellow of the Ministere de l'Enseignement Supérieur et de la Recherche (France) and the Association pour la Recherche sur le Cancer, France.
Authorship
Contribution: D.B. performed research, analyzed data, and wrote the paper; G.M.-A. performed research; F.S. made lentiviral construction, production, and transduction; F.R. supervised the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Filippo Rosselli, FRE2939 du CNRS, Institut Gustave Roussy PR2, 39 rue Camille Desmoulins, 94805 Villejuif Cedex, France; e-mail: rosselli@igr.fr.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal