Abstract
High-molecular-weight kininogen (HK) plays an important role in the assembly of the plasma kallikrein-kinin system. While the human genome contains a single copy of the kininogen gene, 3 copies exist in the rat (1 encoding K-kininogen and 2 encoding T-kininogen). Here, we confirm that the mouse genome contains 2 homologous kininogen genes, mKng1 and mKng2, and demonstrate that these genes are expressed in a tissue-specific manner. To determine the roles of these genes in murine development and physiology, we disrupted mKng1, which is expressed primarily in the liver. mKng1−/− mice were viable, but lacked plasma HK and low-molecular-weight kininogen (LK), as well as ΔmHK-D5, a novel kininogen isoform that lacks kininogen domain 5. Moreover, despite normal tail vein bleeding times, mKng1−/− mice displayed a significantly prolonged time to carotid artery occlusion following Rose Bengal administration and laser-induced arterial injury. These results suggest that a single gene, mKng1, is responsible for production of plasma kininogen, and that plasma HK contributes to induced arterial thrombosis in mice.
Introduction
High-molecular-weight kininogen (HK) is an important member of the plasma kallikrein-kinin system,1,2 the activation of which may contribute to the manifestations of disorders such as hereditary angioedema,3 sepsis,4 ulcerative colitis,5 and Alzheimer disease.6 These effects are mediated, at least in part, by the nonapeptide bradykinin (BK) following its release from domain 4 of HK or low-molecular-weight kininogen (LK) by plasma kallikrein. BK is a potent inflammatory mediator that causes vasodilation and enhanced capillary permeability, induces pain,1 and stimulates the production of nitric oxide and prostacyclin from endothelial cells.7 In the setting of vascular damage, BK stimulates smooth muscle proliferation and intimal hypertrophy.1,2
The human kininogen gene contains 11 exons, and generates both HK and LK mRNA transcripts through alternative splicing. Structurally, both HK and LK contain an identical heavy chain, which consists of domains 1, 2, and 3. However, while HK contains an approximately 56 kDa light chain that consists of domains 5 (D5H) and 6, LK contains a unique light chain (D5L) of only approximately 4 kDa.1 In both proteins, the heavy and light chains are linked by domain 4, which contains the BK nonapeptide.
Plasma kallikrein mediates the release of BK from HK, resulting in the generation of 2-chain high-molecular-weight kininogen (HKa), which contains the HK heavy and light chains joined by a single disulfide bond between Cys10 of domain 1 and Cys604 of domain 6. Rotary shadowing microscopy studies have demonstrated important structural differences between HK and HKa,8 largely reflecting increased exposure of kininogen domain 5 (D5) in the latter. These differences may account for the ability of HKa to induce apoptosis of proliferating endothelial cells and inhibit angiogenesis, a property that resides within specific endothelial binding regions within domain 5.9,10 HK domain 5 and domain 5–derived peptides also block melanoma cell metastasis,11 inhibit platelet adhesion and aggregation,12 and regulate platelet-leukocyte interactions.13
Though HKa is a potent inhibitor of angiogenesis, its precursor, HK, has been reported to stimulate angiogenesis through the release of bradykinin.14 Angiogenesis has been reported to be diminished in Brown–Norway Katholiek rats, in which a spontaneous mutation in the kininogen gene causes deficient kininogen secretion and reduced plasma kininogen levels.15 However, a murine model of kininogen deficiency is not presently available. To develop such a model, we characterized the murine kininogen gene, finding that the murine genome contains 2 homologous copies of this gene located in a head-to-head orientation on chromosome 16. HK mRNA transcripts derived from these 2 genes are differentially expressed in murine tissues. Disruption of one of the 2 kininogen genes, mKng1, leads to the loss of immunologically detectable plasma kininogen. Moreover, mice deficient in plasma kininogen are protected from arterial thrombosis induced by vascular injury, suggesting that plasma HK contributes to intravascular clot formation after vessel injury.
Methods
Detection and cloning of murine kininogen genes
Genomic DNA was isolated from the livers of C57BL/6 and 129J mice using TRIzol (Invitrogen, Carlsbad, CA). A total of 10 μg of purified high-molecular-weight DNA was digested with EcoRV (Promega, Madison, WI), and the DNA fragments were separated using 0.9% agarose gel electrophoresis and transferred to an Immobilon-Ny+ membrane (Millipore, Billerica, MA). Southern blot analysis was performed as previously described,16 using a [α32P] dCTP-labeled 380-bp polymerase chain reaction (PCR) fragment comprising exon 4 through intron 4 of the murine kininogen gene as defined in ENSEMBL17 as a probe.
A murine clone (BAC 452G1) that contained both kininogen genes, derived from the RPCI-23 library and constructed in pBACe 3.6, was purchased from the Research Institute of the Children's Hospital of Oakland, CA. The size of individual introns was determined by PCR analysis using the Expand High Fidelity PCR system (Roche Diagnostics, Basel, Switzerland), or through computer-aided analysis of BAC 452G1.
Tissue specific expression of mKng1- and mKng2-derived HK mRNA
To analyze the expression patterns of HK mRNA derived from each of the kininogen genes, RNA was isolated from murine tissues using TRIzol. Total RNA (1 μg) was reverse transcribed using Moloney murine leukemia virus (MMLV) polymerase, and the product of the first-strand reaction was amplified by PCR using specific primers (forward, 5′-ATGGTCCCGACTGTGAAATGCCAAG-3′; reverse, 5′-CTATCTCTGGGTAGTCTGCTTTACAC-3′; bolded nucleotides in the primer sequences represent start and stop codons for use in studies described in Figure 6, in which in vitro translation of mKng mRNA was performed). These primers were designed to frame gaps present within exon 10 of mKng2 (Figure 1B), yielding mKng1- and mKng2-derived PCR products that differ in size. Concurrent amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward primer, 5′-CCCTTCATTGACCTCAACTACATGGT-3′; reverse primer, 5′-GAGGGGCCATCCACAGTCTTCTG-3′) was performed to ensure that equal amounts of cDNA were added to each PCR reaction.
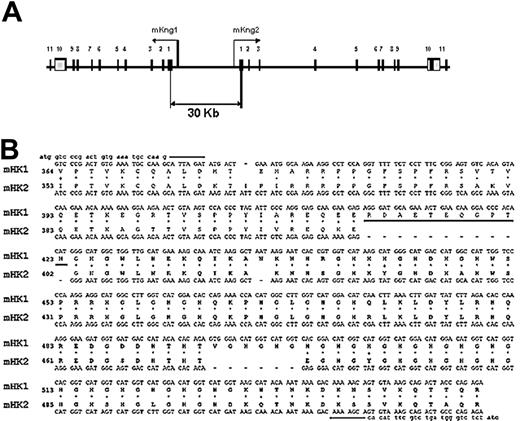
Organization of the kininogen gene in mice. (A) Map of the DNA fragment of murine BAC clone RP23–452G1 containing mKng1 and mKng2. Exons are denoted by numbered boxes, and arrows indicate the direction of gene transcription. (B) Alignment of the mKng1 and mKng2 cDNA regions that were used for RT-PCR amplification. The amplified region spanned the carboxyl-terminal region of domain 3 through the amino-terminal region of domain 6. The 2 gaps in mHK2 responsible for the different sizes of the PCR products from the 2 cDNAs are denoted by dashes. Also shown are the corresponding polypeptide sequences of the mHK1 and mHK2 proteins. The ERDAETEQGPTH sequence used to raise the mHK1-specific antibody is underlined.
Organization of the kininogen gene in mice. (A) Map of the DNA fragment of murine BAC clone RP23–452G1 containing mKng1 and mKng2. Exons are denoted by numbered boxes, and arrows indicate the direction of gene transcription. (B) Alignment of the mKng1 and mKng2 cDNA regions that were used for RT-PCR amplification. The amplified region spanned the carboxyl-terminal region of domain 3 through the amino-terminal region of domain 6. The 2 gaps in mHK2 responsible for the different sizes of the PCR products from the 2 cDNAs are denoted by dashes. Also shown are the corresponding polypeptide sequences of the mHK1 and mHK2 proteins. The ERDAETEQGPTH sequence used to raise the mHK1-specific antibody is underlined.
Assessment of mKng expression by in vitro translation
Liver- and kidney-derived mKng cDNAs were used as templates for amplification of mKng1 and mKng2 sequences encoding Val364 to Arg540 (spanning the C-terminus of domain 3 through the C-terminus of domain 5). PCR products were cloned into pBluescript II KS+, and capped mRNAs were synthesized using the mMessage mMachine T7 kit (Ambion, Austin TX). In vitro translation was performed using the Flexi rabbit reticulocyte lysate system (Promega) in the presence of 35S-Met (555 MBq [15 mCi/mL]; Amersham, Piscataway NJ), and the protein products detected using 15% SDS-PAGE and autoradiography. These recombinant polypeptides were used to characterize an affinity-purified rabbit anti-mHK1 antibody (see Figure 6).
Promoter analysis
MatInspector Professional (Genomatix, München, Germany) was used to analyze putative promoter regions of mKng1 and mKng2. Regions less than 1 kb upstream of the transcription start sites of both genes were assessed using the BAC clone RP23-452G1 sequence (GenBank accession number AC154540). Search parameters for transcription factor binding sites included a core matrix similarity threshold of 1.00 and a matrix similarity threshold of greater than 0.900. Repetitive elements were identified using Repeat Masker (Genetic Information Research Institute, Mountain View, CA).
Disruption of mKng1
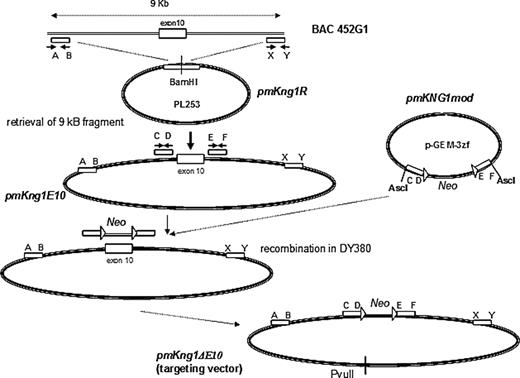
We hypothesized that mKng1, which is highly expressed in liver, was the major source of plasma kininogen, and disrupted this gene. Our strategy for gene disruption was based on recombineering,19 and parallels that of Hu et al.20 This strategy used a 9-kb genomic fragment of BAC clone RP23-452G1 to construct a targeting vector in which an mKng1 knockout allele was created by replacing exon 10 of mKng1 with a PGKNeo cassette (exon 10 contains the splicing site for HK and LK). To construct this vector, BAC clone RP23-452G1 was introduced into DY380 cells by electroporation (Figure 2). A retrieval vector, pmKng1R, was constructed that contained 2 PCR-amplified 400-bp fragments corresponding to the 5′ and 3′ regions of the 9-kb genomic fragment of BAC clone RP23-452G1 cloned into PL253, which contains an Mc1-driven thymidine kinase (TK) cassette for negative selection in embryonic stem (ES) cells. pmKng1R was linearized and electroporated into DY380 cells containing BAC clone RP23-452G1. Induced recombination in DY380 resulted in retrieval of the 9-kb genomic fragment containing mKng1 exon 10 from the bacterial artificial clone (BAC clone RP23-452G1), yielding an intermediate vector called pmKng1E10. Next, a modification cassette was prepared. First, a loxP-PGKNeo-loxP cassette was constructed. In parallel, 2 small 400-bp genomic fragments flanking the 5′ and 3′ ends of exon 10 were amplified and cloned into pGEM-3zf. Insertion of the PGKNeo containing cassette between the 2 genomic fragments gave rise to the plasmid pmKng1mod containing the 5′-arm-loxP-PGKNeo-loxP-3′ arm modification cassette. This cassette was excised from pmHK1mod and introduced into DY380 cells together with pmKng1E10. Induced recombination between the homologous sequences in pmKng1E10 and the modification cassette led to the replacement of exon 10 with the loxP-PGKNeo-loxP cassette and gave rise to the targeting vector pmKng1-ΔE10 (Figure 2). The targeting vector was linearized using PvuII prior to electroporation into murine ES cells.
Construction of an mKng1-targeting vector. Gene disruption was accomplished using recombineering, with details concerning the construction of the mKng1-targeting vector provided in “Methods.” Briefly, the strategy used a 9-kb genomic fragment of BAC452G1 to construct a targeting vector with which an mKng1 knockout allele was created. In this allele, exon 10 of mKng1 was replaced by a PGKNeo cassette (exon 10 contains the splicing site for HK and LK; thus, disruption of this exon should prevent transcription of HK and LK mRNA).
Construction of an mKng1-targeting vector. Gene disruption was accomplished using recombineering, with details concerning the construction of the mKng1-targeting vector provided in “Methods.” Briefly, the strategy used a 9-kb genomic fragment of BAC452G1 to construct a targeting vector with which an mKng1 knockout allele was created. In this allele, exon 10 of mKng1 was replaced by a PGKNeo cassette (exon 10 contains the splicing site for HK and LK; thus, disruption of this exon should prevent transcription of HK and LK mRNA).
Electroporation and selection of mKng1−/− ES cells
pmKng1ΔE10 was electroporated into murine ES cells by the transgenic and targeting service of the Department of Genetics, University Hospitals Research Institute (Cleveland, OH). Briefly, cuvettes containing 0.8 mL cells at a concentration of 7 × 106 cells/mL and 40 μg of linearized DNA were pulsed at 0.240 kV and 500 μF using a Bio-Rad gene pulser (Bio-Rad, Cupertino, CA). After 24 hours, medium containing G418 (200 μg/mL) was added to the cells and changed daily. After 7 days, selected colonies were screened by PCR.
The screening strategy for evaluating homologous recombination was based on the fact that homologous recombination of the targeting construct with chromosomal DNA within the kininogen gene locus caused changes in the DNA digestion profile that were detectable by Southern blotting using a 5′ external probe and a 3′ internal probe. The 5′ probe detected a 6.2-kb fragment after EcoRI digestion of the wild-type allele, but only a 4.6-kb fragment after homologous recombination with the targeting vector, while the 3′ probe detected an approximately 10-kb fragment upon BglII digestion of the wild-type allele, but only an approximately 8.5-kb fragment after recombination.
Generation of mKng1−/− mice
A total of 10 to 15 cloned ES cells containing disrupted mKng1 exon 10 were injected into 3.5-day C57BL/6J blastocysts using blunt tipped pipettes and a Leitz manipulation system (Leitz, Wetzler, Germany). Blastocysts were implanted into 2.5-day-old CD1 pseudopregnant recipients by uterine transfer. The degree of chimerism in the offspring was assessed 2 weeks after birth by identification of agouti coat color. Strong chimeras were tested for germ-line transmission by crossing with CD1 or C57BL/6J mice. Resulting agouti offspring were tested for transmission of the targeted allele by Southern blotting and PCR. Homozygous null offspring were produced by intercrossing heterozygotes.
Detection of HK proteins in murine plasma
We used 2 approaches to detect the high- and low-molecular-weight gene products of mKng1 and mKng2 in plasma and tissue. First, we performed immunoblotting using an antibradykinin antiserum (Biogenesis, Kingston, NH) that recognized the BK sequence in HK and LK, as well as an affinity-purified mHK1-specific polyclonal antibody. The latter was raised against an 11–amino acid peptide sequence (ERDAETEQGPTH) present in mHK1 but not mHK2 (Figure 1B). The specificity of this antibody was confirmed by immunoblotting against in vitro–translated polypeptides corresponding to sequences within mHK1 and mHK2, as described.
We also used plasma clotting assays to measure HK procoagulant activity in wild-type and mKng1-deficient murine plasma. First, we measured the activated partial thromboplastin time (APTT) using plasma from 8 mKng1−/− and 8 wild-type control mice. Next, we constructed an HK procoagulant activity standard curve by adding diluted pooled normal mouse plasma to human kininogen–deficient plasma (Williams plasma, generously donated by the late Mayme Williams, Philadelphia, PA). HK procoagulant activity in individual plasma samples from mKng1−/− mice (strain 129J) and wild-type littermates were then assayed at 1:10 and 1:20 dilutions.
As another approach to assess kininogen levels in murine plasma, we measured plasma bradykinin concentrations by radioimmunoassay. mKng1−/− and wild-type littermate control mice were anesthesized, and blood was drawn by direct cardiac puncture into a solution of EDTA (2 mg/mL, final concentration). Plasma was separated by centrifugation at 13 000g for 5 minutes, and flash-frozen in liquid nitrogen. The concentrations of BK in filtered plasma were measured as previously described; the origin of all samples was blinded at the time of BK measurement.21
Assessment of thrombosis time in mKng1−/− animals
To assess the propensity for mKng1−/− mice to develop thrombosis following arterial injury, animals were studied using the Rose Bengal carotid artery thrombosis model.22,23 Briefly, mice were anesthetized with 40 mg/kg sodium pentobarbital and immobilized on an operating table. A midline incision was made to expose the right common carotid artery and a Doppler flow probe (model 0.5 VB; Transonic Systems, Ithaca, NY) was placed under the vessel. The probe was connected to a flow meter (Transonic model T106) and was interpreted using a computerized data acquisition program (Windaq; DATAQ Instruments, Akron, OH). A total of 120 μL of a 50 mg/kg solution of Rose Bengal (Fisher Scientific, Fairlawn, NJ) was then injected into the tail vein. After injection, a green laser light (Melles Griot, Carlsbad, CA) at a wavelength of 540 nm was applied 6 cm from the carotid artery. Flow was monitored continuously from the onset of injury. The time to occlusion was determined after the vessel remained closed with cessation of blood flow for 20 minutes.
Results
Analysis of the murine kininogen gene locus
The human kininogen gene locus contains 11 exons and generates 2 alternatively spliced transcripts, leading to production of HK and LK.15,24,25 HK and LK each contain a common heavy chain, consisting of domains 1, 2, and 3. The light chain of HK consists of the exon 10–derived product containing domains 5 and 6, while that of LK consists of an alternatively spliced, approximately 4-kDa exon 11 product.1 In both proteins, the heavy and light chains are linked by domain 4, which contains the bradykinin nonapeptide. Since previous work suggested that the murine kininogen gene locus may have a similar organization,25 we analyzed this locus in mice. Southern blot analysis of EcoRV-digested murine genomic DNA using a 380-bp probe spanning exon 4 and part of intron 4 of the murine kininogen gene revealed 2 distinct fragments in DNA from C57BL/6 and 129J mice, suggesting that the gene was duplicated (Figure 3). To define the extent of duplication, we searched the ENSEMBL database17 using the DNA sequences for mouse HK (accession no. D84435) and LK (accession no. D84415), and confirmed the presence of 2 kininogen-like DNA sequences in the Mus musculus chromosome 16 genomic contig (strain C57BL/6J). One sequence contained 11 exons and matched the GenBank18 sequences of murine HK and LK cDNA. The second sequence was 91% identical. An additional search through the high-throughput genome sequence section of GenBank revealed genomic contig RP23–452G1 (accession no. 154450), which contains a consecutive sequence spanning the 2 kininogen genes.
Southern blot analysis of the murine kininogen gene. Genomic DNA from C57Bl/6J and 129J mice was digested with EcoRV and probed using a 32P-dCTP–labeled PCR product spanning intron 4 and exon 4 of the kininogen gene. This probe hybridized to 2 distinct fragments of genomic DNA, suggesting the presence of 2 kininogen genes.
Southern blot analysis of the murine kininogen gene. Genomic DNA from C57Bl/6J and 129J mice was digested with EcoRV and probed using a 32P-dCTP–labeled PCR product spanning intron 4 and exon 4 of the kininogen gene. This probe hybridized to 2 distinct fragments of genomic DNA, suggesting the presence of 2 kininogen genes.
These findings confirmed that the murine kininogen gene was duplicated, and led to the development of a map of the genomic locus (Figure 1A) revealing 2 homologous genes in a head-to-head orientation. The distance between the genes is approximately 30 kb, and both contain open reading frames. The most significant difference in exon sequences between the 2 genes occurs within exon 10, where mKng2 has 2 in-frame gaps of 33 and 18 nucleotides (Figure 1B). In addition, the sizes of individual introns varied markedly between the 2 genes (Table 1), with some introns in mKng2 substantially larger than those in mKng1.
Nuceotide length of mKng1 and mKng2 introns
| Intron no. . | mKng1 . | mKng2 . |
|---|---|---|
| 1 | 1743 | 1558 |
| 2 | 1117 | 2126 |
| 3 | 6001 | 12711 |
| 4 | 1347 | 7954 |
| 5 | 4032 | 3316 |
| 6 | 1189 | 540 |
| 7 | 2463 | 2490 |
| 8 | 103 | 102 |
| 9 | 1403 | 8704 |
| 10 | 2427 | 2177 |
| Intron no. . | mKng1 . | mKng2 . |
|---|---|---|
| 1 | 1743 | 1558 |
| 2 | 1117 | 2126 |
| 3 | 6001 | 12711 |
| 4 | 1347 | 7954 |
| 5 | 4032 | 3316 |
| 6 | 1189 | 540 |
| 7 | 2463 | 2490 |
| 8 | 103 | 102 |
| 9 | 1403 | 8704 |
| 10 | 2427 | 2177 |
Based on these results, we concluded that the mouse kininogen genomic locus spans approximately 95 kb and contains 2 homologous genes in a head-to-head orientation. mKng1 spans approximately 23 kb, while mKng2 spans approximately 43 kb due to larger intron sizes (calculated using the GenBank sequence of BAC DNA: AC154540).
Tissue-specific expression of mHK1- and mHK2-derived HK mRNA transcripts
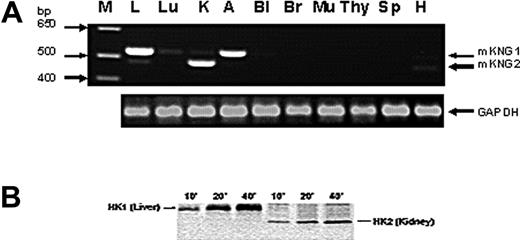
To determine whether HK mRNA derived from the 2 kininogen genes was expressed in a tissue-specific manner, we used reverse transcription (RT)–PCR using primers that spanned exon 10 sequences within mKng1 and mKng2 that encompass the region within this exon in which gaps exist in mKng2 (Figure 1B); this strategy leads to amplification of an mKng1-derived HK mRNA of larger size than that of mKng2. We observed mKng1 HK mRNA expression primarily in the liver and adrenal glands, while mKng2 HK mRNA was most abundant in the kidney (Figure 4A).
Tissue-specific expression of mKng1 and mKng2. (A) RT-PCR of mKng1- and mKng2-derived HK mRNA transcripts in murine tissues. PCR products were resolved using 0.9% agarose gel electropheresis, and stained using ethidium bromide. Arrows indicate the location of PCR products from mKng1- and mKng2-derived HK mRNA. The lane on the left side of the gel (M) contains a DNA ladder. GAPDH mRNA was amplified in parallel to control for the DNA content of individual samples. L indicates liver; Lu, lung; K, kidney; A, adrenal; Bl, bladder; Br, brain; Mu, muscle; Thy, thymus; Sp, spleen; and H, heart. (B) In vitro translation of the regions of liver- and kidney-derived cDNA encoding Val364 to Arg540 of mKng1 (and corresponding region of mKng2). cDNA was amplified using the primers depicted in Figure 3B. PCR products were cloned into pBluescript II KS+, and used to synthesize capped mRNAs. In vitro translation was performed in the presence of 35S-methionine, and labeled polypeptides detected using 15% SDS-PAGE and autoradiography. As demonstrated in this figure, liver cDNA yielded a slightly larger polypeptide than that derived from kidney, consistent with expression of mKng1-derived HK mRNA in the former, and mKng2-derived HK mRNA in the latter.
Tissue-specific expression of mKng1 and mKng2. (A) RT-PCR of mKng1- and mKng2-derived HK mRNA transcripts in murine tissues. PCR products were resolved using 0.9% agarose gel electropheresis, and stained using ethidium bromide. Arrows indicate the location of PCR products from mKng1- and mKng2-derived HK mRNA. The lane on the left side of the gel (M) contains a DNA ladder. GAPDH mRNA was amplified in parallel to control for the DNA content of individual samples. L indicates liver; Lu, lung; K, kidney; A, adrenal; Bl, bladder; Br, brain; Mu, muscle; Thy, thymus; Sp, spleen; and H, heart. (B) In vitro translation of the regions of liver- and kidney-derived cDNA encoding Val364 to Arg540 of mKng1 (and corresponding region of mKng2). cDNA was amplified using the primers depicted in Figure 3B. PCR products were cloned into pBluescript II KS+, and used to synthesize capped mRNAs. In vitro translation was performed in the presence of 35S-methionine, and labeled polypeptides detected using 15% SDS-PAGE and autoradiography. As demonstrated in this figure, liver cDNA yielded a slightly larger polypeptide than that derived from kidney, consistent with expression of mKng1-derived HK mRNA in the former, and mKng2-derived HK mRNA in the latter.
Tissue-specific expression of HK mRNA derived from mKng1 and mKng2 was also analyzed using in vitro transcription/translation. Murine liver or kidney cDNA was used as a template for amplification using the same primers employed to assess tissue- specific HK mRNA expression. RT-PCR products were cloned and used to synthesize capped mRNA, which was translated using a rabbit reticulocyte lysate system. Liver cDNA yielded a larger polypeptide than that derived from kidney cDNA (Figure 4B). Moreover, anti-BK antisera recognized both of these polypeptides. These findings are consistent with predominant expression of mKng1 in the liver, and mKng2, which yields a smaller polypeptide, in the kidney.
Although the mechanisms underlying the tissue specific expression patterns of mKng1 and mKng2 are unknown, analysis of putative promoter regions using the Promoter Inspector program (Genomatix) demonstrated similarities as well as differences between the 2 genes. Potential transcription start sites containing the consensus sequence AGCCCA were located at position −90 to −85 relative to the ATG codon in both genes, with promoter regions extending at least 500 kb upstream. Each putative promoter region contained potential binding sites for GATA binding factor 3, NF-E2, p45, H6 homeodomain HMX3/Nks5.1, and octamer binding site 1 in the −85/−180 region. Several differences in the 2 promoter regions were also noted. These include the presence of binding sites for vErbA and hypoxia inducible factor-1β (HIF-1β) in the −210/−150 region of the mKng1, but not the mKng2 promoter. In contrast, binding sites for AP1 and PAX6 were present in this region of the mKng2, but not the mKng1 promoter. Additional differences, for example, the presence of binding sites for the Farnesoid X activated receptor-binding factor in the mKng1 promoter, were presence in the −210/−350 region. The significance of these differences will require detailed promoter analysis.
Homozygous deletion of mKng1
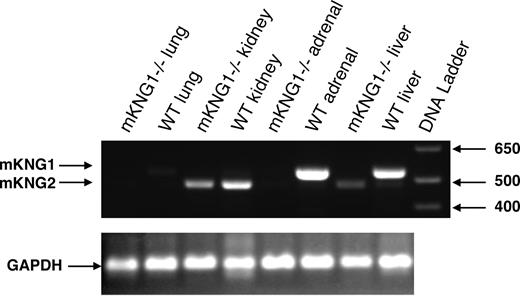
To address the roles of each of the kininogen genes, we focused on the generation of mice in which one or both kininogen genes were deleted. Since the expression pattern of mKng1 in the liver mimics that of human kininogen, we attempted to delete this gene first. We pursued this through disruption of exon 10, which contains a critical alternative splicing site regulating transcription of both HK and LK mRNA. We used recombineering to generate our targeting construct in PL253, using Escherichia coli DY380 as host cells (Figure 2). After electroporation of the targeting construct into ES cells, successful homologous recombination occurred in 5 of 300 clones. Blastocysts injected with one of these clones were implanted in pseudopregnant mice, and viable offspring heterozygous for the absence of mKng1 were obtained. The null allele underwent germ-line transmission, and crosses between F1 heterozygous mKng1-deficient mice yielded homozygous deficient offspring in the expected Mendelian ratio. mKng1−/− mice did not express mKng1-derived HK mRNA in tissues in which the gene was expressed in wild-type littermates (Figure 5), and did not display obvious phenotypic abnormalities.
Absence of mKng1-derived HK mRNA from the liver of mKng1−/− mice. RNA was isolated from liver, kidney, adrenal, and lung of wild-type and mKng1−/− mice as described in “Methods,” and analyzed by RT-PCR. Arrows depict mKng1- and mKng2-derived HK RT-PCR products. GAPDH mRNA was also analyzed by RT-PCR to ensure equal amounts of DNA were added to the reaction mixtures.
Absence of mKng1-derived HK mRNA from the liver of mKng1−/− mice. RNA was isolated from liver, kidney, adrenal, and lung of wild-type and mKng1−/− mice as described in “Methods,” and analyzed by RT-PCR. Arrows depict mKng1- and mKng2-derived HK RT-PCR products. GAPDH mRNA was also analyzed by RT-PCR to ensure equal amounts of DNA were added to the reaction mixtures.
Absence of kininogen antigen and function in the plasma of mKng1−/− mice
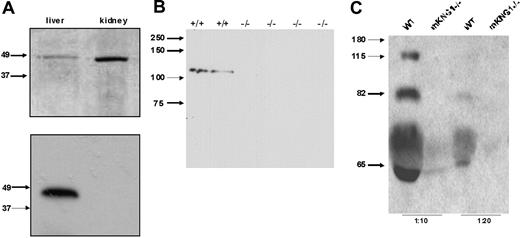
To determine which of the kininogen genes was responsible for the production of plasma kininogen, we analyzed plasma from wild-type and mKng1−/− mice by immunoblotting. Since the HK and LK heavy chains derived from each of these genes are homologous, we focused on differences in the light chains of mKng1 and mKng2 to generate isoforms-specific HK antibodies. Specifically, a rabbit polyclonal antibody was raised against a synthetic peptide, ERDAETEQGPTH, that corresponds to amino acids 412 to 423 of mHK1, a sequence absent in mHK2 (Figure 1A). This antibody specifically recognized the polypeptide derived from in vitro translation of murine liver cDNA, but not murine kidney cDNA (Figure 6A). When used to immunoblot plasma from wild-type mice and mKng1−/− mice, the anti-mHK1 antibody detected an approximately 110 kDa protein exclusively in the plasma of the wild-type mice (Figure 6B). These results confirm the absence of immunologically detectable mHK1 in the plasma of mKng1−/− mice.
Immunoblotting of kininogen in murine plasma. (A) An antibody raised against the mHK1-specific sequence ERDAETEQGPTH was used to blot the in vitro–translated polypeptides encoded by the cloned PCR products derived from liver and kidney cDNA using the primers depicted in Figure 3B. In the top panel, these recombinant polypeptides were separated using 10% SDS-PAGE, and the gel was stained with Coomassie brilliant blue. In the bottom panel, the polypeptides were transferred to PVDF and immunoblotted with the mHK1-specific antipeptide antibody. Only the liver (mKng1)–derived polypeptide is recognized by this antibody, demonstrating specificity. (B) The affinity-purified anti-mHK1 antibody was used to blot plasma from wild-type (+/+) and 4 mKng1-deficient (−/−) mice; mHK1 was present only in the wild-type animals. (C) Immunoblotting of whole mouse plasma (1:10 and 1:20 dilutions) from WT and mKng1−/− mice with an anti-BK antisera. A total of 3 distinct kininogen species are present in plasma from the wild-type mouse, but none are present in the mKng1−/− mice. The upper band recognized by the antisera corresponds to mHK, the intermediate band corresponds to ΔmHK-D5, and the lower band corresponds to mLK.
Immunoblotting of kininogen in murine plasma. (A) An antibody raised against the mHK1-specific sequence ERDAETEQGPTH was used to blot the in vitro–translated polypeptides encoded by the cloned PCR products derived from liver and kidney cDNA using the primers depicted in Figure 3B. In the top panel, these recombinant polypeptides were separated using 10% SDS-PAGE, and the gel was stained with Coomassie brilliant blue. In the bottom panel, the polypeptides were transferred to PVDF and immunoblotted with the mHK1-specific antipeptide antibody. Only the liver (mKng1)–derived polypeptide is recognized by this antibody, demonstrating specificity. (B) The affinity-purified anti-mHK1 antibody was used to blot plasma from wild-type (+/+) and 4 mKng1-deficient (−/−) mice; mHK1 was present only in the wild-type animals. (C) Immunoblotting of whole mouse plasma (1:10 and 1:20 dilutions) from WT and mKng1−/− mice with an anti-BK antisera. A total of 3 distinct kininogen species are present in plasma from the wild-type mouse, but none are present in the mKng1−/− mice. The upper band recognized by the antisera corresponds to mHK, the intermediate band corresponds to ΔmHK-D5, and the lower band corresponds to mLK.
To further examine the effect of mKng1 deletion on total kininogen in murine plasma, plasma was immunoblotted using arabbit antibradykinin antiserum that detects HK and LK, as well as the bradykinin nonapeptide within mHK1 and mHK2. This antibody recognized 3 proteins in the plasma of wild-type mice (Figure 6C). The largest of these (approximately 110 kDa) was mHK, and the lowest (approximately 65 kDa) was mLK. We also observed a band of approximately 82 kDa, which we have attributed to a protein called ΔmHK-D5. Though this polypeptide has not been directly sequenced, its mass corresponds to that of a polypeptide derived from a unique cDNA generated by RT-PCR of murine liver mRNA using primers designed to amplify full-length mHK cDNA (forward primer, 5′-ATGAAGCTCATTACTACACT-3′; reverse primer, 5′-TTAAGAAAGAGCATCAAGGA-3′). The ΔmHK-D5 cDNA encodes a polypeptide with a predicted Mr of approximately 80.2 in which the amino acid sequence spanning Thr400 through Asp482, corresponding to domain 5 and the proximal part of domain 6, is deleted. This sequence was deposited in GenBank18 (accession no. gi:50082913), and appears similar to a sequence reported by Cardoso et al,26 although these investigators did not determine whether the cDNA was translated.
The antibradykinin antibody did not detect any kininogen proteins in the plasma of mKng1−/− mice (Figure 6C), though this antibody did recognize the polypeptides derived from in vitro translation of liver and kidney mRNA, derived from mKng1 and mKng2, respectively, both of which contain the BK sequence.
Measurement of clotting times in mKng1−/− mice
To examine kininogen-dependent clotting activity in the plasma of mKng1−/− mice, we compared APTT values from 8 mKng1−/− mice with those of 8 wild-type littermate controls (129J strain). The mean APTT value of undiluted plasma from 8 wild-type littermate controls was 24.5 (± 2.2) seconds, compared with a mean of 54.1 (± 11.5) seconds in 8 mKng1−/− mice. We also assessed HK-dependent procoagulant activity using human HK-deficient plasma as substrate. The mean procoagulant activity in plasma from 10 individual mKng1−/− mice was 0.14 (± 0.09) U/mL (range, 0.03-0.27 U/mL) compared with a mean of 0.9 (± 0.2) U/mL (range, 0.77-1.33 U/mL) in 12 wild-type littermate controls (Table 2).
In vitro procoagulant activity of wild-type and mKng1−/− plasma
| Mouse . | Wild-type, U/mL . | mKng1−/−, U/mL . |
|---|---|---|
| 1 | 0.8043 | 0.0721 |
| 2 | 0.8875 | 0.0907 |
| 3 | 0.7763 | 0.2889 |
| 4 | 0.7938 | 0.0576 |
| 5 | 0.9111 | 0.2028 |
| 6 | 0.913 | 0.0351 |
| 7 | 1.3317 | 0.1321 |
| 8 | 0.7389 | 0.1502 |
| 9 | 1.0346 | 0.1147 |
| 10 | 0.8196 | 0.2946 |
| 11 | 0.8419 | — |
| 12 | 0.957 | — |
| Mean (SD) | 0.901 (0.159) | 0.144 (0.092) |
| Mouse . | Wild-type, U/mL . | mKng1−/−, U/mL . |
|---|---|---|
| 1 | 0.8043 | 0.0721 |
| 2 | 0.8875 | 0.0907 |
| 3 | 0.7763 | 0.2889 |
| 4 | 0.7938 | 0.0576 |
| 5 | 0.9111 | 0.2028 |
| 6 | 0.913 | 0.0351 |
| 7 | 1.3317 | 0.1321 |
| 8 | 0.7389 | 0.1502 |
| 9 | 1.0346 | 0.1147 |
| 10 | 0.8196 | 0.2946 |
| 11 | 0.8419 | — |
| 12 | 0.957 | — |
| Mean (SD) | 0.901 (0.159) | 0.144 (0.092) |
— indicate no entry.
Measurement of plasma bradykinin levels
As another approach to define whether mKng1−/− mice are truly kininogen deficient, we measured levels of plasma bradykinin.21 Bradykinin was undetectable by radioimmunoassay (threshold level for detection less than 10 pg/mL) in the plasma of 10 of 10 mKng1−/− mice tested (3 129J mice, 7 C57BL/6 mice). Bradykinin levels in the plasma of 7 wild-type mice (all C57BL/6) ranged from 43.7 to 130.8 pg/mL (mean, 80.1 ± 41.1 pg/mL).
mKng1−/− mice are protected from thrombosis
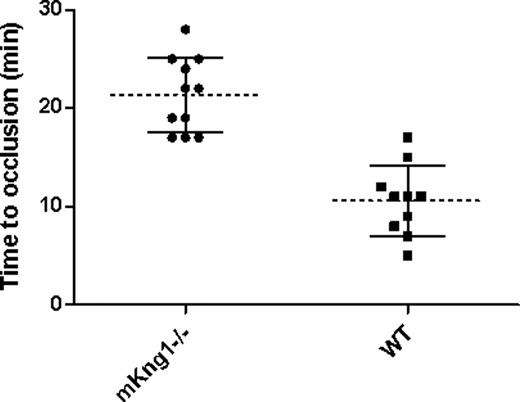
To assess the propensity of mKng1−/− mice to arterial thrombosis, we compared carotid artery thrombosis times in 10 wild-type littermate controls and 11 mKng1−/− mice in which arterial photoinjury was induced by laser-induced photoactivation of Rose Bengal.22,23 The mean time to thrombosis was 21.4 (± 3.8) minutes in mKng1−/− animals, a value significantly longer than the 10.6 (± 3.6) minutes measured in wild-type littermate controls (P < .001; Figure 7).
Time to arterial occlusion in mKng1−/− mice and wild-type littermate controls. Time to occlusion of the carotid artery following Rose-Bengal and laser induced arterial injury was measured as described in “Methods.” mKng1−/− mice displayed significantly longer arterial occlusion times. Dotted lines represent mean values, and error bars represent plus or minus 2 standard deviations from the means.
Time to arterial occlusion in mKng1−/− mice and wild-type littermate controls. Time to occlusion of the carotid artery following Rose-Bengal and laser induced arterial injury was measured as described in “Methods.” mKng1−/− mice displayed significantly longer arterial occlusion times. Dotted lines represent mean values, and error bars represent plus or minus 2 standard deviations from the means.
The mean tail vein bleeding time in 5 mKng1−/− mice was 89.0 (± 8.0) seconds, and was not significantly different than the 76.4 (± 15.1) seconds measured in the wild-type littermate controls (P = .222).
Discussion
Our studies demonstrate several new findings concerning the expression of kininogen in mice. First, we have confirmed previous reports demonstrating that the mouse genome contains 2 homologous kininogen genes. Second, we have demonstrated that these genes are expressed in a tissue-specific manner, with mKng1-derived HK mRNA expressed primarily in the liver and glands adrenal glands and mKng2-derived HK mRNA expressed primarily in the kidney. Third, our studies have defined the role of these genes in the expression of functional protein, and through the selective deletion of mKng1, we have shown that this gene accounts for essentially all detectable HK and LK in plasma. Moreover, the disappearance in mKng1−/− mice of a novel kininogen species, ΔmHK-D5, demonstrates that this kininogen isoform is also derived from mKng1. Finally, we have demonstrated that mKng1−/− mice have normal bleeding times, but a prolonged time to arterial occlusion in the Rose Bengal thrombosis model; though these studies require confirmation in additional models, they suggest that while not necessary for normal hemostasis, kininogen may contribute to the development of thrombosis under pathologic conditions. Whether kininogen plays a similar role in humans is uncertain, as the rarity of such individuals makes clinical correlations difficult to discern.
The finding of 2 kininogen genes in mice contrasts with an original report from Takano et al in which the mouse genome,25 like the human,24 was found to contain only a single kininogen gene. The reason for this discrepancy is uncertain, but may reflect the use of different probes for Southern blots; Takano et al used a full-length HK cDNA, but we used a smaller, 400-nucleotide fragment corresponding to regions of intron 4 and exon 4.
The 2 murine kininogen genes display a similar organization. Both contain 11 exons and 10 introns, and share 91% homology at the exon level. The most significant difference in the exons of the 2 genes is in the coding region of exon 10, in which 2 in-frame gaps of 18 and 33 nucleotides are present in mKng2. In addition, analysis of the sequence of BAC 452G1 demonstrates marked differences in intron size between the 2 genes. This finding may be important, since HK deficiency in humans has been shown to be associated with intronic mutations despite a normal exon sequence.27,28
There are only a few examples of duplicated genes, such as the alpha-tubulin and α/β-globin gene clusters in fish,29,30 that occur in a head-to-head orientation similar to that of the murine kininogen genes. In these genes, the distance between initiation codons usually does not exceed 2 kb, and gene expression is regulated by bidirectional promoters. In contrast, the distance between the 2 murine kininogen genes is approximately 30 kb (Figure 1A), making these genes among the first that maintain a head-to-head orientation despite such a long intervening sequence.
Our findings extend the recent report of Cardoso et al,26 in which 2 murine kininogen genes were also detected by (1) defining the relationship between kininogen gene and protein expression, (2) describing the novel kininogen isoform, ΔmHKD5, which may correspond to kininogen splice variants proposed to exist in humans,28 (3) providing new information on intron sizes of the 2 genes, and (4) defining differences, albeit not yet experimentally validated, in the kininogen gene promoters.
Shesely et al have also described a second kininogen gene in C57BL6/6J and Swiss-Webster mice.31 These investigators reported mKng1-derived HK and LK mRNA and mKng2-derived LK mRNA in the liver, and mKng1-derived LK and mKng2-derived HK and LK mRNA in the kidney. Our results are consistent with these findings, though Shesely et al did not assess the roles of the 2 kininogen genes in production of kininogen protein.31
In our studies, deletion of mKng1 was confirmed by demonstrating the absence of mKng1-derived mRNA in tissues in which the gene is normally expressed (Figure 5). Deletion of this gene allowed us to analyze the contribution of the 2 kininogen genes to the production of plasma kininogen. Our results demonstrate that mKng1-derived mRNA yields 3 distinct isoforms of kininogen in plasma, HK, LK, and ΔmHK-D5, since these proteins were abundant in the plasma of wild-type mice, but not detectable in the plasma of mKng1−/− mice. These results suggest that mKng1 is responsible for the production of plasma kininogen, and that mKng2 mRNA is not translated in sufficient quantity to contribute to plasma kininogen levels, a conclusion supported by our inability to detect BK in mKng1−/− plasma.
Measurement of functional clotting activity in plasma from mKng1−/− mice demonstrated variably low levels of putative HK-dependent procoagulant activity. We were unable to inhibit this activity using monoclonal anti-human kininogen antibodies that neutralize kininogen-dependent clotting activity in human plasma (data not shown), though the reactivity of these antibodies with mouse kininogen is uncertain. Thus, we are uncertain whether the presumed HK procoagulant activity in the mKng1−/− plasma is actually due to kininogen, although the results described here would argue that this is not the case. Other investigators have observed that the measurement of factor-specific procoagulant activity in deficient murine plasma using human plasma as a substrate may yield higher than expected activity despite complete absence of mRNA encoding the factor of interest32 ; the reasons for this phenomenon are not well understood.
The finding that mKng1−/− mice had no gross phenotype under nonstressed conditions was not unexpected given that the few kininogen-deficient humans recognized have not demonstrated major primary illnesses such as a bleeding diathesis. However, mKng1−/− mice did display a significantly prolonged time to vessel occlusion in an arterial thrombosis model. This finding may potentially be consistent with recent studies that demonstrate a similar phenotype in factor XII–deficient mice,33 and suggests the possible involvement of the kallikrein-kinin system in experimentally induced thrombosis due to vascular injury.34 However, direct comparison of those studies with ours is difficult, as factor XII–null animals were studied using different vascular injury models that provide a stronger thrombogenic stimulus than Rose Bengal, potentially leading to different thrombotic responses. Further studies will be required to assess the contribution of HK to induced arterial and venous thrombosis in additional models, and if such a role is confirmed, to discern whether the mechanism reflects direct enhancement of thrombus stability or other mechanisms.22,35
Several abnormalities have been observed in the Brown Norway Katholiek rat, a model of spontaneous kininogen deficiency (kininogen levels 3%-5% of normal) resulting from an H163D mutation in kininogen. These animals display decreased angiogenesis in an inflammatory sponge model,15 develop aortic aneurysms,36 and may have heightened thrombosis risk37 and dampened inflammatory responses.38 While we have not yet determined whether similar phenotypic responses occur in mKng1−/− mice, these observations demonstrate the broad range of physiologic activities affected by the kallikrein-kinin system, and suggest that mKng1−/− mice may provide a new and important model for studying the role of the this system in numerous biological processes.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants HL076810 (K.R.M.), HL052779 (A.H.S.), and HL073750 (F.J.C.).
National Institutes of Health
Authorship
Contribution: S.M.M. designed, prepared and characterized mKng1−/− mice. W.M.Z. assisted with coagulation, thrombosis, and bradykinin assays. A.A.K. assisted with in vitro translation assays and promoter analyses. A.H.S. designed and assisted in interpretation of coagulation and thrombosis assays. E.B. and X.L. assisted in preparing the targeting vector. Y.Z. performed coagulation and thrombosis assays. T.I. performed BK assays. F.J.C. assisted in design and interpretation of BK assays. G.B.L. assisted in the design of the targeting vector for mKng1−/− deletion. K.R.M. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Keith R. McCrae, Hematology-Oncology, WRB 2-132, Case Western Reserve School of Medicine, 10900 Euclid Ave, Cleveland, OH 44106; e-mail: keith.mccrae@case.edu.