Multiple myeloma (MM) is characterized by accumulation and dissemination of malignant plasma cells (PCs) in the bone marrow (BM). Gene expression profiling of 2 MM cell lines (OH-2 and IH-1) indicated that expression of PRL-3, a metastasis-associated tyrosine phosphatase, was induced by several mitogenic cytokines. Cytokine-driven PRL-3 expression could be shown in several myeloma cell lines at both the mRNA and protein levels. There was significantly higher expression of the PRL-3 gene in PCs from patients with monoclonal gammopathy of undetermined significance (MGUS), smoldering myeloma (SMM), and myeloma than in PCs from healthy persons. Among 7 MM subgroups identified by unsupervised hierarchical cluster analysis, PRL-3 gene expression was significantly higher in the 3 groups denoted as “proliferation,” “low bone disease,” and “MMSET/FGFR3.” PRL-3 protein was detected in 18 of 20 BM biopsies from patients with MM. Silencing of the PRL-3 gene by siRNA reduced cell migration in the MM cell line INA-6, but had no detectable effect on proliferation and cell-cycle phase distribution of the cells. In conclusion, PRL-3 is a gene product specifically expressed in malignant plasma cells and may have a role in migration of these cells.
Introduction
Multiple myeloma (MM) is a B-cell lineage malignancy characterized by widespread dissemination of mature plasma cells (PCs) throughout the bone marrow. MM remains incurable, and the need for better therapy targeting the weak points of MM is urgent
It is generally believed that myeloma cells are dependent on growth factors for proliferation and survival. The observation that many cytokines1,,,–5 have redundant effects on growth of myeloma cells, led us to hypothesize that the intracellular signals from cytokines converge and regulate transcription of a set of genes that are common targets for several growth factors. For future treatment, it may be futile to directly target the cytokines or their receptors because of redundancy in signaling but, if this hypothesis is correct, one could attack selected genes that are common downstream targets for all the cytokines. Using gene expression analysis, we identified PRL-3 as one of the molecules that was up-regulated in the cytokine-dependent myeloma cell lines OH-2 and IH-1 after stimulation with growth factors. The PRL-3 gene is known as a metastasis-associated phosphatase,6,,,,,,–13 and there are several reports showing its importance in cancer cell invasion and migration,7,14,–16 especially in colon cancer.13,14 This, together with the knowledge that cell migration is one of the processes fundamental to myeloma cell invasion and dissemination, led us to look closer into its expression and function in MM.
Phosphatases of regenerating liver (PRL phosphatases) constitute a class of small (20 kDa) phosphatases17 with possible oncogenic activity. To date, the exact cellular role, the substrates, and involvement in signaling pathways have not been determined for any of the PRLs (PRL-1, -2, 3). PRL phosphatases belong to dual-specificity phosphatases, which are able to dephosphorylate tyrosine, serine, and threonine residues as well as inositol phospholipids in some cases. There is a high level of amino acid sequence identity among PRL family members: PRL-1 and PRL-3 share 78% identity, and PRL-2 and PRL-3 share 86% identity. They possess a unique COOH-terminal prenylation motif, and prenylation seems to be important for subcellular location. There are some data supporting that the catalytic domain of PRL-3 is important for its metastatic quality.18
In the present study we wanted to explore whether PRL-3 plays a role in MM, which by its nature is diffuse and disseminated. We found that PRL-3 expression was higher in bone marrow (BM) plasma cells from patients with newly diagnosed monoclonal gammopathies than in plasma cells from healthy donors. Down-regulation of PRL-3 expression by siRNA impaired SDF-1–induced migration of MM cells, but we could not show any influence on cell-cycle distribution or cell proliferation.
Methods
Cell lines and culture condition
The human myeloma cell lines IH-1,3 OH-2,19 ANBL-6 (gift from Dr D. Jelinek, Mayo Clinic, Rochester, MN), INA-6 (gift from Dr M. Gramatzki, Erlangen, Germany), JJN-3 (gift from J. Ball, University of Birmingham, United Kingdom), CAG (gift from Dr J. Epstein, Little Rock, AR), RPMI-8226 and U266 (both from American Type Culture Collection [ATCC], Rockville, MD) were cultured as previously described.20 The nonmyeloma hematologic cell lines we used were HL-60, IM9, Jurkat, U937, MC-CAR, Daudi, ARH-77, K562 (all from ATCC), Monomac (gift from Dr L. Ziegler-Heitbroch, Gauting, Germany), DOHH2–2, and CRO-AP5 (both from DSZM, Brauschweig, Germany). All cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. We washed the cells 4 times in Hanks balanced salt solution (HBSS; Gibco, Grand Island, NY) to deplete them of cytokines before performing experiments.
In the gene expression profiling studies done at the University of Arkansas for Medical Sciences (UAMS), 45 different myeloma cell lines used to determine the expression of PRL-3 mRNA included the following, as described previously21,–23 : RPMI-8226, ARP1, 11278R, CAG, Dp6p43, Hoffman4, KHM1B, KMS12BM, KMS12BM, KMS28BM, KMS28PE, KMS34, Kas6, p11p2, OCI-MY7, SACHI, XG2, CRAIG, Delta47, EJM, FLAM76, FR4, H1112, H929, INA6, JIM3, JJN3, KARPAS620, KHM11, KMM1, KMS11, KMS12PE, KMS18, L363, LP1MM-M1, MM1-144, NwU266, OCI-MY5, OPM1, OPM2, SKMM1, SKMM2, UTMC2, XG1, and XG7.
Antibodies and cytokines
Hepatocyle growth factor (HGF) was purified from medium conditioned by the human myeloma cell line JJN-3 as described previously.24 All other cytokines were recombinant and human. IL-6 was from Biosource (Camarillo, CA); IGF-1, IL-15, and bone morphogenetic protein 4 (BMP-4) were from R&D Systems (Minneapolis, MN); TNF-α was from Genentech (South San Francisco, CA); and SDF-1α was from Peprotech (London, United Kingdom). IL-21 was a gift from R. Holly (ZymoGenetics, Seattle, WA). PRL-3 antibodies were from Novus Biologicals (Littleton, CO) when used for confocal microscopy (polyclonal goat anti-PRL3) and from Zymed Laboratories (South San Francisco, CA) when used in immunohistochemistry (polyclonal rabbit anti–PRL-3). Monoclonal anti–PRL-3 (gift from Dr Q. Zeng)25 was used for Western blots. Monoclonal anti-GAPDH (Abcam, Cambridge, United Kingdom) was used for loading controls on Western blots.
Patients and healthy individuals, isolation of myeloma cells
The patients were recruited from UAMS (Little Rock, AR) and from St Olavs University Hospital (Trondheim, Norway). The use of the BM samples was approved by the institutional review board of UAMS and the regional ethics committee in Trondheim. Written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki.
At UAMS, expression of PRL-3 in CD138+ myeloma cells from 256 patients with newly diagnosed MM was compared with expression in CD138+ cells from BM of 44 patients with monoclonal gammopathy of undetermined significance (MGUS), 12 patients with smoldering myeloma (SMM), and 22 healthy individuals. The cases are characterized in references.26,–28 BM biopsies from 20 random patients (St Olavs University Hospital), were selected for immunohistochemistry, and RNA was isolated from 12 random patients for quantification of PRL-3 mRNA.
PCs were separated from BM aspirate by retrieval of CD138+ cells.29 The mononuclear cells were first collected using Lymphoprep (Axis-Shield, Oslo, Norway), and then incubated with Macs CD138 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's protocol. The purity of the plasma cells obtained by this method was at least 97%.
RNA isolation and real-time PCR of PRL-3 mRNA
Total RNA was isolated with an RNeasy midi kit (Qiagen, Crawley, United Kingdom). Total cDNA was synthesized using Taqman Reverse transcriptase reagents from Applied Biosystems (Foster City, CA), following the manufacturer's protocol. Quantitative polymerase chain reaction (PCR) of PRL-3 cDNA was performed using an MJ Opticon 2 thermal cycler (MJ Research, Waltham, MA). PCR reactions were performed using forward primer (5′-gggacttctcaggtcgtgtc-3′), reverse primer (5′-agccccgtacttcttcaggt-3′), and a Taqman probe (5′-tggaggtgagctacaaacacatgcg-3′). The reaction mix consisted of 1× GeneAmp PCR Gold buffer (Applied Biosystems), MgCl2 (1.5 mM), dNTP (0.4 mM), 6 μM of each primer, Taqman probe (0.2 μM), and 1 U AmpliTaq Gold DNA polymerase in a total volume of 25 μL. All reactions were done 4 times. The relative expression of PRL-3 from each sample was normalized to the expression of the gene coding for β-actin in that sample. The fold change was calculated and normalized to the expression in the cell line that showed lowest relative expression of PRL-3.
Immunoblotting
Cells were harvested after 18 hours, washed with ice-cold phosphate-buffered saline (PBS) and resuspended in 50 μL lysis buffer (Tris-HCL [50 mM] pH 7.5, NaCl [150 mM], NP-40 1%, a protease inhibitor mixture [Complete mini tablets; Roche, Basel, Switzerland], NaF [50 mM], Na3VO4 [1 mM]). After 30 minutes on ice, the nuclei were removed by centrifugation at 12 000g, 4°C for 20 minutes. Supernatants were stored at −80°C until additional processing. Aliquots (25 μL) were mixed with 4× LDS sample buffer (Invitrogen, Oslo, Norway) with 100 mM DTT, heated for 2 minutes at 98°C, separated on 12% NuPAGE Bis-Tris gels (Invitrogen), and finally electrophoretically transferred to 0.45-μm nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were blocked with 5% nonfat dried milk in Tris-buffered saline with 0.05% Tween 20, and incubated with antibodies as indicated overnight at 4°C. Detection was performed with horseradish peroxidase–conjugated antibodies (DAKO Cytomation, Copenhagen, Denmark) and chemiluminescence (ECL; Amersham Biosciences, Amersham, United Kingdom).
Gene-expression profiling
Gene-expression profiling was performed with Affymetrix U133Plus2.0 microarray on CD138-enriched BM plasma cells from 256 newly diagnosed patients with myeloma, 44 patients with MGUS, 12 patients with SMM, and 22 healthy donors. In addition, the constitutive expression level of the PRL-3 gene was examined in 45 human myeloma cell lines. The data were processed with Affymetrix Microarray Suite GCOS1.1; Affymetrix signal intensity was log base 2–transformed for each sample.26,–28
FISH analysis
Cytospin slides (2 × 104-3 × 104 cells per slide) of various cell lines were used for fluorescent in situ hybridization (FISH) analysis. A chromosome 8 centromeric probe, SpectrumAqua CEP-8 (Vysis, Downers Grove, IL) was used to estimate the copy number of chromosome 8. The PRL-3 probe was prepared from BAC clone RPCI-11953B20 containing the whole PRL-3 gene (Invitrogen). Plasmid was isolated using the Qiagen Large-Construct Kit (Qiagen) and labeled with SpectrumOrange dUTP using a nick translation kit (Vysis). FISH hybridization was performed using standard procedure (Vysis). After hybridization the cells were counterstained with DAPI (Vysis). Cells were scored using a Nikon Eclipse 90i epifluorescence microscope with PlanApo VC 60×/1.4oel (Nikon Instruments Europe, Badhoevedorp, the Netherlands), and software from Applied Imaging (CytoVision version 3.7 Build 58, 2005; San Jose, CA).
Immunohistochemistry
Sections (4 μm) of formalin-fixed BM biopsies, decalcified in EDTA and paraffin-embedded, were deparaffinized. Antigen retrieval was done in a steamer for 12 minutes, and the biopsies were allowed to cool at room temperature for at least 30 minutes. Sections were incubated with polyclonal rabbit anti–PRL-3 diluted 1:30 in Tris-buffered saline (TBS), 0.25% BSA, 0.25% Tween 20 (pH 7.6) for 1 hour at room temperature in a DAKO autostainer. Immunohistochemical reactions were visualized with DAKO Cytomation EnVisionDAB, K5007. The sections were counterstained with hematoxylin. Images were captured using a Nikon Eclipse 80i microscope and Nikon dig SIGHT DS5-M camera (Nikon Instruments Europe).
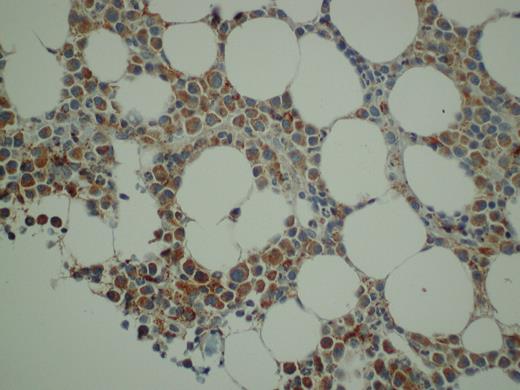
The biopsies were considered positive for PRL-3 when there were more than 20% positive tumor cells.
Confocal microscopy
To examine the cell distribution of PRL-3, 105 cells were fixed for 15 minutes on ice in 2% paraformaldehyde (PFA), washed 3 times in PBS with 5% fetal calf serum (FCS), and 0.4% saponin before staining for 30 minutes with a primary antibody against PRL-3. After further washing, secondary staining was done for 45 minutes on ice with FITC-conjugated rabbit anti–goat Ig (Vector Laboratories, Burlingame, CA). Nuclei were visualized with Draq 5 (Biostatus, Shepshed, United Kingdom). In the experiments with cell-cycle separation, the cells were propidium iodide (PI)–stained (25 μg/mL) after fixation in 2% PFA on ice for 15 minutes, and washed in PBS 5% FCS, 0.4% saponin. The cells were analyzed and sorted by flow cytometry with a Coulter Epics Elite ESP (Beckman-Coulter, Hialeah, FL), using a 15 mW argon laser (488 nm), a 640-nm dichroic long-pass filter, and a 610/10-nm band-pass filter. According to the dot plot of forward versus side scatter and PI linear versus PI peak signals, aggregates and debris were identified and excluded from the analysis. Instrumental settings for sorting were 10 psi, 20.4 kHz, 3 droplet sort option, and the sort was performed at 4°C at the speed of 3000 cells per second. Examination of the cells was done with a LSM 510 Zeiss confocal microscope (Zeiss, Jena, Germany). In the cell-cycle analysis, 100 to 200 cells per cycle were evaluated.
Transfection of INA-6 cells
Cells were transfected by electroporation using the Nucleofector device and corresponding kits (Amaxa Biosystems, Cologne, Germany). First, we optimized the transfection program, selecting the solution R and program X-001, which resulted in the highest transfection efficiency with the lowest mortality (green fluorescent protein [GFP]–positive, PI-negative by FACS). Due to transfection efficacy in the range of 20% to 40%, the cells were cotransfected with the selection marker pcDNA3 CD4 (kind gift from Dr M. Janz, Berlin, Germany), and after 24 hours the transfected cells were isolated using Dynabeads CD4 (Dynal). By this procedure we regularly obtained a population with approximately 80% transfected and viable cells. Cells were then diluted and seeded for proliferation and migration assay and for analysis by immunoblotting.
Synthetic double-stranded siRNA was commercially obtained from Dharmacon (Diagen, Rygge, Norway): SMART Pool, siRNA PRL-3, siCONTROL Non-Targeting siRNA Pool, siCONTROL Cyclophilin B siRNA. All siRNA concentrations used were 1 μM (lowest effective concentration knocking down the PRL-3 protein). For Western blotting, cells were grown in RPMI with 10% FCS and IL-6 (0.5 ng/mL) at a density of 104 cells/mL for 48 to 72 hours after transfection.
Proliferation assay
Cells were seeded in 96-well plastic culture plates (Corning Costar, Corning, NY) at a density of 104 to 2 × 104 cells per well in 200 μL RPMI with 10% FCS and IL-6 (1 ng/mL). After 48 hours, cells were pulsed with 0.037M8q methyl- [3H]-thymidine (NEN Life Science Products, Boston, MA) per well, and harvested 4 to 6 hours later with a Micromate 196 cell harvester (Packard, Meriden, CT). Beta radiation was measured with a Matrix 96 beta counter (Packard).
Migration assay
Three days after the transfection, the INA-6 cells were washed in Hanks balanced salt solution (HBSS) to remove the paramagnetic beads, then suspended in RPMI-1640 supplemented with 0.1% bovine serum albumin (Sigma-Aldrich, Oslo, Norway) and IL-6 (0.1 ng/mL). Cells were seeded (2 × 105 cells in 100 μL) in the upper compartment of polycarbonate membrane Transwell (pore size: 5 μm) from Corning. SDF-1α (75 ng/mL) was added to the lower compartment (600 μL). All samples were performed in duplicate. After 22 to 24 hours, at 37°C and 5% CO2, the number of cells that had migrated through the membrane to the lower compartment was determined by a Coulter Counter Z1 (Beckman Coulter).
Cell-cycle analysis
Transfection and CD4 isolation was done as described in “Transfection of INA-6 cells.” Cells (5 × 105) were incubated with DRAQ5 (15 μM; Biostatus Limited, Leicestershire, United Kingdom) for 15 minutes in RPMI with IL-6 (1 ng/mL) and 10% FCS at 37°C in a humidified atmosphere containing 5% CO2. Thereafter, cellular DNA content was analyzed by flow cytometry, and histograms were analyzed with MultiCycle (Phoenix Flow System, San Diego, CA).
Statistical analysis
Signal differences in expression of PRL-3 among plasma cells from patients with myeloma and normal plasma cells, MGUSs, SMMs, and MMCLs were analyzed with one-way analysis of variance (ANOVA) procedure using SPSS 12.0 (SPSS, Chicago, IL). The chi-square test was used to compare the parameter of detection of PRL-3 among these groups.
Results
Cytokine-induced expression of the PRL-3 gene
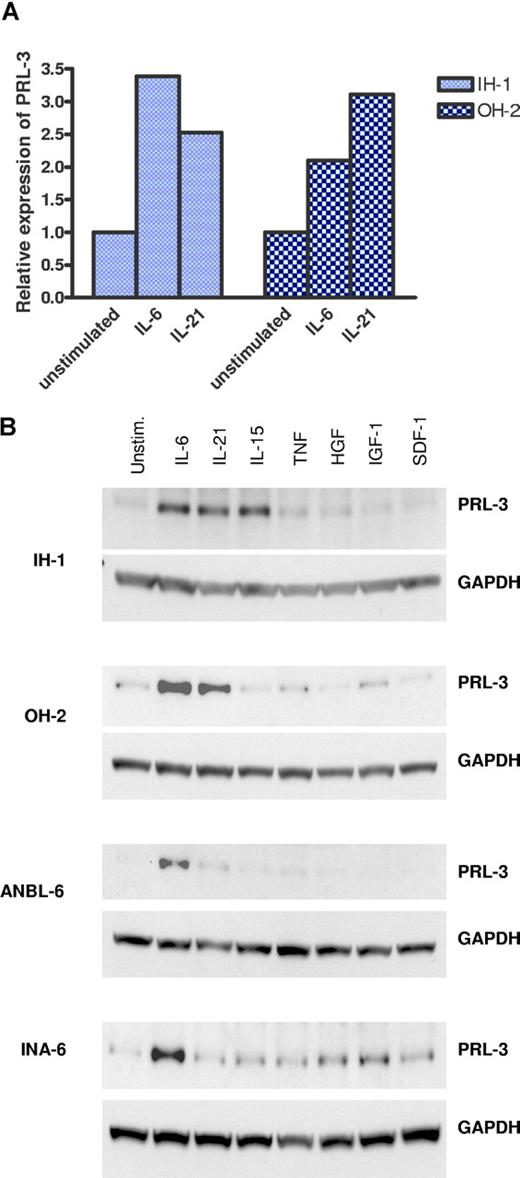
The human serum– and cytokine-dependent cell lines IH-1 and OH-2 proliferate in response to various growth-promoting cytokines, including IL-6, TNF, and IL-21.2,3,19 We found that PRL-3 was among the genes whose expression was significantly increased in both cell lines when stimulated with cytokines (M.B., unpublished data, July 2007). IH-1 cells and OH-2 cells expressed PRL-3 mRNA as analyzed by reverse transcription (RT)–PCR (Figure 1A). In both cell lines the expression was between 2- and 3.5-fold higher than in nonstimulated cells after 24 hours stimulation with IL-6 or IL-21.
Expression of PRL-3 mRNA and protein after cytokine stimulation. (A) Quantification of PRL-3 mRNA by real-time RT-PCR was performed after stimulation of IH-1 and OH-2 cells either with or without IL-6 and IL-21 for 24 hours. The ΔCt value between PRL-3 and β-actin from the unstimulated cells reflects the expression of PRL-3 relative to its internal control β-actin. The ΔCt value for the unstimulated IH-1 cells was 2.68 with a standard variation of 0.17 and for the unstimulated OH-2 cells it was 22.49 with a standard variation of 0.31. Thus, standard variation for all ΔCt values from RT-PCR reactions was between 1% and 2% of its ΔCt value. The relative expression level of PRL-3 to β-actin in the unstimulated cells was represented by 2−ΔCt and was arbitrarily set to 1. The relative expression of PRL-3 to β-actin in the stimulated cell culture, 2−ΔCt, was then normalized to that of the unstimulated cell culture, 2−ΔCt(unstim) / 2−ΔCt(stim), and is illustrated on the y-axis. (B) PRL-3 expression was determined by Western analysis after an 18-hour incubation with various cytokines. Cytokine concentrations were as follows: IL-6, 5 ng/mL; IL-21, 20 ng/mL; IL-15, 20 ng/mL; TNF, 10 ng/mL; HGF, 150 ng/mL; IGF-1, 100 ng/mL, and SDF-1, 75 ng/mL. The loading control was GAPDH (bottom panels).
Expression of PRL-3 mRNA and protein after cytokine stimulation. (A) Quantification of PRL-3 mRNA by real-time RT-PCR was performed after stimulation of IH-1 and OH-2 cells either with or without IL-6 and IL-21 for 24 hours. The ΔCt value between PRL-3 and β-actin from the unstimulated cells reflects the expression of PRL-3 relative to its internal control β-actin. The ΔCt value for the unstimulated IH-1 cells was 2.68 with a standard variation of 0.17 and for the unstimulated OH-2 cells it was 22.49 with a standard variation of 0.31. Thus, standard variation for all ΔCt values from RT-PCR reactions was between 1% and 2% of its ΔCt value. The relative expression level of PRL-3 to β-actin in the unstimulated cells was represented by 2−ΔCt and was arbitrarily set to 1. The relative expression of PRL-3 to β-actin in the stimulated cell culture, 2−ΔCt, was then normalized to that of the unstimulated cell culture, 2−ΔCt(unstim) / 2−ΔCt(stim), and is illustrated on the y-axis. (B) PRL-3 expression was determined by Western analysis after an 18-hour incubation with various cytokines. Cytokine concentrations were as follows: IL-6, 5 ng/mL; IL-21, 20 ng/mL; IL-15, 20 ng/mL; TNF, 10 ng/mL; HGF, 150 ng/mL; IGF-1, 100 ng/mL, and SDF-1, 75 ng/mL. The loading control was GAPDH (bottom panels).
Up-regulation of PRL-3 protein as determined by immunoblotting
Western blot analysis in the cytokine-dependent cell lines ANBL-6, IH-1, OH-2, and INA-6 (Figure 1B) showed that the expression of PRL-3 protein was induced by mitogens. In ANBL-6, IH-1, and OH-2 cells the protein was induced with several of the cytokines used. IL-6 and IL-21, strong growth factors for IH-1 and OH-2,3,19 were the most potent inducers of PRL-3. In the INA-6 cell line the PRL-3 protein was induced after IL-6, HGF, and IGF-1 stimulation. Up-regulation of PRL-3 with IL-6 stimulation could also be observed in cell lines that are not strictly dependent on IL-6 for proliferation, such as RPMI-8226 and JJN-3 (data not shown). In U266 and CAG there was no detectable PRL-3 on Western blots even in the presence of IL-6 (data not shown), in line with the RT-PCR data (Figure 2).
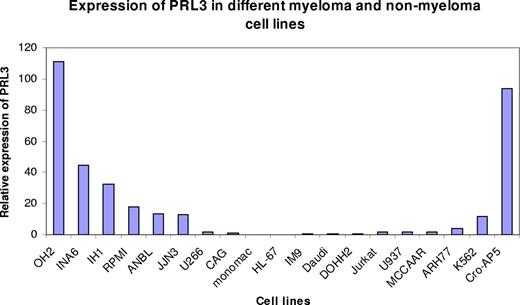
Expression of PRL-3 mRNA in myeloma and nonmyeloma cell lines. The ΔCt between PRL-3 and β-actin for CAG was 8.1 (± 0.58). The relative expression level of PRL-3 to β-actin in the CAG cells, 2−ΔCt, was arbitrarily set to 1. The relative expression level of other cell lines was normalized to that of CAG cells and was represented by 2−ΔΔCt, as is illustrated in this figure.
Expression of PRL-3 mRNA in myeloma and nonmyeloma cell lines. The ΔCt between PRL-3 and β-actin for CAG was 8.1 (± 0.58). The relative expression level of PRL-3 to β-actin in the CAG cells, 2−ΔCt, was arbitrarily set to 1. The relative expression level of other cell lines was normalized to that of CAG cells and was represented by 2−ΔΔCt, as is illustrated in this figure.
Expression of PRL-3 mRNA in a panel of myeloma cell lines and nonmyeloma hematologic cell lines
We examined the expression of PRL-3 mRNA in 4 IL-6–dependent and 4 IL-6–independent myeloma cell lines by use of real-time PCR (Figure 2). The myeloma cell lines cultivated in IL-6 ranked 1, 2, 3, and 5 in expression level. We also examined 11 nonmyeloma cell lines. With the exception of CRO-AP5, all had either undetectable levels or lower levels of PRL-3 mRNA than all the IL-6–dependent myeloma cell lines (Figure 2).
PRL-3 gene expression in normal PCs, MM cell lines, and PCs from patients with MGUS, SMM, and MM
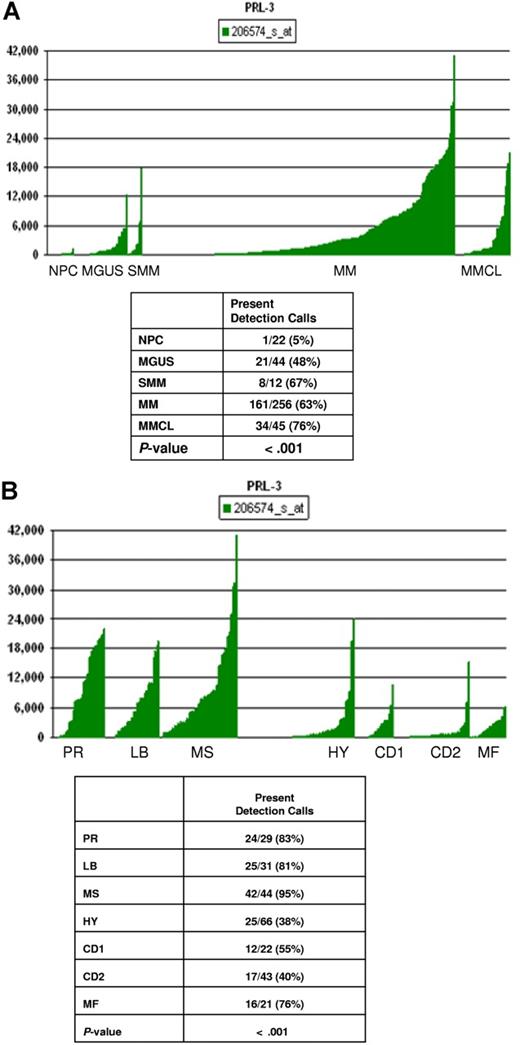
Next, we wanted to explore the PRL-3 gene expression pattern in primary myeloma cells and compare with expression in BM PCs from healthy donors, and from patients with MGUS or SMM. This was done in a cohort of 256 patients, earlier described.27 There was significantly (P < .001) higher expression of the PRL-3 gene in PCs from patients with MGUS, SMM, and myeloma than in PCs from healthy subjects (determined with chi-square test of genes defined as present or absent) (Figure 3A). The level of PRL-3 expression did not influence patient survival (data not shown).
PRL-3 gene expression in normal PCs, MM cell lines, and PCs from patients with MGUS, SMM, and MM. (A) PRL-3 gene expression in PCs from 22 control subjects with normal BM, 44 patients with MGUS, 12 patients with SMM, 256 patients with MM, and in 45 myeloma cell lines. The Affymetrix signal, a quantitative measure of gene expression, is indicated on the y-axis. The level of expression of PRL-3 in each sample is indicated by the height of the bar. Samples are ordered from the lowest to highest level of expression of the PRL-3 gene, from left to the right on the x-axis. The table shows the results of the chi-square test by comparison of the Affymetrix Detection signal among the groups. The numbers in boxes above panels A and B indicate the Affymetrix annotation number of the PRL-3 gene. (B) PRL-3 gene expression in MM subgroups. The correlation of the Affymetrix signal (expression level: vertical axis) of PRL-3 with 7 myeloma subgroups from the 256 cases is shown on the top panel. The expression levels for PRL-3 are proportional to the height of each bar (representing a single patient sample). PRL-3 was significantly overexpressed in the proliferation (PR), MMSET/FGFR3, and low-bone disease (LB) groups. The table in the bottom panel shows the chi-square test for Affymetrix detection of PRL-3 among the 7 subgroups.
PRL-3 gene expression in normal PCs, MM cell lines, and PCs from patients with MGUS, SMM, and MM. (A) PRL-3 gene expression in PCs from 22 control subjects with normal BM, 44 patients with MGUS, 12 patients with SMM, 256 patients with MM, and in 45 myeloma cell lines. The Affymetrix signal, a quantitative measure of gene expression, is indicated on the y-axis. The level of expression of PRL-3 in each sample is indicated by the height of the bar. Samples are ordered from the lowest to highest level of expression of the PRL-3 gene, from left to the right on the x-axis. The table shows the results of the chi-square test by comparison of the Affymetrix Detection signal among the groups. The numbers in boxes above panels A and B indicate the Affymetrix annotation number of the PRL-3 gene. (B) PRL-3 gene expression in MM subgroups. The correlation of the Affymetrix signal (expression level: vertical axis) of PRL-3 with 7 myeloma subgroups from the 256 cases is shown on the top panel. The expression levels for PRL-3 are proportional to the height of each bar (representing a single patient sample). PRL-3 was significantly overexpressed in the proliferation (PR), MMSET/FGFR3, and low-bone disease (LB) groups. The table in the bottom panel shows the chi-square test for Affymetrix detection of PRL-3 among the 7 subgroups.
In pursuit of individually tailored therapy, the molecular classification of multiple myeloma27 is an important step forward. We found that there was a significant difference (P < .001) in PRL-3 gene expression across 7 subgroups of MM identified by unsupervised hierarchical cluster analysis of gene expression profiles from this large cohort of patients (Figure 3B). Thus, the groups denoted as proliferation (PR), low bone disease (LB), and MMSET/FGFR3 (MS) express higher levels of PRL-3.
In another patient and control population, we could confirm the same pattern of PRL-3 expression as in the gene expression experiments. RT-PCR on PCs from 12 patients showed high levels of PRL-3 in a subgroup of patients compared with the level in a patient with a benign disorder with only 3% to 4% PCs in the BM. There was no expression of PRL-3 in PCs from 5 healthy subjects (data not shown).
FISH analysis of the PRL-3 gene in myeloma cell lines
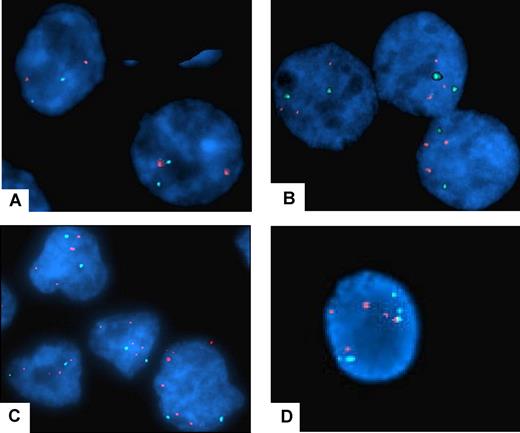
The enhanced PRL-3 expression in some samples of colorectal cancer is derived from increased copy number of the PRL-3 gene.6,12,32 We wanted to explore if the overexpression of PRL-3 in some cell lines used in this study correlated with the copy number of the PRL-3 gene. FISH analysis of the PRL-3 gene was performed on the myeloma cell lines OH-2, INA-6, IH-1, RPMI-8226, ANBL-6, JJN-3, U266, CAG, and on 2 nonmyeloma cell lines, K562 and Cro-AP5. In OH-2 cells, a cell line with a high PRL-3 level, both the PRL-3 probe and the centromeric probe gave 2 distinct signals inside the cells (Figure 4A), indicating that the number of PRL-3 genes was not increased. Therefore, the high level of PRL-3 in OH-2 cells (as shown in Figure 2) is unlikely to be caused by an increased copy number of the gene. RPMI-8226 cells (Figure 4B) showed 2 signals for probe CEP-8 and 3 for the PRL-3 probe. The extra copy might cause the increase in mRNA level (Figure 2). The other MM cell lines, as exemplified by IH-1 (Figure 4D) were shown to have 3 to 5 copies of chromosome 8. IH-1 had high levels of PRL-3 whereas CAG with 4 chromosomes 8 had very low levels of PRL-3 (Figure 2). Taken together, this indicates that there is no clear correlation between mRNA levels of PRL-3 and chromosome copy numbers in MM cell lines. However, an increased copy number of the PRL-3 gene (4 to 5 copies/cell, despite only 2 copies of chromosome 8) was detected in Cro-AP5 (Figure 4C), which is a primary effusion lymphoma cell line that expresses a high level of PRL-3 mRNA.
Fluorescent in situ hybridization with centromer and PRL-3 probe on MM cell lines and CRO-AP5. The results of FISH analysis with PRL-3 probe (SpectrumOrange) and centromer probe (SpectrumAqua). The red signals mark PRL-3 (8q24.3) and the aqua marks the centromer on chromosome 8. The cell nucleus is stained with DAPI. Original magnification is ×1000. (A) The OH-2 cell line has a normal chromosome 8 with 2 centromer signals and 2 PRL-3 signals. (B) The RPMI-8226 cell line has 2 centromer signals and 3 PRL-3 signals. (C) The CRO-AP5 cell line has 2 centromer signals and 4 to 5 PRL-3 signals, indicating that the area where the PRL-3 gene is located is amplified. (D) The IH-1 cell line has 5 PRL-3 signals and 5 centromer signals, demonstrating polyploidy of chromosome 8.
Fluorescent in situ hybridization with centromer and PRL-3 probe on MM cell lines and CRO-AP5. The results of FISH analysis with PRL-3 probe (SpectrumOrange) and centromer probe (SpectrumAqua). The red signals mark PRL-3 (8q24.3) and the aqua marks the centromer on chromosome 8. The cell nucleus is stained with DAPI. Original magnification is ×1000. (A) The OH-2 cell line has a normal chromosome 8 with 2 centromer signals and 2 PRL-3 signals. (B) The RPMI-8226 cell line has 2 centromer signals and 3 PRL-3 signals. (C) The CRO-AP5 cell line has 2 centromer signals and 4 to 5 PRL-3 signals, indicating that the area where the PRL-3 gene is located is amplified. (D) The IH-1 cell line has 5 PRL-3 signals and 5 centromer signals, demonstrating polyploidy of chromosome 8.
Immunohistochemical detection of PRL-3 in BM biopsies from patients with myeloma
We next examined the expression of PRL-3 protein in BM biopsies from 20 randomly selected patients. In 18 of 20 MM biopsies there was positive staining located almost exclusively in the PCs. The staining was predominantly cytoplasmic. Representative results of PRL-3 staining are shown in Figure 5. Normal BM did not show detectable PRL-3 staining (data not shown).
Expression of PRL-3 in malignant PCs from myeloma patients evaluated by immunohistochemistry. We stained BM biopsies from 20 patients, and found that 18 patients were PRL-3–positive to different degrees. In 11 of the biopsies more than 50% of the PCs were stained. A representative biopsy is illustrated. Original magnification ×400.
Expression of PRL-3 in malignant PCs from myeloma patients evaluated by immunohistochemistry. We stained BM biopsies from 20 patients, and found that 18 patients were PRL-3–positive to different degrees. In 11 of the biopsies more than 50% of the PCs were stained. A representative biopsy is illustrated. Original magnification ×400.
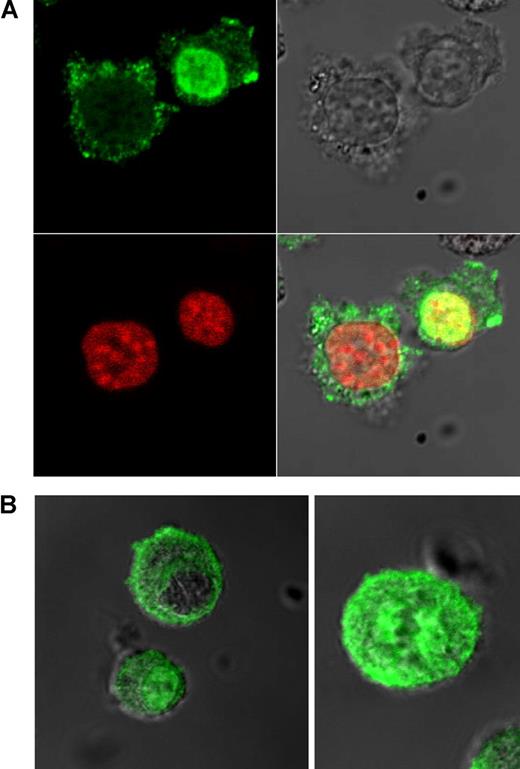
Confocal microscopy demonstrated specific localization of PRL-3 in myeloma cells
Interestingly, anti–PRL-3 stained cells in the interphase in a specific pattern. The PRL-3 protein seemed to cycle between cytosol and nucleus in a cell cycle–dependent way, as exemplified by the OH-2 cell line in Figure 6A. OH-2 cells were sorted in G0/G1, S, and G2M phases and stained against PRL-3. The nuclear localization dominated in the G0/G1 phase, whereas the distribution of PRL-3 in the S and G2M phases was more equal between the nucleus and cytoplasm (Table 1). Exclusive staining of the cytoplasm was seen only in the G2M phase (data not shown). When the OH-2 cells were treated with BMP-4, which is known to increase the proportion of cells in G0/G1,33 we observed an increase in nuclear PRL-3 staining. On the other hand, cells treated with paclitaxel (Sigma-Aldrich), which arrests cells in G2M, resulted in an increased number of cells without nuclear staining (data not shown) compared with controls. In cells from patients with myeloma there was a similar staining pattern with PRL-3 localized either exclusively in the nucleus or in the cytoplasm (Figure 6B).
Staining of OH-2 cells in interphase and patient cells in interphase and metaphase with PRL-3. Goat polyclonal antibody against PRL-3 is shown in green. Nuclei visualized with Draq 5 are shown in red. (A) OH-2 cells in interphase. (B) Patient cells in interphase and metaphase. Examination by confocal microscopy.
Staining of OH-2 cells in interphase and patient cells in interphase and metaphase with PRL-3. Goat polyclonal antibody against PRL-3 is shown in green. Nuclei visualized with Draq 5 are shown in red. (A) OH-2 cells in interphase. (B) Patient cells in interphase and metaphase. Examination by confocal microscopy.
| OH-2 cell cycle . | PRL-3 nuclear localization . | PRL-3 in cytoplasm . | Ratio nuclear /cytoplasm . |
|---|---|---|---|
| G0G1 | 94% | 6% | 15.50 |
| S phase | 78.2% | 21.8% | 3.60 |
| G2M | 67.3% | 32.7% | 2.05 |
| OH-2 cell cycle . | PRL-3 nuclear localization . | PRL-3 in cytoplasm . | Ratio nuclear /cytoplasm . |
|---|---|---|---|
| G0G1 | 94% | 6% | 15.50 |
| S phase | 78.2% | 21.8% | 3.60 |
| G2M | 67.3% | 32.7% | 2.05 |
The OH-2 cells were cell cycle–separated and localization of PRL-3 was determined. A total of 100 to 200 cells per cycle were evaluated for nuclear and cytoplasmic staining and the ratio nuclear/cytoplasma was calculated.
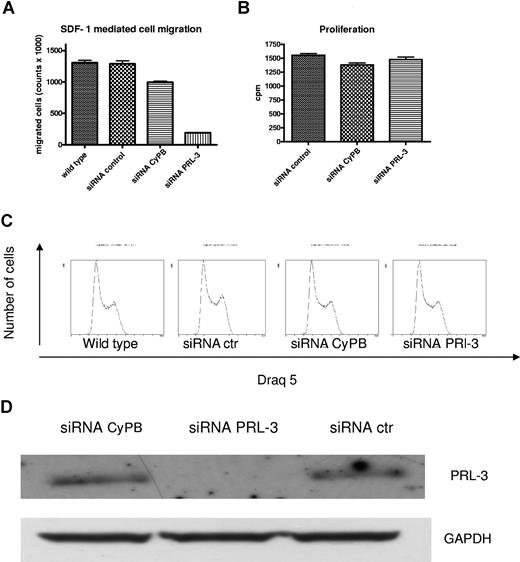
siRNA-mediated down-regulation of PRL-3 reduced migration of INA-6 cells
There are several reports that PRL-3 may play a role in cell migration and invasion.6,7,10,–12,14,15 We transfected INA-6 cells with siRNA against PRL-3, and verified the down-regulation on Western blots after 2 and 3 days (Figure 7D). Phenotype changes were evaluated with assays measuring thymidine incorporation, cell-cycle analysis, and SDF-1–induced migration in INA-6 cells. Interestingly, reduction of PRL-3 to levels undetectable on Western blots did not affect proliferation or cell-cycle distribution, but gave a significant reduction in SDF-1–induced migration (Figure 7A-D).
The effect of down-regulation of PRL-3 expression on migration, proliferation, and cell cycle in INA-6 cells. (A) Transfection of PRL-3 siRNA reduced SDF-1–induced migration in INA-6 cells as compared with transfection with control siRNAs. Wild-type INA-6 cells are cells stimulated with SDF-1 but not transfected. (B) PRL-3 siRNA as well as control siRNAs did not influence the proliferation of INA-6 cells. (C) PRL-3 siRNA did not induce cell-cycle arrest. The percentages of cells in G0/G1, S, and G2M in wild-type INA-6 were 35.2%, 44.9%, and 19.9%, respectively. In transfected cells, the percentages were 32.3%, 44.4%, and 23.4% in siRNA-negative control; 29.7%, 46.2%, and 24.1% in positive control siRNA cyclophilin B; and 32.2%, 46.6%, and 21.3% in siRNA PRL-3, respectively. (D) The down-regulation of the PRL-3 protein was verified on Western blot. Similar results were shown in 3 independent experiments. Error bars represent +1 standard deviation of 5 (migration) and 4 (thymidine incorporation) measurements.
The effect of down-regulation of PRL-3 expression on migration, proliferation, and cell cycle in INA-6 cells. (A) Transfection of PRL-3 siRNA reduced SDF-1–induced migration in INA-6 cells as compared with transfection with control siRNAs. Wild-type INA-6 cells are cells stimulated with SDF-1 but not transfected. (B) PRL-3 siRNA as well as control siRNAs did not influence the proliferation of INA-6 cells. (C) PRL-3 siRNA did not induce cell-cycle arrest. The percentages of cells in G0/G1, S, and G2M in wild-type INA-6 were 35.2%, 44.9%, and 19.9%, respectively. In transfected cells, the percentages were 32.3%, 44.4%, and 23.4% in siRNA-negative control; 29.7%, 46.2%, and 24.1% in positive control siRNA cyclophilin B; and 32.2%, 46.6%, and 21.3% in siRNA PRL-3, respectively. (D) The down-regulation of the PRL-3 protein was verified on Western blot. Similar results were shown in 3 independent experiments. Error bars represent +1 standard deviation of 5 (migration) and 4 (thymidine incorporation) measurements.
Discussion
We have described here for the first time that PRL-3 expression was increased in several MM cell lines stimulated with different growth-promoting cytokines. The increased expression is demonstrated at the gene, mRNA, and protein levels. We have also shown in a large cohort of patients that this gene was expressed at a higher level in PCs from MGUS, SMM, and MM than in normal PCs and that the protein was expressed in BM samples from patients with MM.
The PRL-3 expression in normal tissue in humans is not well characterized. So far it is only known to be expressed in the heart, skeletal muscle, and pancreas.6,13,34,–36 In contrast, the expression of PRL-3 in tumor tissue and tumor cell lines is far more abundant (reviewed in Stephens et al13 ).
Several recent studies6,10,12 have shown that PRL-3 is expressed at higher levels and at a greater frequency in colorectal cancer metastases than in primary colorectal tumors and normal colon tissue. The same pattern has been shown in human gastric carcinomas10 and there is also a report showing that PRL-3 was overexpressed in liver carcinoma cells compared with normal liver cell tissue.15 PRL-3 has also been detected in breast cancer tissue as well as in invasive breast tumor vasculature.37,38 It was also found to be highly expressed in the Hodgkin lymphoma cell line L1236, compared with a nonmalignant counterpart.39
In contrast to the more ubiquitous expression of PRL-1 and PRL-2 in various tissues (reviewed in Stephens et al13 ), PRL-3 seems to be a cancer-related gene, sparsely expressed in normal tissues, and expressed in just 1 of 22 normal plasma cell samples. This suggests that PRL-3 expression is one of the fundamental changes associated with the malignant transformation of PCs.
We have previously shown, based on gene expression signatures in a large population of samples from patients with myeloma, that the disease can be classified into 7 distinct molecular entities27 defined for the most part on recurrent translocations and hyperdiploidy. When looking at PRL-3 gene expression in 256 cases,27 we found a highly significant (P < .001) difference across the 7 subgroups. PR and MS groups, representing high-risk entities in the tandem transplant setting, and the LB group, harbored the highest expression level of PRL-3. As the PR group is derived from the transformation of the other groups,27 it is interesting to speculate if the PRL-3–positive cases within the PR group are derived from the MS and LB subtypes that have converted, or if the PRL-3 gene is activated in progression of other subtypes that do not express the gene as part of an initiating event. All patients were given the same treatment. However, we could not demonstrate that the level of PRL-3 expression had prognostic impact. This is not surprising given the fact that the PRL-3 gene is elevated both in low-risk (LB) and high-risk (MS) cases. A better test is to show whether LB patients with high PRL-3 are at increased risk of converting. Longer follow-up will be required to answer this question.
Our findings are in line with data from an accumulating list of different human cancers showing increased expression of PRL-3 and, in some instances, correlation with progression and survival.6,10,14,38,40,–42
Migration is a fundamental process for the myeloma tumor cells when they invade the BM and disseminate, and data indicate that SDF-1 is instrumental in attracting the cells to the BM.30,43,44 We showed that siRNA-mediated down-regulation of the PRL-3 gene expression reduced SDF-1–induced migration of INA-6 cells, suggesting a role for PRL-3 in myeloma cell migration.
This is in line with data from several other malignancies, among them colorectal carcinoma and breast cancer,14,15,45,46 showing reduced migration/invasion after down-regulating PRL-3. Stable expression of wild-type, active PRL-3 has been shown to enhance cell migration to a great extent, whereas the catalytically inactive PRL-3 (C104S) mutant reduced cell migration,16 indicating that cell migration is dependent on the phosphatase activity. Even though PRL-3 expression was increased by cytokines known to promote cell proliferation in myeloma cells, we were not able to show that PRL-3 played a role in cell proliferation, nor did knockdown of the PRL-3 gene induce cell-cycle arrest. This was somewhat surprising given the observed cell cycle–dependent localization of PRL-3, which indicates a role for the protein in cell-cycle progression. Data from other groups47,48 support a role for PRL-3 in proliferation and transformation. The different results in different malignancies, after knock-down of the gene,14,45,47 may reflect different roles for the phosphatase depending on cell type. Another possible explanation could be differences in the methods used for evaluation.
The regulation of PRL-3 expression is not well understood. We show here that PRL-3 mRNA was up-regulated by cytokines, and that the same principal pattern was confirmed at the protein level in myeloma cell lines. Our findings showing cytokine-induced expression of PRL-3 at the mRNA level in the OH-2 and IH-1 cell lines are in line with previously published array lists that show up-regulation of PRL-3 mRNA by IL-6 in the myeloma cell lines INA-6 and ANBL-6.49,50 Interestingly, we also showed up-regulation of PRL-3 after IL-6 stimulation in RPMI-8226 and JJN-3, 2 cell lines that do not need IL-6 for proliferation.
Taken together, this underscores the significance of exogenous stimuli and may be an indication of the importance of the microenvironment for expression of the PRL-3 gene. Whether the difference in PRL-3 expression between normal and malignant PCs reflects genetic aberrations in malignant PCs or is a reflection of perturbations in the cytokine network in the BM of patients with monoclonal gammopathies is an unresolved question. However, the observation that PRL-3 is present in PCs from many patients in the MGUS group, in which an aberrant cytokine network is presumably not a common feature, argues in favor of malignant PCs being more prone to express PRL-3 upon stimulation than healthy PCs.
We did FISH to explore if the overexpression in our cell lines could be at least partly due to amplification. Our data indicated that amplification of the PRL-3 gene was not the main cause of overexpression in the myeloma cell lines, but that it might play a role in the relatively high PRL-3 expression in RPMI-8226, one of the cytokine-independent cell lines. OH-2, which showed the highest expression of PRL-3 mRNA, had a normal copy number of the PRL-3 gene. These results indicate that a high chromosome number does not correlate with gene expression levels in MM cell lines. Most likely, mechanisms other than gene amplifications due to chromosome copy number are involved in most of the up-regulation of PRL-3 expression, and our results suggest that microenvironment factors like mitogenic cytokines are instrumental. PRL-3 gene amplification was found in the nonmyeloma cell line with highest PRL-3 expression, the primary effusion lymphoma cell line CRO-AP5. Interestingly, primary effusion lymphoma cells share common characteristics with myeloma cells as they express plasma cell-related markers such as CD138, and seem to be derived from post–germinal center B cells that have undergone preterminal differentiation.51
We still know little about the substrates and the signaling pathways for PRL-3 but Peng et al52 recently showed that integrin-α1 is a protein interacting with PRL-3. Integrins are involved in adhesion and migration, and the importance of integrin-β1 in adhesion in INA-6 cells was published earlier by our group.53 Fiordalisi7 showed that PRL-3 promotes the activation of the Rho family GTPases RhoA and RhoC, molecules that are known to have a profound effect on the actin cytoskeleton and thereby on cell migration.7,54,55 A recent report56 showed that PRL-3 down-regulates phosphatase and tensin homolog (PTEN) expression and signals through PI3K to promote epithelial-mesenchymal transition (EMT) in the cell line DLD-1. So far nothing is known about substrates and signaling of PRL-3 in MM. However, SDF-1 activates RhoA (R.U.H., unpublished data, July 2007) in the INA-6 cell line, and together with our present siRNA data on SDF-1–induced migration, this may suggest a role for Rho GTPases in PRL-3–mediated migration in multiple myeloma. Further research to explore this possibility is needed.
Taken together, these data suggest that PRL-3 is one of the proteins produced by the myeloma cells in response to several growth-promoting cytokines, and that it may have a role in migration also in myeloma cells. Recently, nuclear magnetic resonance (NMR) structure data on PRL-3 were published,8,9 an important prerequisite for developing candidate inhibitors. Several protein tyrosine phosphatases (PTPs) seem to be attractive drug targets,57 and PRL-3 could be a molecular target in subgroups of patients with myeloma.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Prof Qi Zeng for the kind gift of the monoclonal PRL-3 antibody. We thank Berit Størdal, Borgny Ytterhus, and Hanne Hella for excellent technical help.
This work is supported by grants from the Norwegian Cancer Society, the Cancer Fund of St Olavs Hospital, Trondheim, the Research Council of Norway, Familien Blix' fond‘, Rakel og Otto Kr Bruuns legat, the Lebow Fund to Cure Myeloma, the Nancy and Stephen Grand Philanthropic Fund, and National Institutes of Health grant P01 CA55819 from the National Cancer Institute (F.Z., B.B., and J.D.S.).
National Institutes of Health
Authorship
Contribution: U.M.F. designed and performed research, analyzed data, and wrote the paper; R.U.H., T.H., T.K.V., K.W.E., H.Y.D., M.B., and H.A. performed research; A.W. and B.B. provided clinical samples; J.D.S., F.Z., and B.B. performed microarrays and analyzed the data; J.D.S. contributed to writing the paper; A.S. and M.B. designed research, analyzed data, and supervised writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Unn-Merete Fagerli, Department of Cancer Research and Molecular Medicine, Norwegian University of Science and Technology, MTFS, N-7489 Trondheim, Norway; e-mail: unn-merete.fagerli@ntnu.no.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal