ALK1 belongs to the type I receptor family for transforming growth factor-β family ligands. Heterozygous ALK1 mutations cause hereditary hemorrhagic telangiectasia type 2 (HHT2), a multisystemic vascular disorder. Based largely on in vitro studies, TGF-β1 has been considered as the most likely ALK1 ligand related to HHT, yet the identity of the physiologic ALK1 ligand remains controversial. In cultured endothelial cells, ALK1 and another TGF-β type I receptor, ALK5, regulate angiogenesis by controlling TGF-β signal transduction, and ALK5 is required for ALK1 signaling. However, the extent to which such interactions between these 2 receptors play a role in pathogenesis of HHT is unknown. We directly addressed these issues in vivo by comparing the phenotypes of mice in which the Alk1, Alk5, or Tgfbr2 gene was conditionally deleted in restricted vascular endothelia using a novel endothelial Cre transgenic line. Alk1-conditional deletion resulted in severe vascular malformations mimicking all pathologic features of HHT. Yet Alk5- or Tgfbr2-conditional deletion in mice, or Alk5 inhibition in zebrafish, did not affect vessel morphogenesis. These data indicate that neither ALK5 nor TGFBR2 is required for ALK1 signaling pertinent to the pathogenesis of HHT and suggest that HHT might not be a TGF-β subfamily disease.
Introduction
Hereditary hemorrhagic telangiectasia (HHT) is an autosomal-dominant vascular disorder characterized by recurrent nosebleeds, mucocutaneous telangiectases, and arteriovenous malformations (AVMs) in the brain, lungs, liver, and gastrointestinal tract.1,2 It has been shown that heterozygous mutations in ENDOGLIN (ENG) and Activin receptor-like kinase 1 (ALK1) cause HHT1 and HHT2, respectively.2,–4 Both of these genes are expressed predominantly in endothelial cells.5,6 Because ENG and ALK1 are transforming growth factor-β (TGF-β) type III and type I receptors, respectively, it has been postulated that HHT is caused by impaired signaling of a common TGF-β family ligand that interacts with these 2 receptors. Recent finding of mutations in the common downstream mediator of TGF-β family signals, SMAD4, in a subset of HHT patients also support this hypothesis.7
Despite the identification of these genes responsible for HHT, the underlying mechanisms for the pathogenesis of HHT remain obscure. One of the chief contributing factors underlying this obscurity is the complexity of the transduction pathway of ENG, ALK1, and SMAD4. The TGF-β superfamily consists of more than 40 ligands that can be classified into several subfamilies, including TGF-β, Activin, and bone morphogenetic protein (BMP).8 TGF-β family cytokines exert their effects by binding to heteromeric complexes of 2 types of transmembrane serine/threonine kinase receptors.9 The type II receptors function primarily as the binding receptors. On binding their ligand(s), type II receptors associate with and phosphorylate the type I receptors, which in turn activate downstream SMAD proteins. Each TGF-β ligand interacts with one or more type II and type I receptors, but TGFBR2 is the only type II receptor that has been shown to interact with TGF-β subfamily ligands (TGF-β1, -β2, and -β3).
ENG can interact with multiple TGF-β family members, such as TGF-β1/β3, Activin-A, BMP2, and BMP7, in the presence of a suitable ligand-binding type II or type I receptor.10 Likewise, ALK1 can also interact with a variety of TGF-β ligands, including TGF-β1, TGF-β3, BMP-9, and BMP-10.11,–13 TGF-β1, a common ligand for both ENG and ALK1, is widely considered to be the most likely TGF-β family ligand relevant to HHT pathogenesis. TGF-β1 uses ALK5 (TGFBR1) as a type I receptor in most cells. However, in cultured vascular endothelial cells (ECs), ALK5, ALK1, and ENG can bind to TGF-β1 in the presence of TGFBR2.10,14 Downstream of receptor activation, the ALK5 and ALK1 pathways diverge: activated ALK5 phosphorylates SMAD2/3, whereas activated ALK1 phosphorylates SMAD1/5/8.14,15 Therefore, it was hypothesized that TGF-β may elicit an opposing cellular outcome, in terms of controlling proliferation and migration of ECs, depending on which type I receptor and thus which SMAD signaling pathway is predominant (Figure 1A).14
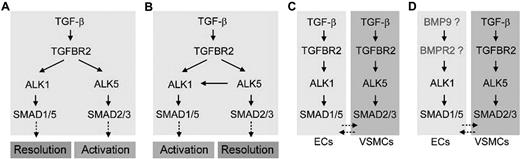
Multiple versions of TGF-β signaling pathways in endothelial and smooth muscle cells. During the activation phase of angiogenesis, endothelial cells degrade their vascular basement membranes, migrate into extracellular spaces, proliferate, and form vascular lumens. During the resolution phase, endothelial cells cease to migrate and to proliferate and instead reconstitute their basement membranes. The maturation and remodeling of the vessels also occur in this phase, as mesenchymal cells are recruited for endothelial tube ensheathment. (A,B) It has been reported that both ALK1 and ALK5 are TGF-β subfamily type I receptors in ECs: that is, they are both activated by TGF-β subfamily ligands binding to TGFBR2. As ALK1 and ALK5 signal through different SMAD proteins, it has been suggested that the opposing activities of these 2 type I receptors regulate angiogenesis. However, whereas some studies have suggested a role or ALK1 in resolution and ALK5 in activation,14,17 others have suggested opposite roles, with ALK5 being necessary for ALK1 function.16,20 (C) Both balance models (A,B) are called into question by an expression study in mice showing that, whereas Alk1 is endothelial-specific, Alk5 is expressed not in the endothelium but in neighboring smooth muscle cells.21 (D) Data presented do not support a role for TGF-β subfamily ligands and TGFBR2 in ALK1 function, suggesting that TGF-β superfamily ligands outside of the TGF-β subfamily may be physiologic ligands for ALK1 in endothelial cells. This hypothesis is supported by recent biochemical data demonstrating that BMP9 serves as an ALK1 ligand.12,13
Multiple versions of TGF-β signaling pathways in endothelial and smooth muscle cells. During the activation phase of angiogenesis, endothelial cells degrade their vascular basement membranes, migrate into extracellular spaces, proliferate, and form vascular lumens. During the resolution phase, endothelial cells cease to migrate and to proliferate and instead reconstitute their basement membranes. The maturation and remodeling of the vessels also occur in this phase, as mesenchymal cells are recruited for endothelial tube ensheathment. (A,B) It has been reported that both ALK1 and ALK5 are TGF-β subfamily type I receptors in ECs: that is, they are both activated by TGF-β subfamily ligands binding to TGFBR2. As ALK1 and ALK5 signal through different SMAD proteins, it has been suggested that the opposing activities of these 2 type I receptors regulate angiogenesis. However, whereas some studies have suggested a role or ALK1 in resolution and ALK5 in activation,14,17 others have suggested opposite roles, with ALK5 being necessary for ALK1 function.16,20 (C) Both balance models (A,B) are called into question by an expression study in mice showing that, whereas Alk1 is endothelial-specific, Alk5 is expressed not in the endothelium but in neighboring smooth muscle cells.21 (D) Data presented do not support a role for TGF-β subfamily ligands and TGFBR2 in ALK1 function, suggesting that TGF-β superfamily ligands outside of the TGF-β subfamily may be physiologic ligands for ALK1 in endothelial cells. This hypothesis is supported by recent biochemical data demonstrating that BMP9 serves as an ALK1 ligand.12,13
Two independent groups tested this hypothesis using similar approaches, but their respective data were in disagreement.16,17 Lamouille et al showed that ALK1 signaling inhibited the proliferation and migration of ECs (Figure 1A),17 whereas Goumans et al showed that ALK1 signaling promoted proliferation and migration of ECs (Figure 1B).16 The role of ENG in ALK1 signaling is another conflicting issue. Whereas Lebrin et al demonstrated that ENG promoted TGF-β/ALK1 signaling and EC proliferation,18 Pece-Barbara et al reported that loss of ENG function resulted in increased ALK1 signaling and EC proliferation.19 To complicate matters further, it has also been shown in cultured endothelial cells that TGF-β1 induces heteromeric complex formation of ALK1 and ALK5 and that ALK5 is required for TGF-β1 signaling via ALK1 (Figure 1B).20 Taken together, these data paint an unclear picture of the roles of ALK1 and ALK5 in endothelial cells; importantly, the in vivo relevance of these data has only begun to be addressed. We have recently shown that in mice, Alk1 expression was found predominantly in the endothelium, whereas Alk5 expression was detected primarily in vascular smooth muscle layers and not in the endothelium.5,21 This finding challenges the ALK1/ALK5 balance model and leads us to hypothesize that ALK1 is the sole TGF-β type I receptor in ECs and that ALK5 does not play a direct role in TGF-β family signaling within these cells (Figure 1C).
To test this hypothesis directly in vivo, we generated mice in which Alk1, Alk5, or Tgfbr2 was conditionally deleted in endothelial cells. We also inhibited Alk5 activity in wild-type and alk1-mutant zebrafish. We predicted that, if the preceding hypothesis were correct, endothelium-specific Alk1- and Tgfbr2-knockout mice would exhibit similar vascular phenotypes, whereas endothelium-specific Alk5-knockout mice or zebrafish treated with an Alk5 inhibitor would exhibit no vascular phenotype. Our results uphold our hypothesis that ALK1 activity does not require ALK5 activity in endothelial cells in the developing embryo, and demonstrate that ALK5 activity is not critical for development of the vascular intima (Figure 1D). Furthermore, results suggest that TGFBR2 is not relevant to ALK1 signaling in endothelial cells in vivo and imply that impaired signaling through TGF-β superfamily ligands outside of the TGF-β subfamily may be most important in HHT pathogenesis (Figure 1D).
Methods
The Alk1loxP and Tg(Alk1-cre) mice were generated at the University of Florida. R26R and Tg(EIIa-cre) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Establishment of Alk5loxP and Tgfbr2loxP lines was described previously.22,23 All mouse procedures performed were reviewed and approved by the University of Florida Institutional Animal Care and Use Committee.
Generation of Alk1loxP mice
For the construction of the Alk1 conditional knockout vector, a modified pflox vector (pflox-Δtk) was created by 2 modifications. First, one of the 2 NotI sites that is located next to the SalI site was eliminated. Second, EcoRI fragment containing the herpes simplex virus thymidine kinase (HSV-tk) gene was removed. The following mouse Alk1 genomic fragments were obtained as previously described5 and were subcloned into the designated unique restriction enzyme sites in the pflox-Δtk vector: the 2 kb HindIII-HindIII fragment containing exon 3 into NotI site, and the 1.2 kb HindIII-HindIII fragment containing exons 4-6 into the BamHI site as 5′ arm; and the 5 kb BamHI-BamHI fragment containing exons 7-9 into the XbaI site as 3′ arm (Figure 2A). A diphtheria toxin A selection marker24 was subcloned into SalI site for negative selection. After electroporation of the linearized construct, approximately 400 G418-resistant colonies were screened by Southern blot analysis using a 5′ external probe (5′-probe in Figure 2A,B). One positive clone was microinjected into C57BL/6J (B6) blastocysts to generate chimeras, which were crossed with B6 mice to establish and maintain the Alk13loxP mouse line on a mixed 129/B6 hybrid background.
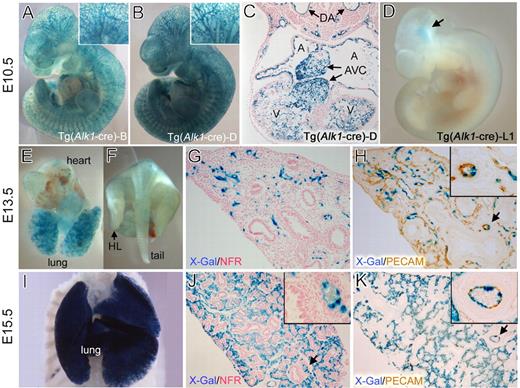
Tg(Alk1-cre)-L1 mice express the Cre recombinase predominantly in the pulmonary vascular endothelial cells. The cells in which Cre–mediated recombination has occurred were visualized by staining the Tg(Alk1-cre);R26R bigenic embryos with X-gal for the β-gal activity at E10.5 (A-D), E13.5 (E-H), and E15.5 (I-K) stages. (A,B) X-gal-positive staining is visible in the blood vessels throughout embryos in the Tg(Alk1-cre)-B (A) and -D (B) lines. (C) Transverse sections of X-gal stained Tg(Alk1-cre)-D embryo showing lacZ expressions in the vascular ECs, endocardial cells in atria and ventricles, and mesenchymal cells in the atrioventricular cushion (AVC). (D) In contrast with that in the B and D lines, almost no lacZ expression was detected in the L1cre embryos; only a spotty staining pattern in the head region (arrow). (E,F) Dorsal aorta (DA) view of the heart and lungs of L1cre:R26R embryos stained with X-gal, showing a strong lacZ expression in the lung in comparison with a patch staining in the heart (E) and body trunk (F). (G,H) Histologic sections of the X-gal stained lung was counterstained with NFR (G) or costained with anti-PECAM antibodies (H). The inset in panel H is a magnified view of the area indicated by the arrow. Note that X-gal-positive cells resided in pulmonary ECs, but only in a subpopulation of PECAM-positive cells. (I) Ventral view of the X-gal stained L1cre:R26R lung attached to the body trunk. The heart was removed for clarity of the view. Note a strong X-gal staining in the lung but not in the body trunk. (J,K) Histologic sections demonstrate that most PECAM-positive cells are positive for X-gal staining in E15.5 embryonic lungs, yet no X-gal-positive cells were detected in airway epithelial and smooth muscle cells. Insets are magnified views of the areas indicated by the arrow in each panel.
Tg(Alk1-cre)-L1 mice express the Cre recombinase predominantly in the pulmonary vascular endothelial cells. The cells in which Cre–mediated recombination has occurred were visualized by staining the Tg(Alk1-cre);R26R bigenic embryos with X-gal for the β-gal activity at E10.5 (A-D), E13.5 (E-H), and E15.5 (I-K) stages. (A,B) X-gal-positive staining is visible in the blood vessels throughout embryos in the Tg(Alk1-cre)-B (A) and -D (B) lines. (C) Transverse sections of X-gal stained Tg(Alk1-cre)-D embryo showing lacZ expressions in the vascular ECs, endocardial cells in atria and ventricles, and mesenchymal cells in the atrioventricular cushion (AVC). (D) In contrast with that in the B and D lines, almost no lacZ expression was detected in the L1cre embryos; only a spotty staining pattern in the head region (arrow). (E,F) Dorsal aorta (DA) view of the heart and lungs of L1cre:R26R embryos stained with X-gal, showing a strong lacZ expression in the lung in comparison with a patch staining in the heart (E) and body trunk (F). (G,H) Histologic sections of the X-gal stained lung was counterstained with NFR (G) or costained with anti-PECAM antibodies (H). The inset in panel H is a magnified view of the area indicated by the arrow. Note that X-gal-positive cells resided in pulmonary ECs, but only in a subpopulation of PECAM-positive cells. (I) Ventral view of the X-gal stained L1cre:R26R lung attached to the body trunk. The heart was removed for clarity of the view. Note a strong X-gal staining in the lung but not in the body trunk. (J,K) Histologic sections demonstrate that most PECAM-positive cells are positive for X-gal staining in E15.5 embryonic lungs, yet no X-gal-positive cells were detected in airway epithelial and smooth muscle cells. Insets are magnified views of the areas indicated by the arrow in each panel.
Generation of Tg(Alk1-cre) lines
Procedures for generation of the transgene constructs are detailed in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Inserts of pBSmA1SIACre and pBSAlk1eGFPCreTg vectors were injected into male pronuclei of FVB strain. F1 mice from each founder were intercrossed with R26R mice, and their bigenic offspring at various embryonic stages were subject to X-gal staining. Mice showing consistent expression patterns among their progeny were maintained as stable transgenic lines.
Genotyping
PCR genotyping primer sequences for Alk1, Alk5, and Tgfbr2 alleles are listed in Document S1.
X-gal staining, dye injections, histology, and immunostaining
Tissue samples were dissected in 1- to 2-mm slices to facilitate penetration of the X-gal solution, and stained overnight as described previously.5 Black India ink (Universal 3080-F; Koh-I-Noor, Bloomsbury, NJ) was mixed with heparin (100 unit/mL) in 1:1, and filtered through 0.22 μm Millipore filter before injection into the vitelline artery with pulled capillary glass pipettes. The stained samples were subsequently fixed with 10% formalin, 4% paraformaldehyde or zinc fixative, and either embedded in paraffin for histology or cleared with organic solvent (benzyl alcohol:benzyl benzoate = 1:1; Sigma, St Louis, MO) for whole-mount imaging. [Whole mount specimens immersed in phosphate buffered saline were viewed with a Leica MZ8 inverted microscope (Leica Microsystems, Wetzlar, Germany) using an optic 10×/21B and objective plain 1.0. The images were acquired using a Leica DFC 320 digital camera with Leica Application Suite version 2.4.0 R1 software and processed with Adobe Photoshop CS3 software (Adobe Systems, San Jose, CA).] The embedded samples were cut into 6- to 7-μm slices. Each lung sample was transversely cut with 5μm thickness and subjected to hematoxylin and eosin staining and immunostaining with antibody against anti–α-smooth muscle actin (SMA; clone: 1A4, 1:800; Sigma). The X-gal stained sections were counterstained with nuclear fast red (NFR) or immunostained with anti-αSMA, or anti–platelet endothelial cell adhesion molecule (PECAM; clone: Mec13.3, 1:200; BD Pharmingen, San Diego, CA) antibodies. The standard ABC method was used with a Vector M.O.M. staining kit (Vector Laboratories, Burlingame, CA). After the secondary antibody reaction, color development was carried out with DAB+ substrate chromogenic solution (Vector Laboratories). [Sectional slide were inspected using Zeiss Axiophot II compound microscope (Carl Zeiss). The images were captured using Retiga Qimage CCD camera (QIMAGING, Burnaby, BC) and OpenLab 5.0.3 imaging software (Improvision, Waltham, MA) and processed with Adobe Photoshop CS3 software (Adobe Systems).]
Zebrafish methods
Adult zebrafish were maintained and embryos raised and staged using standard methods.25 Using bioinformatics approaches, portions of alk5a (GenBank Accession number EF462417) and alk5b (GenBank Accession number EF462418) were identified in the zebrafish genome on chromosomes 2 and 24, respectively, and full-length cDNAs were isolated by PCR and RACE (SMART RACE, BD Biosciences, San Jose, CA). Chromosome assignment was confirmed using the LN54 radiation hybrid mapping panel,26 and in situ hybridization was performed as previously described.27 [Brightfield images were captured using an Olympus MVX-10 MacroView macro zoom microscope equipped with an MV PLAPO 1×/0.25 NA objective, 2× magnification changer, and an Olympus DP71 camera (DP controller software, version 3.1; Olympus America, Center Valley, PA).] To determine the role of Alk5 in Alk1 signaling in the endothelium, embryos were generated from an alk1+/−; TG(flk1:GFP)la116 intercross and exposed to 100 μM SB-431542 (Tocris Bioscience, Bristol, United Kingdom) or 1% dimethyl sulfoxide (DMSO) beginning at the 8- to 10-somite stage. Medium was changed every 8 hours. Vessels were viewed with a Fluoview500 laser scanning confocal microscope (Olympus America) using a UMPLFL20×W/0.50 NA water immersion objective, and Z-series were projected in 2 dimensions. All images were compiled using Adobe Photoshop 9.0.2 (Adobe Systems). To assess efficacy of SB-431542 against zebrafish Alk5 in vivo, constitutively active28 alk5a and alk5b were generated using QuikChange (Stratagene), converting threonine 201 or 206, respectively, to aspartic acid. mRNA was generated using mMessage mMachine (Ambion, Austin, TX); 1 pg was injected into 1- to 4-cell stage wild-type embryos, and embryos were exposed to 100 μM SB-431542 or 1% DMSO at the 8-cell stage. To assess efficacy of late SB-431542 exposure, 85 pg 3TP-lux (Smad2/3-responsive reporter), BRE-lux (Smad1/5-responsive reporter), or pGL2 basic (vector backbone) was injected into zebrafish embryos at the 1-cell stage, embryos were exposed to 100 μM SB-431542 or 1% DMSO beginning at the 8- to 10-somite stage, and reporter activity was analyzed at 32 hours after fertilization (hpf) using the Luciferase Assay System (Promega, Madison, WI).
Results
Generation of a novel endothelium-specific Cre-expressing transgenic line
We have previously shown that a 9.2-kb Alk1 regulatory sequence is sufficient to drive transgene expression in arterial endothelium and pulmonary vessels.29 Using this 9.2-kb regulatory fragment, we have generated Cre transgenic lines, named, Tg(Alk1-cre). Because the transgene expression is often dependent on endogenous regulatory elements near the chromosomal site of transgene integration, we characterized multiple independent founder lines. The spatiotemporal Cre expression was monitored by crossing several F1 mice from each founder line with the ROSA26-lacZ reporter line (R26R), in which β-galactosidase (β-gal) is designed to be expressed only in cells expressing the Cre recombinase.30 Cre-mediated gene excision was assessed by staining tissues/organs from Cre:R26R bigenic mice at various embryonic and postnatal stages with X-gal.
The Tg(Alk1-cre) founder lines could be categorized into 3 groups. In the first group of several founder lines, the β-gal activity was found in ectopic cell types, such as neuronal cells or some epithelial cells, in addition to endothelial cells (data not shown). The second group of mice, designated Tg(Alk1-cre)-B and -D, displayed the X-gal staining pattern consistent with our expectation based on the endogenous Alk1 expression pattern.5 As shown in Figure 3A,B, the β-gal activity was detected in blood vessels throughout the embryos. Histologic sections revealed that the X-gal–positive cells were restricted to vascular endothelial cells (Figure 3C). Progeny of endocardial cells, such as atrioventricular (AV) cushions, were also X-gal–positive (Figure 3C). The third group, designated Tg(Alk1-cre)-L1 (L1cre hereafter), displayed a peculiar temporal Cre expression pattern. Unlike Tg(Alk1-cre)-B and -D lines, the β-gal activity was almost undetectable in the embryo proper of embryonic day (E) 10.5 L1cre;R26R embryos (Figure 3D). As development proceeded, the β-gal activity was detected predominantly in the lungs (Figure 3E,I), whereas all other systemic organs and tissues had some patchy staining in the blood vessels (Figure 3F,I). In E13.5 lungs, most X-gal–positive cells were restricted to PECAM-positive ECs, but not all PECAM-positive ECs were X-gal positive (Figure 3G,H). However, by E15.5, almost all lung ECs exhibited β-gal activity (Figure 3J,K). β-gal activity was also detected in extraembryonic vessels of L1cre;R26R mice from E9.5 (Figure S1). No X-gal staining was detected in endocardial, cardiac muscle, or endocardial cushion cells of L1cre;R26R mice at any stages examined (data not shown).
Generation of Alk1-conditional alleles. (A) Schematic diagram of the Alk1 wild-type allele, Alk1-conditional targeting vector, and Alk13loxP, Alk12loxP, and Alk11loxP alleles. Exons and loxP sequences are indicated by boxes and arrowheads, respectively. Locations of primer pairs used for amplifying specific regions containing a loxP sequence are also indicated. (B) Genomic Southern blot analysis from EcoRI digested DNA isolated from several ES clones, showing the homologous recombination of the Alk13loxP vector into the Alk1 locus. (C) Representative PCR genotyping results from intercrosses of Alk1+/3loxP (top) and Alk1+/2loxP (bottom). The arrowheads indicate the PCR amplicon containing the loxP sequence; Alk13loxP/3loxP (lanes 2 and 5); Alk1+/3loxP (lanes 1, 4); Alk1+/+ (lane 3); Alk1+/2loxP (lanes 6 and 8); and Alk12loxP/2loxP (lanes 7, 9, and 10). (D) PCR detection of the Alk11loxP allele from genomic DNA isolated from multiple organs/tissues of E16.5 L1cre(+)Alk13loxP/3loxP (top), L1cre(+)Alk1+/3loxP (middle), and L1cre(−);Alk13loxP/3loxP (bottom) fetuses, demonstrating tissue-specific Cre activities. The arrowheads indicate the Alk11loxP-specific PCR amplicon.
Generation of Alk1-conditional alleles. (A) Schematic diagram of the Alk1 wild-type allele, Alk1-conditional targeting vector, and Alk13loxP, Alk12loxP, and Alk11loxP alleles. Exons and loxP sequences are indicated by boxes and arrowheads, respectively. Locations of primer pairs used for amplifying specific regions containing a loxP sequence are also indicated. (B) Genomic Southern blot analysis from EcoRI digested DNA isolated from several ES clones, showing the homologous recombination of the Alk13loxP vector into the Alk1 locus. (C) Representative PCR genotyping results from intercrosses of Alk1+/3loxP (top) and Alk1+/2loxP (bottom). The arrowheads indicate the PCR amplicon containing the loxP sequence; Alk13loxP/3loxP (lanes 2 and 5); Alk1+/3loxP (lanes 1, 4); Alk1+/+ (lane 3); Alk1+/2loxP (lanes 6 and 8); and Alk12loxP/2loxP (lanes 7, 9, and 10). (D) PCR detection of the Alk11loxP allele from genomic DNA isolated from multiple organs/tissues of E16.5 L1cre(+)Alk13loxP/3loxP (top), L1cre(+)Alk1+/3loxP (middle), and L1cre(−);Alk13loxP/3loxP (bottom) fetuses, demonstrating tissue-specific Cre activities. The arrowheads indicate the Alk11loxP-specific PCR amplicon.
Because TGF-β/BMP signaling molecules are involved in AV cushion development, deletion of Alk5 or Tgfbr2 in endocardial cells may result in cardiac defects, making vascular phenotype analysis complicated. Likewise, deletion of Alk1 in all endothelial cells would be expected to result in early embryonic lethality, similar to Alk1−/− embryos. To bypass effects on cardiac development and global vascular development, we chose to use the L1cre line as the common Cre line for investigating the role of ALK1, ALK5, and TGFBR2 in vascular ECs.
Generation of Alk1-conditional allele
To investigate the role of ALK1 signaling in the later stages of vascular development, we generated a conditional Alk1 knockout allele in which 3 loxP sequences were inserted into intronic regions of the Alk1 gene. The 5′ most loxP sequence and the neomycin cassette flanked by loxP sequences were inserted into the third and the sixth introns, respectively; thus, exons 4-6 could be deleted by Cre–mediated recombinations (Figure 2A). Because exon 5 encodes the transmembrane (TM) domain, the deletion of exons 4-6 would produce a null allele. The heterozygous Alk1+/3loxP mice (“3loxP” refers to 3 loxP sites in the Alk1 locus) were produced from chimeric mice containing the correctly targeted ES cells (Figure 2B). By the weaning age, the Alk1+/3loxP interbreeding produced Alk13loxP/3loxP, Alk1+/3loxP, and Alk1+/+ mice in a Mendelian ratio (Figure 2C). Alk1lacZ/3loxP mice from the intercrossing of Alk1+/3loxP and Alk1+/lacZ mice5 also appeared to be viable, indicating normal ALK1 production from the Alk13loxP allele. The Alk1+/3loxP mice were further intercrossed with Tg(EIIa-cre) males31 to segregate partially deleted floxed alleles in the germ cells, and we successfully isolated Alk1+/1loxP and Alk1+/2loxP mice. As expected, Alk12loxP/2loxP mice are viable (Figure 2C), whereas Alk11loxP/1loxP mice die at E10.5. The E9.5 Alk11loxP/1loxP embryos were morphologically indistinguishable from other Alk1-null embryos previously characterized (Figure S2).5,14 These results demonstrate that Alk13loxP and Alk12loxP are conditional alleles, whereas Alk11loxP is a null allele.
Endothelial-specific Alk1 deletion resulted in severe vascular malformations
To investigate the role of ALK1 in ECs, we intercrossed L1cre;Alk1+/3loxP males with Alk13loxP/3loxP or Alk1+/3loxP females. The Cre activity from L1cre mice was monitored by scrutinizing the Alk11loxP allele from genomic DNA isolated from various tissues and organs of E16.5 L1cre(+);Alk13loxP/3loxP, L1cre(+);Alk1+/3loxP, and L1cre(−);Alk13loxP/3loxP mice by PCR. Consistent with the X-gal staining pattern in L1cre(+);R26R, the Alk11loxP-specific amplification was detected strongly in lung, intestines, yolk sac, and placenta; very weakly in brain, kidney, and heart; but not in liver, limbs, and skin (Figure 2D). This indicates that Cre was expressed in specific vascular beds as we anticipated.
From their progeny, no viable L1cre(+);Alk13loxP/3loxP mice were recovered at the newborn stage. Timed mating studies revealed that L1cre(+);Alk13loxP/3loxP fetuses died by E18.5 (Table 1). All L1cre(+);Alk13loxP/3loxP fetuses at E15.5-E17.5 were readily distinguishable from their littermate controls, ie, L1cre(−);Alk13loxP/3loxP and L1cre(+);Alk1+/3loxP, by the bulged blood vessels in the yolk sac (Figure 4A,B). In control fetuses, the umbilical vessels were typically larger than vitelline vessels, and veins were larger than arteries (Figure 4C). In L1cre(+);Alk13loxP/3loxP fetuses, however, the vitelline vessels appeared to be larger than umbilical vessels (Figure 4D). Transverse sections of these extraembryonic vessels revealed by the markedly increased lumen diameter and decreased wall thickness that the vitelline artery of the mutant fetuses lost its arterial character and resembled a vitelline vein of control fetuses (Figure 4E,F). This result indicates that endothelial ALK1 plays an important role for the morphogenesis of arterial vessels.
L1cre (+); Alk13loxp/3loxP fetuses resulted in embryonic lethality at late gestational stages
| Cross* . | Stage . | Progeny, total no. . | Mutants [L1cre(+);Alk13loxP/3loxP], no. . | Embryos exhibiting phenotype* among the mutants, no. . | Embryos found dead among the mutants, no. . |
|---|---|---|---|---|---|
| L1cre(+); Alk1+/3loxP | E14.5 | 13 | 4 | 4† | 0 |
| × Alk13loxP/3loxP or | E15.5 | 24 | 10 | 10 | 0 |
| Alk1+/3loxP | E16.5 | 91 | 17 | 14 | 3 |
| E17.5 | 78 | 9 | 3 | 6 | |
| E18.5 | 22 | 8 | 0 | 8 |
| Cross* . | Stage . | Progeny, total no. . | Mutants [L1cre(+);Alk13loxP/3loxP], no. . | Embryos exhibiting phenotype* among the mutants, no. . | Embryos found dead among the mutants, no. . |
|---|---|---|---|---|---|
| L1cre(+); Alk1+/3loxP | E14.5 | 13 | 4 | 4† | 0 |
| × Alk13loxP/3loxP or | E15.5 | 24 | 10 | 10 | 0 |
| Alk1+/3loxP | E16.5 | 91 | 17 | 14 | 3 |
| E17.5 | 78 | 9 | 3 | 6 | |
| E18.5 | 22 | 8 | 0 | 8 |
Phenotype in L1cre(+);Alk13loxP/3loxP refers to the bulged yolk sac blood vessels similar to those in Figure 4B.
The mutant yolk sac vessels looked enlarged, but not as distinctive as the ones in E15.5 and later stages.
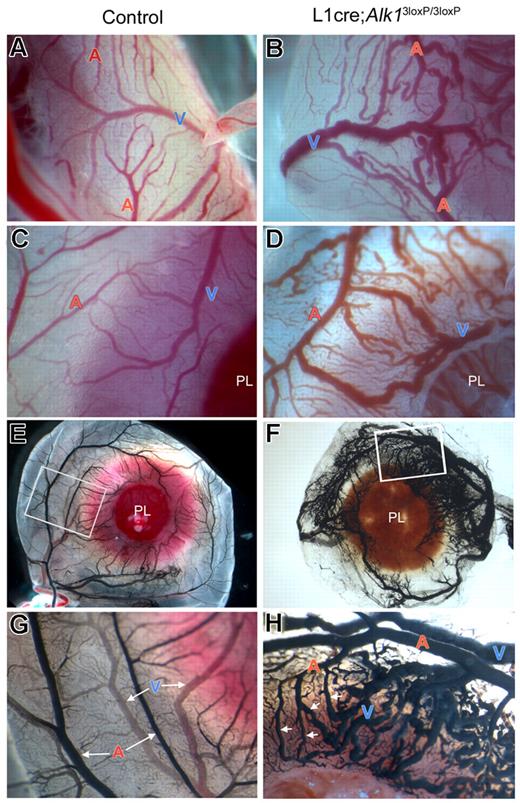
Alk1 deletion resulted in abnormal extraembryonic vasculature in E16.5 L1cre(+);Alk13loxP/3loxP fetuses. (A,B) Gross morphology of control and L1cre(+);Alk13loxP/3loxP mutant fetuses enclosed in the yolk sac attached to the placenta (PL). Note bulged arteries (A) and veins (V) in the mutant yolk sac. The inset in panel B shows magnified view of typical dilated, tortuous vitelline vessels in the mutants. (C,D) Umbilical arteries (UA) and veins (UV) are connected to the placenta, whereas vitelline arteries (VA) and veins (VV) are connected to the yolk sac. Note markedly enlarged VA and AVMs (circled; see enlarged view in Figure 5D) in the mutants. (E,F) Cross-sectional view of the extraembryonic vessels indicated by the scissors symbols in panels C,D demonstrates marked dilation and thinning of mutant VA (F), which has a similar morphology as control VV (E).
Alk1 deletion resulted in abnormal extraembryonic vasculature in E16.5 L1cre(+);Alk13loxP/3loxP fetuses. (A,B) Gross morphology of control and L1cre(+);Alk13loxP/3loxP mutant fetuses enclosed in the yolk sac attached to the placenta (PL). Note bulged arteries (A) and veins (V) in the mutant yolk sac. The inset in panel B shows magnified view of typical dilated, tortuous vitelline vessels in the mutants. (C,D) Umbilical arteries (UA) and veins (UV) are connected to the placenta, whereas vitelline arteries (VA) and veins (VV) are connected to the yolk sac. Note markedly enlarged VA and AVMs (circled; see enlarged view in Figure 5D) in the mutants. (E,F) Cross-sectional view of the extraembryonic vessels indicated by the scissors symbols in panels C,D demonstrates marked dilation and thinning of mutant VA (F), which has a similar morphology as control VV (E).
L1cre(+);Alk13loxP/3loxP mice exhibit arteriovenous malformations
The vitelline vessels in the mutant yolk sac exhibited not only dilated, convoluted and tortuous morphologies, but also abnormal direct connections between arteries and veins without connecting capillary beds. As shown in Figure 5 as representative examples, numerous AVMs appeared in the yolk sac of mutants, typically in the area where the yolk sac attaches to the placental plate (Figures 4D and 5B,D). To better assess the AVMs, we injected India ink into the vitelline artery. In control fetuses, the injected dye stayed mostly in the arterial vessels and displayed artery-vein connections through capillaries (Figure 5E,G). In the mutants, however, the dye crossed readily to venous vessels probably via multiple AVMs and stained the whole yolk sac vascular bed evenly (Figure 5F,H).
Alk1 deletion results in multiple AVM formations. Dissection microscopic views of representative arteries (A) and veins (V) in control (A,C,E,G) and L1cre(+);Alk13loxP/3loxP (B,D,F,H) E16.5 embryonic yolk sacs. In the control yolk sac (A,C), arteries and veins are intercalating and not connected directly to one another. In the mutants (B,D), however, there are numerous regions where dilated and tortuous arteries and veins are directly connected without the connecting capillaries. (E-H) India ink was injected into the vitelline artery to visualize the yolk sac vessels. In control (E,G), arteries and veins were easily distinguishable and were connected by capillaries. In mutants (F,H), numerous AVMs (indicated by arrows) were formed between arteries and veins.
Alk1 deletion results in multiple AVM formations. Dissection microscopic views of representative arteries (A) and veins (V) in control (A,C,E,G) and L1cre(+);Alk13loxP/3loxP (B,D,F,H) E16.5 embryonic yolk sacs. In the control yolk sac (A,C), arteries and veins are intercalating and not connected directly to one another. In the mutants (B,D), however, there are numerous regions where dilated and tortuous arteries and veins are directly connected without the connecting capillaries. (E-H) India ink was injected into the vitelline artery to visualize the yolk sac vessels. In control (E,G), arteries and veins were easily distinguishable and were connected by capillaries. In mutants (F,H), numerous AVMs (indicated by arrows) were formed between arteries and veins.
Because Cre recombinase is extensively expressed in pulmonary ECs of L1cre mice, we examined the lungs of the viable E16.5-E17.5 controls and L1cre(+);Alk13loxP/3loxP fetuses. On visual inspection of the pulmonary blood vessels, an organized pattern of vascular trees was observed in control lungs (Figure 6A inset), whereas dilated, tortuous, and irregular blood vessels were often recognized in the mutant lungs (Figure 6B inset, arrows). Histologic sections revealed that the mutant pulmonary vessels were markedly dilated and irregular in size and shape (Figure 6B,E). Anti–α-SMA antibody staining demonstrated thinning and irregularity of vascular smooth muscle cell (VSMC) layers in the mutant pulmonary vessels (Figure 6F). Elastin staining showed more compact VSMC layers in the dorsal aorta of mutants in comparison with that of controls, yet the difference was not as apparent as in pulmonary or vitelline vessels (data not shown).
Alk1 deletion resulted in abnormal pulmonary vasculature in E17.5 L1cre(+);Alk13loxP/3loxP fetuses. Transverse sections of the left lobe of the control (A,C,D) and mutant (B,E,F) lungs. Blood vessels are readily identifiable by the red blood cells in them. Dissection microscopic views of the left lung are shown as insets. The control lung displayed organized vascular trees (A, inset), whereas the mutant lung exhibited dilated, tortuous, and irregular blood vessels (B, inset, arrows). In the control lungs (A,C), bronchial trees and blood vessels are coordinated, and blood vessels are well defined as a circular shape. Br indicates bronchus; PA, pulmonary artery; and H&E, hematoxylin and eosin. In the mutant lungs (B,E), the bronchial lumens are not expanded as much as control lungs, and blood vessels are noticeably enlarged and irregular, presumably resulting from fusions between neighboring vessels (arrowheads in E). (D,F) Immunostaining with anti-αSMA antibodies revealed thinning of blood vessel walls with irregular thickness of smooth muscle layers (arrowheads in F).
Alk1 deletion resulted in abnormal pulmonary vasculature in E17.5 L1cre(+);Alk13loxP/3loxP fetuses. Transverse sections of the left lobe of the control (A,C,D) and mutant (B,E,F) lungs. Blood vessels are readily identifiable by the red blood cells in them. Dissection microscopic views of the left lung are shown as insets. The control lung displayed organized vascular trees (A, inset), whereas the mutant lung exhibited dilated, tortuous, and irregular blood vessels (B, inset, arrows). In the control lungs (A,C), bronchial trees and blood vessels are coordinated, and blood vessels are well defined as a circular shape. Br indicates bronchus; PA, pulmonary artery; and H&E, hematoxylin and eosin. In the mutant lungs (B,E), the bronchial lumens are not expanded as much as control lungs, and blood vessels are noticeably enlarged and irregular, presumably resulting from fusions between neighboring vessels (arrowheads in E). (D,F) Immunostaining with anti-αSMA antibodies revealed thinning of blood vessel walls with irregular thickness of smooth muscle layers (arrowheads in F).
L1cre(+);Alk5loxP/loxP and L1cre(+);Tgfbr2loxP/loxP mice were viable and did not exhibit vascular malformations
To determine whether ALK5 is necessary for ALK1 signaling in vascular endothelium, we generated L1cre(+); Alk5loxP/loxP mice to determine whether these Alk5 mutants would develop vascular phenotypes similar to those in L1cre(+); Alk13loxP/3loxP. The L1cre(+);Alk5loxP/loxP mice appeared to be viable over 3 months (n = 25). As observed in the L1cre(+);Alk13loxP mice, the null allele-specific PCR amplicon in the L1cre(+);Alk5loxP/loxP mice was detected in the lung but not in the liver or tail DNA samples (Figure 7B). We also detected a homogeneous X-gal staining pattern in pulmonary ECs of L1cre(+);Alk5loxP/loxP;R26R mice (Figure 7F). These results indicate that the lack of vascular phenotype in these Alk5 mutants is not the result of lack of Cre activities and that ALK5 is not required for ALK1 signaling. To our surprise, L1cre(+);Tgfbr2loxP/loxP mice also appeared to be viable over 3 months (n = 24). Histologic sections of 2-month-old lungs of these Alk5 and Tgfbr2 mutants exhibited no apparent pathologic signs (Figure 7H-J). These results suggest that TGFBR2 is also not required for ALK1 signaling.
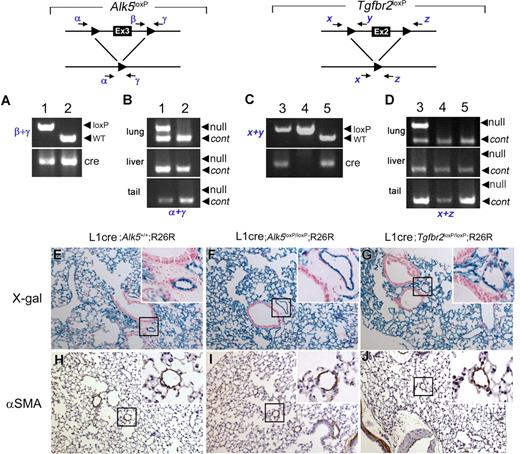
The Alk5 and Tgfbr2 are deleted in the lungs of L1cre(+);Alk5loxP/loxP and L1cre(+);Tgfbr2loxP/loxP mice, respectively, yet no noticeable pathologic signs were observed in the lungs of 2-month-old mutants. (A) PCR genotyping with primers β and γ (top) and Cre (bottom), showing L1cre(+);Alk5loxP/loxP (lane 1) and L1cre(+);Alk5+/+ (lane 2) mice. WT, wild-type allele (B) PCR amplification of the Alk5 null allele is specific for the lung. Genomic DNA isolated from the lung (top), liver (middle), and tail (bottom) were used as template to amplify the null allele by primers α and γ. Another primer set detecting a diploid genome (ie, Alk1 = control) was also included in the PCR reaction to demonstrate equal loading of the template. (C) PCR genotyping with primers x and y (top) and cre (bottom), showing L1cre(+);Tgfbr2loxP/loxP (lane 3), L1cre(−);Tgfbr2loxP/loxP (land 4), and L1cre(+);Tgfbr2+/+ (lane 5) mice. (D) PCR amplification of the Tgfbr2 null allele is specific for the lung. Genomic DNA isolated from the lung (top), liver (middle), and tail (bottom) was used as template to amplify the null allele by primers x and z. Another primer set detecting a diploid genome (ie, Alk1) was also included in the PCR reaction to demonstrate equal loading of the template. (E-J) Histologic sections of the lungs of 2-month-old control (ie, L1cre(+);Alk5+/+;R26R; E,H), L1cre(+);Alk5loxP/loxP;R26R (F,I), and L1cre(+);Tgfbr2loxP/loxP;R26R (G,J) mice. (E-G) Histologic sections of the X-gal stained lungs were counterstained with NFR, demonstrating that the Cre–mediated recombination has occurred in these lungs as expected. Insets are high magnification views showing that lacZ expression is restricted to pulmonary ECs, not in bronchial epithelial or smooth muscle cells. (H-J) Anti-αSMA antibody staining of the control and mutant lungs showing no specific pathologic signs. Insets show that similar thickness of the VSMC layers of distal arteries.
The Alk5 and Tgfbr2 are deleted in the lungs of L1cre(+);Alk5loxP/loxP and L1cre(+);Tgfbr2loxP/loxP mice, respectively, yet no noticeable pathologic signs were observed in the lungs of 2-month-old mutants. (A) PCR genotyping with primers β and γ (top) and Cre (bottom), showing L1cre(+);Alk5loxP/loxP (lane 1) and L1cre(+);Alk5+/+ (lane 2) mice. WT, wild-type allele (B) PCR amplification of the Alk5 null allele is specific for the lung. Genomic DNA isolated from the lung (top), liver (middle), and tail (bottom) were used as template to amplify the null allele by primers α and γ. Another primer set detecting a diploid genome (ie, Alk1 = control) was also included in the PCR reaction to demonstrate equal loading of the template. (C) PCR genotyping with primers x and y (top) and cre (bottom), showing L1cre(+);Tgfbr2loxP/loxP (lane 3), L1cre(−);Tgfbr2loxP/loxP (land 4), and L1cre(+);Tgfbr2+/+ (lane 5) mice. (D) PCR amplification of the Tgfbr2 null allele is specific for the lung. Genomic DNA isolated from the lung (top), liver (middle), and tail (bottom) was used as template to amplify the null allele by primers x and z. Another primer set detecting a diploid genome (ie, Alk1) was also included in the PCR reaction to demonstrate equal loading of the template. (E-J) Histologic sections of the lungs of 2-month-old control (ie, L1cre(+);Alk5+/+;R26R; E,H), L1cre(+);Alk5loxP/loxP;R26R (F,I), and L1cre(+);Tgfbr2loxP/loxP;R26R (G,J) mice. (E-G) Histologic sections of the X-gal stained lungs were counterstained with NFR, demonstrating that the Cre–mediated recombination has occurred in these lungs as expected. Insets are high magnification views showing that lacZ expression is restricted to pulmonary ECs, not in bronchial epithelial or smooth muscle cells. (H-J) Anti-αSMA antibody staining of the control and mutant lungs showing no specific pathologic signs. Insets show that similar thickness of the VSMC layers of distal arteries.
Inhibition of Alk5 activity does not affect zebrafish vascular development
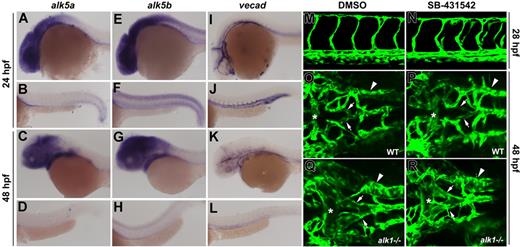
To further investigate the role of Alk5 in Alk1 signaling in the endothelium, we cloned 2 zebrafish alk5 genes, which probably arose as a consequence of a genome duplication that occurred after teleost fish diverged from tetrapods.32 At 24 and 48 hpf, both alk5a (chromosome 2) and alk5b (chromosome 24) are expressed predominantly in neural tissue, with alk5a in the eye and brain, and alk5b in the eye, brain, and entire spinal cord (Figure 8A-H). Both genes are also expressed in pharyngeal arches and weakly in endoderm, whereas only alk5b is expressed in ventral somites at 24 hpf. Neither pattern is suggestive of endothelial expression, in whole mount (compare with vascular endothelial cadherin expression, 8I-L) or in histologic sections at these or earlier stages (data not shown). Given the limitations in sensitivity and resolution of whole-mount in situ hybridization, we further investigated the role of Alk5 in Alk1 signaling in the endothelium by inhibiting Alk5 activity in the developing embryo and assaying vascular anatomy. Zebrafish alk1 mutants (violet beauregarde, or vbg) exhibit enlarged cranial arteries that abnormally connect directly to veins,33 a similar phenotype to that seen in Alk1 null mice14,34 and human HHT2 patients.35 Thus, we reasoned that, if Alk5 were necessary for Alk1 signaling, a similar vessel phenotype would be observed in the absence of Alk5 activity.
Zebrafish alk5a and alk5b are not expressed in the endothelium and activities are not necessary for vessel development.alk5a is expressed in the eye, brain, pharyngeal arches, and endoderm at 24 and 48 hpf (A-D). alk5b is also expressed in these tissues, as well as in the spinal cord and ventral somites (E-H). Neither seems to be expressed in blood vessels (compare A-H with I-L, vecad expression). Exposure of phenotypically wild-type zebrafish embryos to 100 μM SB-431542 beginning at the 8- to 10-somite stage had no effect on trunk (M,N) or cranial (O,P) vascular anatomy at 24 or 48 hpf, respectively. This same exposure regimen did not exacerbate the cranial vascular phenotype in alk1−/− embryos (Q,R). Comparing panels Q and R with O and P, note enlargement of basal communicating artery (asterisk), posterior connecting segments (arrows), and primordial hindbrain channel (arrowhead). (A-L) In situ hybridization, lateral views, anterior to the left. First and third rows, head; second and fourth rows, trunk and tail. M-R, 2-dimensional reconstructions of laser scanning confocal Z-series of TG(flk1:GFP)la116 embryos. (M,N) Lateral views of the trunk, anterior to the left. (O-R) Dorsal views of the head, anterior to the left.
Zebrafish alk5a and alk5b are not expressed in the endothelium and activities are not necessary for vessel development.alk5a is expressed in the eye, brain, pharyngeal arches, and endoderm at 24 and 48 hpf (A-D). alk5b is also expressed in these tissues, as well as in the spinal cord and ventral somites (E-H). Neither seems to be expressed in blood vessels (compare A-H with I-L, vecad expression). Exposure of phenotypically wild-type zebrafish embryos to 100 μM SB-431542 beginning at the 8- to 10-somite stage had no effect on trunk (M,N) or cranial (O,P) vascular anatomy at 24 or 48 hpf, respectively. This same exposure regimen did not exacerbate the cranial vascular phenotype in alk1−/− embryos (Q,R). Comparing panels Q and R with O and P, note enlargement of basal communicating artery (asterisk), posterior connecting segments (arrows), and primordial hindbrain channel (arrowhead). (A-L) In situ hybridization, lateral views, anterior to the left. First and third rows, head; second and fourth rows, trunk and tail. M-R, 2-dimensional reconstructions of laser scanning confocal Z-series of TG(flk1:GFP)la116 embryos. (M,N) Lateral views of the trunk, anterior to the left. (O-R) Dorsal views of the head, anterior to the left.
To test this hypothesis, we exposed zebrafish embryos to the ALK4/5/7 inhibitor, SB-431542,36 at the 8- to 10-somite stage (onset of angioblast differentiation), and assayed vessel architecture at 24 and 48 hpf. This exposure regimen avoided drug–induced inhibition of early Nodal/Alk4 signaling, which leads to failed mesendoderm formation37 ; 100 μM SB-431542 had no effect on the cranial or trunk vasculature of wild-type, alk1+/−, or alk1−/− embryos compared with DMSO-treated controls (Figure 8M-R), suggesting that Alk5 is dispensable for zebrafish vascular development and that alk1 and alk5 do not genetically interact. This lack of effect could not be attributed to inability of the drug to inhibit zebrafish Alk5 activity in vivo, as 100 μM SB-431542 (8-cell stage exposure) completely abrogated the ability of constitutively active alk5a and alk5b to induce Smad2/3–mediated goosecoid expression at shield stage (6 hpf; Figure S3A-F). This lack of effect also could not be attributed to ineffective drug penetration at later developmental stages, because 8- to10-somite stage exposure abrogated endogenous left-sided Nodal/Alk4–mediated pitx2c expression38 in the dorsal diencephalon and gut at 24 hpf and also significantly decreased activity of a Smad2/3- (but not Smad1/5-) responsive transgene (Figure S3G-J). Taken together, these results support our findings in Alk5-deficient mouse endothelium and uphold our conclusion that ALK5 activity is not necessary for ALK1 signaling in the endothelium.
Discussion
In this article, we report a novel strain of transgenic mice expressing Cre recombinase in restricted vascular endothelia and a novel conditional Alk1 knockout line. Using the transgenic mouse line as a common Cre expressor, we deleted Alk1, Alk5, or Tgfbr2 gene in restricted vascular endothelium. We show that affected Alk1-deficient blood vessels exhibit vascular malformations mimicking all pathologic features of HHT vascular lesions. In contrast, Alk5- or Tgfbr2-deficient blood vessels did not show any defects. The in vivo data presented here indicate that neither ALK5 nor TGFBR2 is required for ALK1 signaling relevant for pathogenesis of HHT.
The Cre/LoxP system has become an extremely valuable approach for studying functions of particular genes in specific tissues. For this system, characterization of tissue-specific promoters and generation of tissue-specific Cre expressors are imperative tasks. For vascular biology, multiple transgenic or knockin lines showing EC-specific Cre expression have been developed, including Flk1Cre,39 Tg(Tie1-cre),40 Tg(Tie2-cre),41 and Tg(Vecad-cre)42 lines. One limitation of each of these lines is that they drive Cre expression in endocardium at the heart tube stage (ie, E8-E9). If a gene of interest has a dual function in vascular ECs and endocardial progenies, deletion of the gene in endocardial cells can potentially impair heart development. Thus, it would be difficult to determine whether the resultant phenotype is due solely to functional loss of the gene in vascular endothelia or contributed to by cardiac dysfunction. For instance, Flk1Cre– or Tg(Tie1-cre)–mediated Alk3 (Bmpr1a) deletion resulted in embryonic lethality with AV cushion defect.43,44 The L1cre line is therefore a valuable and unique resource with which function of particular genes in endothelia can be assessed without affecting cardiogenesis. Our data suggest that this transgenic Cre expressor would be particularly useful for studying function of a gene in pulmonary vascular endothelia.
HHT is a dominantly inherited genetic disorder, and haploinsufficiency (reduced amount of functional protein) is the probable cause of associated vessel malformations.45,46 An intriguing feature of HHT is variability of disease manifestations. Even among kin, the severity, age of onset, and locations of the vascular lesions are extremely variable from one patient to another.47 In addition, only selective vascular beds develop telangiectasis or AVM lesions, whereas other such areas remain normal in an HHT patient. Therefore, it is clear that there are other factors (or “second hits”), in addition to the haploinsufficiency of ALK1 or ENG, involved in the development of vascular lesions in HHT patients. This variable phenotype expression is a major impediment to studying pathogenesis of HHT. The Alk1+/− and Eng+/− mice develop HHT-like vascular lesions with unpredictable age of onset, severity, and location, similar to human cases of HHT.48,–50 These heterozygous knockout mice are excellent resources for investigating genetic or environmental factors influencing the disease manifestations but are difficult to use for studying molecular pathogenetic mechanisms that underlie the vascular malformations.
In contrast, the L1cre(+);Alk13loxP/3loxP mice displayed the hallmarks of HHT vascular phenotypes (dilation of lumen, thinning of vascular walls, loss of capillaries, development of excessive tortuous vessels, and AVMs) in a consistent and predictable manner. These mice would provide a valuable resource for identification of key molecular pathways involved in the progression of vascular malformations. Based on localization of ALK1 and ENG within endothelial cells, it has been inferred that the primary cellular defect leading to HHT vascular malformations lay within endothelial cells. Data presented here are the first direct in vivo evidence demonstrating that endothelial-specific Alk1 deletion is sufficient to cause HHT-like vascular lesions. Based on the fact that the HHT-like vascular phenotype occurs consistently in L1cre(+);Alk13loxP/3loxP mice, but not in L1cre(+);Alk1+/3loxP or Alk1+/− mice,50 it is tempting to speculate that a loss of heterozygosity (LOH) of ALK1 during development might be a cause of some AVM vascular lesions in HHT2 patients. It was reported that the normal copy of ENG was present and a reduced level of ENG was detected in AVM lesions of HHT1 patients, suggesting that a LOH of ENG is not required for AVM formation in HHT1.51 It has yet to be investigated whether a LOH of ALK1 is involved in AVM lesions of HHT2 patients.
The vitelline artery of the Alk1 mutants exhibited a vein-like morphology, and pulmonary arteries were also dilated and had thin and irregular vessel walls. This result suggests that ALK1 may play a pivotal role in arteriogenesis. It is noteworthy that Cre activity was detected in almost all vascular beds in the yolk sac, yet not every yolk sac blood vessel in L1cre(+);Alk13loxP/3loxP mice develops the characteristic morphologic changes. Whether hemodynamic factors, such as shear or pressure, are essential for the vascular malformations of Alk1-deficient vessels remains to be investigated.
If ALK5 is required for TGF-β/ALK1 signaling as previously reported,20 an overlapping vascular phenotype between L1cre(+);Alk13loxP/3loxP and L1cre(+);Alk5loxP/loxP mice was predicted. However, L1cre(+);Alk5loxP/loxP mice appeared to be viable and did not exhibit any vascular malformations observed in L1cre(+);Alk13loxP/3loxP mice. These results suggest that ALK5 is not required for ALK1 signaling and that the proposed balance of ALK1 and ALK5 in ECs has a limited role in vascular development. Our findings are consistent with our prediction based on the undetectable level of Alk5 expression in ECs of mice.21 However, our conclusions contradict the report that EC-specific Tg(Tie1-cre)–mediated Alk5 deletion resulted in embryonic lethality similar to Alk5-null embryos, suggesting that endothelial ALK5 has an essential function in vascular development.52 It is possible that endothelial ALK5 indeed plays an essential role in early stages of mouse vascular development not uncovered in our L1cre(+);Alk5loxP/loxP model, although the lack of Alk5 expression in ECs21 argues against this possibility. An alternative explanation for this discrepancy is that ALK5 has an important function in the endocardial cell lineage for the development of the heart and that Tg(Tie1-cre);Alk5loxP/loxP embryos might die as a result of impaired cardiogenesis.44
To address the role of ALK5 in vessel development in an alternative vertebrate model system, we inhibited Alk5 activity using the ALK4/5/7 inhibitor, SB-431542. We chose this chemical genetic approach over classical forward and reverse genetic approaches for 2 reasons. First, we identified 2 alk5 genes in zebrafish, alk5a and alk5b, and to our knowledge, neither of these genes is represented by an existing mutant line. And second, morpholino–mediated knockdown of alk5a and alk5b beginning at the 1-cell stage produced global effects on head structure and body axis, at least some of which seemed to be nonspecific,53 which precluded accurate analysis of vessel development (data not shown). With our chemical genetic approach, we were able to inhibit both Alk5 orthologs and bypass effects of early loss of Alk5a and Alk5b. This treatment produced no effects on vessel patterning or architecture at either 24 or 48 hpf, consistent with findings in Alk5lacZ/lacZ mice21 and L1cre(+);Alk5loxP/loxP mice. It should be noted that, whereas defects in vascular smooth muscle investment were apparent in Alk5lacZ/lacZ mice, zebrafish vessels do not have a smooth muscle sheath at this time (Ramiah Subramanian, St George's, University of London, oral communication, July 14, 2002). In light of the controversy surrounding the interplay between ALK1 and ALK5 in vascular endothelium, we also inhibited Alk5 activity in alk1+/−and alk1−/− embryos. Alk5 inhibition had no effect on the vasculature of these embryos, suggesting that there is no genetic interaction between alk1 and alk5 in zebrafish endothelial cells. Taken together with results from studies in L1cre(+);Alk5loxP/loxP mice, our data strongly suggest that ALK5 is not required within vertebrate endothelial cells for proper vessel formation.
Given that TGF-β1 has been shown to bind to and activate ALK1 in concert with TGFBR2,11,14,16,20 we were surprised to find that L1cre(+);Tgfbr2loxP/loxP mice were viable and did not exhibit any apparent vascular abnormality. Since the emergence of modern vascular biology, innumerable papers have described the diverse cellular functions of TGF-β subfamily proteins in endothelial cells. However, the precise in vivo function of TGF-β in endothelia for vascular development remains undefined. Although our conditional knockout approach has limitations in addressing the role of endothelial TGF-β signaling in overall vascular development and maintenance, our results indicate that endothelial TGFBR2 is dispensable for yolk sac and pulmonary vascular development. Our data also demonstrate that TGFBR2 is not required for ALK1 signaling, suggesting that TGF-β subfamily proteins may not be the ALK1 ligands pertinent to HHT and that HHT may not be a TGF-β subfamily disease. Both ALK1 and ENG can interact with other TGF-β family ligands in the presence of their corresponding ligand-binding receptors.10,11 Presence of an ALK1 ligand distinct from TGF-β1 and TGF-β3 was observed in serum,11 and recently it has been shown that BMP9 or BMP10 can specifically bind to and signal through ALK1 and BMPR2.12,13 Although whether BMP9 and/or BMP10 is an in vivo ligand for ALK1 pertinent to HHT is an open question, a recent report substantiated this possibility by showing that Bmpr2-specific short hairpin RNA (shRNA) transgenic mice exhibited gastrointestinal bleeding and HHT-like vascular lesions.54
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Rebecca Howard for editorial assistance; Mike Rule in University of Florida Shands Cancer Center Transgenic Core for the generation of Tg(Alk1-cre) lines; Harold L. Moses (Vanderbilt University) and Stefan Karlsson (Lund University) for Tgfbr2- and Alk5-conditional knockout mouse lines, respectively; and Jau-Nian Chen (University of California, Los Angeles) for TG(flk1:GFP)la116 transgenic zebrafish.
This work was supported by National Institutes of Health (NIH) grant HL64024, American Heart Association (AHA) grant 0455336B, and a research grant from Novartis Institutes for BioMedical Research (S.P.O); NIH grant HL079108 (B.L.R.); an AHA postdoctoral fellowship (S.O.P); AHA Scientist Development Grant (S.D.G., Y.J.L., and T.S.); and an AHA predoctoral fellowship (K.-H.H.).
National Institutes of Health
Authorship
Contribution: S.O.P. performed experiments, analyzed data, and wrote the paper; Y.J.L., T.S., K.-H.H., N.F., Z.J., A.P., and X.W. performed experiments; V.K. contributed a vital reagent; and B.L.R. and S.P.O. designed research, performed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: S. Paul Oh, Department of Physiology and Functional Genomics, University of Florida, 1376 Mowry Rd, Rm 456, Gainesville, FL 32610; e-mail: ohp@phys.med.ufl.edu.