Abstract
EPO functions primarily as an erythroblast survival factor, and its antiapoptotic actions have been proposed to involve predominantly PI3-kinase and BCL-X pathways. Presently, the nature of EPO-regulated survival genes has been investigated through transcriptome analyses of highly responsive, primary bone marrow erythroblasts. Two proapoptotic factors, Bim and FoxO3a, were rapidly repressed not only via the wild-type EPOR, but also by PY-deficient knocked-in EPOR alleles. In parallel, Pim1 and Pim3 kinases and Irs2 were induced. For this survival gene set, induction failed via a PY-null EPOR-HM allele, but was restored upon reconstitution of a PY343 STAT5–binding site within a related EPOR-H allele. Notably, EPOR-HM supports erythropoiesis at steady state but not during anemia, while EPOR-H exhibits near wild-type EPOR activities. EPOR-H and the wild-type EPOR (but not EPOR-HM) also markedly stimulated the expression of Trb3 pseudokinase, and intracellular serpin, Serpina-3G. For SERPINA-3G and TRB3, ectopic expression in EPO-dependent progenitors furthermore significantly inhibited apoptosis due to cytokine withdrawal. BCL-XL and BCL2 also were studied, but in highly responsive KitposCD71highTer119neg erythroblasts, neither was EPO modulated. EPOR survival circuits therefore include the repression of Bim plus FoxO3a, and EPOR/PY343/STAT5-dependent stimulation of Pim1, Pim3, Irs2 plus Serpina-3G, and Trb3 as new antiapoptotic effectors.
Introduction
In response to anemia, erythropoietin (EPO) is expressed by interstitial kidney and fetal liver cells via hypoxia-inducible transcription factor pathways.1,2 As a secreted monomeric sialoglycoprotein, EPO then targets developing erythroblasts, and is essential for red cell formation during definitive bone marrow and fetal liver erythropoiesis.3-8 Prospective roles for EPO in promoting primitive red cell formation in yolk sac also have recently been described.9 Beyond this, recombinant EPO has been demonstrated in ischemia and other cell damage models to provide cytoprotective effects for injured renal, cardiac, retinal, and neuronal tissues.10,11 Taken together, these considerations have heightened interest in the specific nature of key EPO action mechanisms, especially those associated with progenitor cell survival
EPO's prime effects are mediated via interactions with its dimeric single-transmembrane receptor (EPOR).3-8,12 These interactions appear to evoke EPOR conformational events,13 which are relayed to an upstream Janus kinase, JAK214 (and JAK2 likewise may preassemble with EPOR dimers at a juxtamembrane box1 domain).15,16 JAK2, as activated via a Y1007 phosphorylation loop,17 next stimulates 2 separable signal transduction pathways. First, JAK2 interestingly can support steady-state erythropoiesis via EPOR-PY–independent routes that, in part, may involve MEK1,2 and ERK1,2 stimulation.18 Second, JAK2 also mediates the phosphorylation of 8 conserved EPOR cytoplasmic PY sites, which can then form a scaffold for the binding of up to 20 SH2- or PTB-domain encoding signal transduction factors and molecular adaptors.6-8,19-21
Among conserved EPOR cytoplasmic PY sites, 2 appear to act predominately to engage pathways to progenitor cell survival. First, EPOR-PY479 is a key binding site for p85-alpha and the corecruitment of PI3-kinase's p110 catalytic subunit.22 PI3-kinase's antiapoptotic actions also involve AKT activation, and AKT's subsequent effects on mTOR, GSK3, and FOXO transcription factors.23-26 In keeping with this notion, PI3-kinase inhibi-tors have been shown to attenuate the development of CD34pos progenitor-derived primary human erythroblasts ex vivo.27 Ectopic expression of a constitutively active AKT mutant in transduced murine Jak2−/− fetal erythroblasts also can at least partially compensate for JAK2 deficiency.26 Second, it has been argued that an EPOR-proximal PY343 site that selectively recruits STAT518,28 in cell line models mediates Bcl-x gene expression.29 BCL-XL, itself, is known to be expressed at high levels in late-stage erythroblasts, and conditional disruption of the Bcl-x gene leads to cell death at a late erythroblast stage.30,31 Proposed EPO-dependent routes to Bcl-x modulation, however, remain a matter of some controversy.32
To advance an understanding of key EPO-regulated survival pathways, we presently used global transcriptome analyses to identify EPO-modulated (anti)apoptotic factors in developmentally staged bone marrow–derived erythroblasts. To define EPOR subdomains that might regulate distinct subsets of survival genes, such investigations were further extended to primary erythroblasts expressing minimal PY-null EPOR-HM and PY343-containing EPOR-H alleles. Studies overall reveal that EPO modulates the expression of 8 (anti)apoptotic genes. Four factors previously associated with PI3K and/or AKT kinase pathways were EPO regulated: the signal transduction docking factor Irs2,33,34 the forkhead transcription factor FoxO3a,24,25 and 2 pseudokinases Trb3 and Trb2.35,36 By comparison, only one Bcl2 family gene was EPO modulated: Bim, a proapoptotic BH3-only factor.37,38 Two antiapoptotic S/T kinases also were upmodulated, Pim1 and Pim3,39 as was an atypical serpin protease inhibitor, Serpina3G.40,41 Notably, efficient EPO upmodulation of each survival-reinforcing factor proved to depend upon an intact EPOR-PY343 (and STAT5) signaling axis, an axis that can play key roles during anemia.42,43 In contrast, downmodulation of Bim, Fox03a, and Trb2 was mediated via an EPOR-PY–independent “JAK2-only” axis. Finally, both TRB3 and SERPINA-3G were discovered through functional analyses to significantly protect erythroid progenitor cells from apoptosis due to EPO withdrawal. The nature and possible action mechanism of these new EPO-regulated survival factors are discussed further in the context of a new mechanistic model for proerythroblast survival.
Methods
Mouse models and primary erythroblast preparations
Mice expressing knocked-in EPOR-H and EPOR-HM alleles (and congenic controls) were as described.18,42 All mice and all associated procedures were Institutional Animal Care and Use Committee (IACUC) approved. For mice at age 8 to 12 weeks, marrow cells were extruded into Iscove modified Dulbecco medium (IMDM; Invitrogen, Carlsbad, CA), 2% FBS. Cells then were passed through a 40-μm sieve, washed in IMDM, and resuspended in 1 mL phosphate-buffered saline (PBS; Invitrogen). Red cells then were lysed, and progenitors were collected through 50% FBS in PBS.42,44 For ex vivo expansions, cells were plated at 8 × 105 cells/mL (7 mL per 100-mm dish) in StemPro-34 (Invitrogen) supplemented with 2.5 U/mL EPO, 100 ng/mL mSCF, 1 μM dexamethasone, 1 μM beta-estradiol, 75 μg/mL h-transferrin, (Sigma-Aldrich, St Louis, MO), 0.5% BSA (StemCell Technologies, Vancouver, BC), 0.1 mM 2-mercaptoethanol, 100 U/mL penicillin G, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin B (1XPSF) (Invitrogen), and 1.5 mM l-glutamine (ie, “SP34-EX” medium).45 At 24 hours, 4 mL fresh medium was added. At 48 hours, cells were collected, resuspended in 1 mL conditioned medium, combined with 9 mL fresh medium, and cultured for an additional 24 hours.
Isolation of KitposCD71highTer119neg erythroblasts and analyses of EPO-modulated candidate survival genes
In the isolation of KitposCD71highTer119neg erythroblasts, Linpos cells first were depleted from ex vivo cultures using biotinylated antibodies to CD5 (Ly-1), CD45R/B220, CDllb (Mac1), Ter119, and Ly6G (Gr1; StemCell Technologies). KitposCD71highTer119neg cells then were retrieved at purities of more than 99.9% via CD117 magnetic activated cell separation (Miltenyi Biotech, Auburn, CA). For profiling, these (pro)erythroblasts were incubated for 6 hours in 50 μg/mL transferrin, 15 ng/mL insulin, 0.1 mM 2-mercaptoethanol, 0.5% BSA, in IMDM. Cells then were exposed to EPO (± 5 U/mL) for 90 minutes, and RNA was isolated.45 RNA (4 μg) was used for biotin-cRNA syntheses, and hybridizations were to Affymetrix 430-2.0 arrays (Santa Clara, CA). GeneChip 3000 scanning and GCOS software (Affymetrix) were used to process signals. Data analyses used SAM (significance analysis of microarrays), Array Assist software 5.0 (Stratagene, La Jolla, CA), and exploratory visual analysis (EVA).
Reverse transcription and quantitative PCR
Reverse-transcription reactions used TURBO DNase (Ambion, Austin, TX) and Superscript III (Invitrogen). For polymerase chain reaction (PCR), primer pairs (SuperArray Bioscience, Frederick, MD) were as follows: Irs-2, XM 357863; Trb3, NM 021562; Trb2, NM 144551; Foxo3A, NM 019740; Bim, NM 009754; Pim1, NM 008842; Pim3, NM 145478; and S3g, NM 009251. Quantitative PCR was performed using iQ SYBR Green and an i-Cycler (Bio-Rad, Hercules, CA). In time course experiments, purified KitposCD71high cells were incubated for 6 hours in transferrin (50 μg/mL), insulin (15 ng/mL), 0.5% BSA, and 0.1 mM 2-mercaptoethanol, in IMDM. Cells then were exposed to EPO (2.5 U/mL) for 0, 30, 60, 90, and 270 minutes. RNA was isolated, reverse transcribed, and used in quantitative PCR analyses.
Flow cytometry and apoptosis assays
In flow cytometry, 106 cells were washed, and resuspended in 0.2 mL PBS, 0.1% BSA, and 1 μg rat IgG. Following a 15-minute incubation at 4°C, cells were stained with allophycocyanin (APC)–CD117 (1 μg) plus fluorescein (FITC)–CD71 (1 μg) (BD Biosciences, San Jose, CA). Equivalent numbers of gated events were analyzed using a BD Biosciences FACScalibur flow cytometer. For cytospin preparations, 105 cells were used (10g,15 minutes; Hettich Universal 16A cytocentrifuge, Tuttlingen, Germany). Staining was with Dip-Stain reagent (Volu-sol, Salt Lake City, UT). In apoptosis assays, FITC–annexin-V– or APC–annexin-V–binding assays were performed in 140 mM NaCl, 2.5 mM CaCl2, 10 mM HEPES, pH 7.4, for 30 minutes at 25°C.
Western blotting
Purified KitposCD71highTer119neg erythroblasts (107) were isolated, washed, and incubated for 5.5 hours in 50 μg/mL transferrin, 15 ng/mL insulin, 0.1 mM 2-mercaptoethanol, 0.5% BSA in IMDM. Cells then were exposed to EPO (2.5 U/mL) for the time courses indicated. Cells then were washed in 2°C PBS and lysed in 1% Igepal, 150 mM NaCl, 50 mM NaF, 2 mM Na2EDTA, 0.1 mM NaVO3, 1 mM dithiothreitol, 10 mM sodium pyruvate, 25 mM beta-glycerol phosphate, 10% glycerol, 50 mM HEPES (pH 7.5) plus 0.25 mg/mL phenylmethylsulfonylfluoride, 1 × protease inhibitor and 1 × phosphatase inhibitor cocktails (Sigma-Aldrich). Then 1% Triton-X 100, 0.5% sodium deoxycholate, 0.1% SDS, 112.5 mM NaCl, 37.5 mM Tris-HCL (pH 7.4) was added (0.15 mL). Alternatively, cytoplasmic and nuclear extracts were prepared as described by Menon et al.18 Extracellular medium was concentrated (10-fold) using Centricon-10 cartridges (Amicon, Billerica, MA). Cleared extracts were assayed for protein content, denatured, electrophoresed, and blotted. Antibodies used were to phospho-GAB2 (no. 3881; Cell Signaling); GAB2 (no. 06-967; Upstate, Lake Placid, NY); phospho-AKT and AKT (nos. 9271 and 9272, respectively; Cell Signaling); anti-Flag M2 antibody (no. F1804; Sigma-Aldrich); and BCL2 and BCL-XL (nos. 2762 and 2876, respectively; Cell Signaling). In chemiluminescence, HRP-conjugated antibodies (Jackson Immunoresearch, West Grove, PA) and Super-Signal West-Dura reagent (Pierce Biotechnology, Rockford, IL) were used.
Cell lines and viral constructs
UT7epo cells45 were maintained in 10% FBS, 0.1 mM 2-mercaptoethanol plus EPO at 2 U/mL in IMDM. G1E/JC4 cells were maintained in 10% FBS, EPO (2 U/mL), SCF (100 ng/mL), 0.15 mM monothioglycerol in IMDM. A MIGR1 vector encoding a Flag epitope-tagged SERPINA-3G cDNA was prepared, and was used to produce amphotropic retrovirus via transient cotransfection of 293FT cells with retroviral plus Ecopak plasmids.45 Lentiviruses encoding TRB3 were prepared similarly using an EF1alpha plus IRES-GFP vector template, and pVSVg. Cells were transduced with retroviruses or lentiviruses, and stably transduced GFPpos populations were isolated by fluorescence-activated cell sorting (FACS, FACS Aria; BD Biosciences).
Results
Bone marrow erythroblast preparations, EPO survival responses, and transcriptome-based discovery of EPO-modulated (anti)apoptotic genes
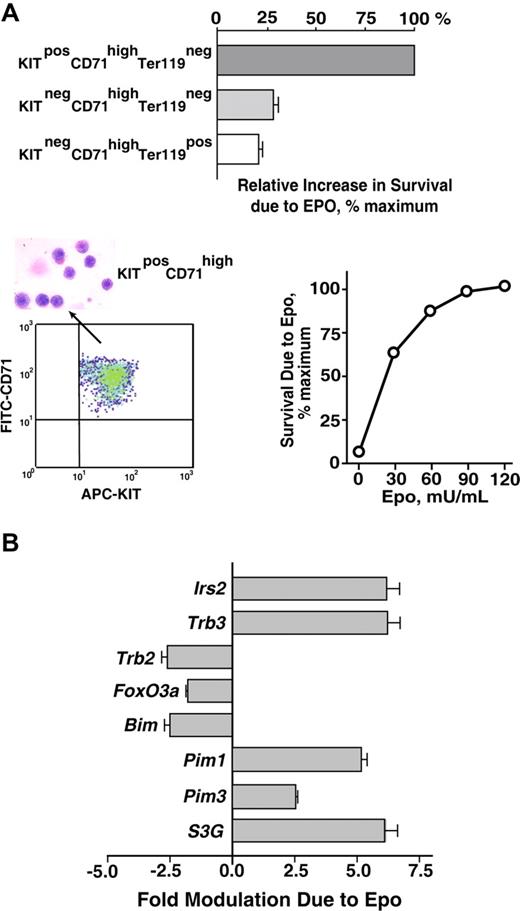
As a first step toward analyzing EPO survival responses in primary bone marrow–derived erythroblasts, a serum-free expansion system was used to generate cohorts of erythroblasts at KitposCD71highTer119neg, KitnegCD71highTer119neg, and KitnegCD71highTer119pos stages. Each population then was purified (by magnetic-activated cell sorting [MACS] and/or FACS), and the capacity of EPO to inhibit cell death in these early-, mid-, and late-stage erythroblasts was assessed. Specifically, each population was cultured for 18 hours in the presence of EPO at a limiting dose (0.1 U/mL). Frequencies of apoptotic cells were then assayed by FITC–annexin-V staining (Figure 1A). EPO's effects on survival were most marked for early-stage KitposCD71highTer119neg erythroblasts. For later-stage KitnegCD71highTer119neg or KitnegCD71highTer119pos cells, protection against apoptosis was afforded, but at several-fold lower efficiencies. In subsequent investigations of antiapoptotic pathways, KitposCD71highTer119neg erythroblasts therefore were prepared, isolated, and uniformly used.
Bone marrow erythroblast preparations, EPO-dependent survival responses, and transcriptome-based identification of EPO-regulated survival genes. (A) Bone marrow preparations were expanded for 3 days in SP34-EX medium to yield KitposCD71highTer119neg, KitnegCD71highTer119neg, and KitnegCD71highTer119pos erythroblast populations. Each was then purified by MACS and/or FACS, and increases in survival as afforded by EPO were determined. Specifically, cells were cultured in SP34-EX medium in the absence of SCF and presence of EPO at 0.1 U/mL. At 18 hours, frequencies of apoptotic cells were assayed using FITC–annexin-V and flow cytometry. Values are normalized means plus or minus SE. KitposCD71highTer119neg erythroblasts as purified by multiparameter MACS also were visualized in cytospin preparations and were assayed for protection against programmed cell death at a range of EPO doses. In the lower left panel, flow cytometry analyses also depict the homogeneity of isolated KitposCD71highTer119neg cells. (B) KitposCD71highTer119neg erythroblasts were cultured for 6 hours in the absence of hematopoietic cytokines, and were then exposed to EPO (± 5 U/mL) for 90 minutes. This included parallel processing of bone marrow–derived erythroblasts from n = 4 independent mice. After EPO exposure, RNA was isolated directly; 4 μg was used in biotin-cRNA syntheses and Affymetrix 430-2.0 array hybridizations. Among candidate (anti)apoptosis-related genes, 8 proved to be modulated by EPO at a confidence interval more than 99%. This included Irs2, Trb3, Trb2, Foxo3a, Bim, Pim1, Pim3, and Serpina3G (S3G). Values are mean-fold modulation plus or minus SD.
Bone marrow erythroblast preparations, EPO-dependent survival responses, and transcriptome-based identification of EPO-regulated survival genes. (A) Bone marrow preparations were expanded for 3 days in SP34-EX medium to yield KitposCD71highTer119neg, KitnegCD71highTer119neg, and KitnegCD71highTer119pos erythroblast populations. Each was then purified by MACS and/or FACS, and increases in survival as afforded by EPO were determined. Specifically, cells were cultured in SP34-EX medium in the absence of SCF and presence of EPO at 0.1 U/mL. At 18 hours, frequencies of apoptotic cells were assayed using FITC–annexin-V and flow cytometry. Values are normalized means plus or minus SE. KitposCD71highTer119neg erythroblasts as purified by multiparameter MACS also were visualized in cytospin preparations and were assayed for protection against programmed cell death at a range of EPO doses. In the lower left panel, flow cytometry analyses also depict the homogeneity of isolated KitposCD71highTer119neg cells. (B) KitposCD71highTer119neg erythroblasts were cultured for 6 hours in the absence of hematopoietic cytokines, and were then exposed to EPO (± 5 U/mL) for 90 minutes. This included parallel processing of bone marrow–derived erythroblasts from n = 4 independent mice. After EPO exposure, RNA was isolated directly; 4 μg was used in biotin-cRNA syntheses and Affymetrix 430-2.0 array hybridizations. Among candidate (anti)apoptosis-related genes, 8 proved to be modulated by EPO at a confidence interval more than 99%. This included Irs2, Trb3, Trb2, Foxo3a, Bim, Pim1, Pim3, and Serpina3G (S3G). Values are mean-fold modulation plus or minus SD.
From day-3 expansion cultures, KitposCD71highTer119neg erythroblasts could be isolated via stepwise linpos depletion plus Kitpos cell selection, and purities of more than 99.9% routinely were achieved (Figure 1A lower left panel). In addition, these purified erythroblasts retained sharp dependence on EPO for survival (Figure 1A lower right panel). Toward uncovering new candidate EPO-modulated regulators of progenitor cell survival, global transcriptome analyses next were used. Here, erythroblasts were expanded for 3 days from bone marrow preparations from n = 4 independent mice. For each, KitposCD71highTer119neg erythroblasts were purified, subdivided, and cultured for 6 hours in the absence of hematopoietic cytokines (and in 50 μg/mL transferrin, 15 ng/mL insulin, 0.1 mM 2-mercaptoethanol, 0.5% BSA in IMDM). Cells then were exposed to EPO (± 5 U/mL) for 90 minutes, and RNA was isolated. Biotin-cDNA was prepared, and was hybridized to Affymetrix 430–2.0 arrays. Target genes that were modulated more than 2-fold by EPO at a confidence level of more than 99% were identified via significance analysis of microarrays, Student t test, and Array Assist algorithms. Based on EPO's key role as an erythroblast survival factor, prime attention then was paid to EPO-regulated antiapoptotic factors.
Among greater than 1000 prospective (anti)apoptotic transcripts represented on 430–2.0 assays, only 8 proved to be significantly modulated by EPO (Figure 1B). These included 4 that previously have been associated with PI3K and/or ATK pathways: Irs2, Trb3, Trb2, and FoxO3a. Specifically, Irs2 and Trb3 were up-regulated more than 5-fold, while Trb2 and FoxO3a were repressed several fold. IRS2 is a pleckstrin homology and phosphotyrosine binding domain adaptor protein that associates with p85alpha.33,34,46 TRB3 and TRB2 are pseudokinases (and candidate E3 ubiquitin ligase adaptors) that also have been demonstrated to modulate AKT.35,36,47-49 FOXO3a is a forkhead transcription factor that can repress proapoptotic gene expression (eg, Trail and FasL) and is negatively regulated by AKT.50,51
Among Bcl2-related genes, only the proapoptotic BH3 domain factor Bim37,38 proved to be EPO regulated, and was rapidly downmodulated several fold. Two PIM kinases, Pim1 and Pim3,39 which act in parallel to AKT (and mTOR), however, were induced up to 5-fold by EPO. Finally, an atypical serpin (and putative protease inhibitor),40,41 which recently has been implicated in helper T-cell development,52 was observed to be strongly EPO induced, Serpina-3G (S3G).
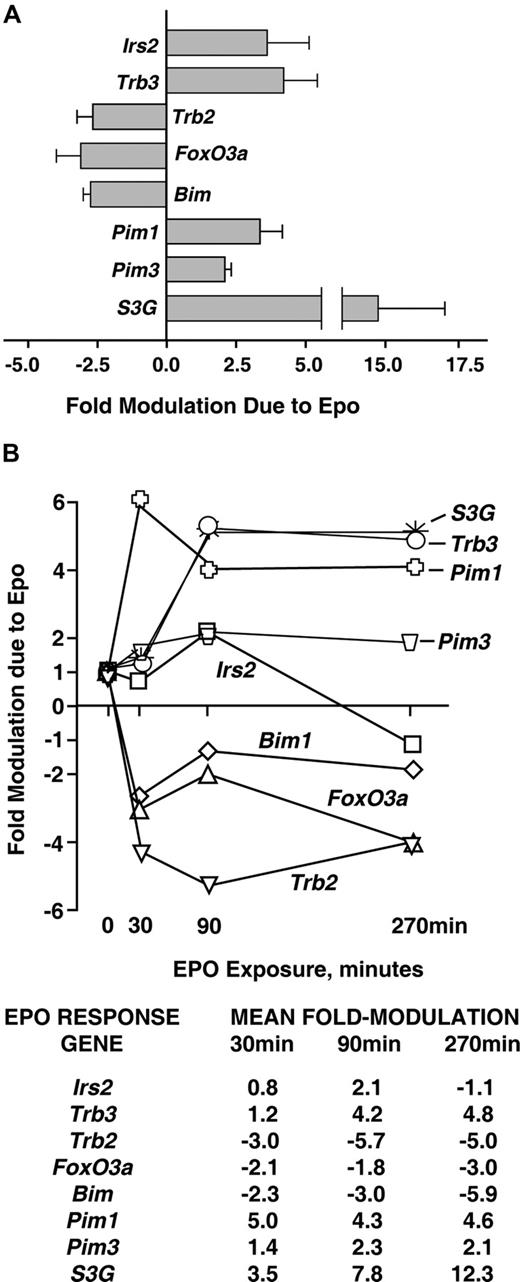
Analyses of EPO-regulated (anti)apoptotic transcriptional circuits were next extended via reverse-transcription and quantitative PCR (RT-QPCR) analyses. These experiments confirmed EPO-regulation of each of the 8 candidate (anti)apoptotic factors discovered via transcriptome analyses in primary bone marrow KitposCD71highTer119neg erythroblasts (Figure 2A). Kinetics of transcript modulation were examined at 0, 30, 90, and 270 minutes of EPO exposure. Outcomes were of interest first in that each up-regulated factor (Irs2, Trb3, Pim1, Pim3, and S3G) was induced in a time course consistent with an immediate-early response (Figure 2B; as indicated via direct comparison with an EPO-regulated immediate-early gene, Egr1; data not shown). Second, the downmodulated transcripts Trb2, FoxO3a, and Bim1 were affected rapidly by EPO (ie, within 30 minutes). Third, effects were both most prominent and most sustained for Trb3 and Serpina-3G as new candidate EPO-regulated survival factors.
Quantitative RT-PCR analyses of EPO modulation of Irs2, Foxo3a, Trb2, Trb3, Bim, Pim1, Pim3, and S3G expression. (A) For each of these (anti)apoptotic factors, multifold modulation by EPO was confirmed by RT-QPCR within KitposCD71highTer119neg erythroblasts as exposed to EPO for 90 minutes. Values are means plus or minus SD for n = 3 independent samples. (B) For each factor, time courses of modulation by EPO also were analyzed. Here, KitposCD71highTer119neg erythroblasts were isolated, cultured for 6 hours in the absence of hematopoietic cytokines, and exposed to EPO (2.5 U/mL). At the indicated intervals, RNA was prepared, reverse-transcribed, and used in quantitative PCR analyses (vs beta-actin as a normalizing control). For an independent sample set, time course analyses were repeated. Results are illustrated for 2 such analyses as mean fold modulation due to EPO for each EPO-modulated candidate survival factor.
Quantitative RT-PCR analyses of EPO modulation of Irs2, Foxo3a, Trb2, Trb3, Bim, Pim1, Pim3, and S3G expression. (A) For each of these (anti)apoptotic factors, multifold modulation by EPO was confirmed by RT-QPCR within KitposCD71highTer119neg erythroblasts as exposed to EPO for 90 minutes. Values are means plus or minus SD for n = 3 independent samples. (B) For each factor, time courses of modulation by EPO also were analyzed. Here, KitposCD71highTer119neg erythroblasts were isolated, cultured for 6 hours in the absence of hematopoietic cytokines, and exposed to EPO (2.5 U/mL). At the indicated intervals, RNA was prepared, reverse-transcribed, and used in quantitative PCR analyses (vs beta-actin as a normalizing control). For an independent sample set, time course analyses were repeated. Results are illustrated for 2 such analyses as mean fold modulation due to EPO for each EPO-modulated candidate survival factor.
Coupling of EPO-modulated transcriptional response circuits to EPO receptor functional subdomains
JAK2 is essential for erythropoiesis,13,53 and has been argued to be sufficient for EPOR-directed red cell formation.26,54 This notion is supported by the ability of mice expressing a PY-null EPOR-HM allele to support steady-state, but not stress, erythropoiesis.42 Stress erythropoiesis, in contrast, is efficiently rescued upon the restoration of a single EPOR-PY343 STAT5–binding site.42,43 It therefore was of interest to investigate the extent to which EPOR coupling to modulated survival genes might depend upon PY343, distal EPOR-PY sites, or perhaps an EPOR juxtamembrane box-1 domain for JAK2 binding.15,16
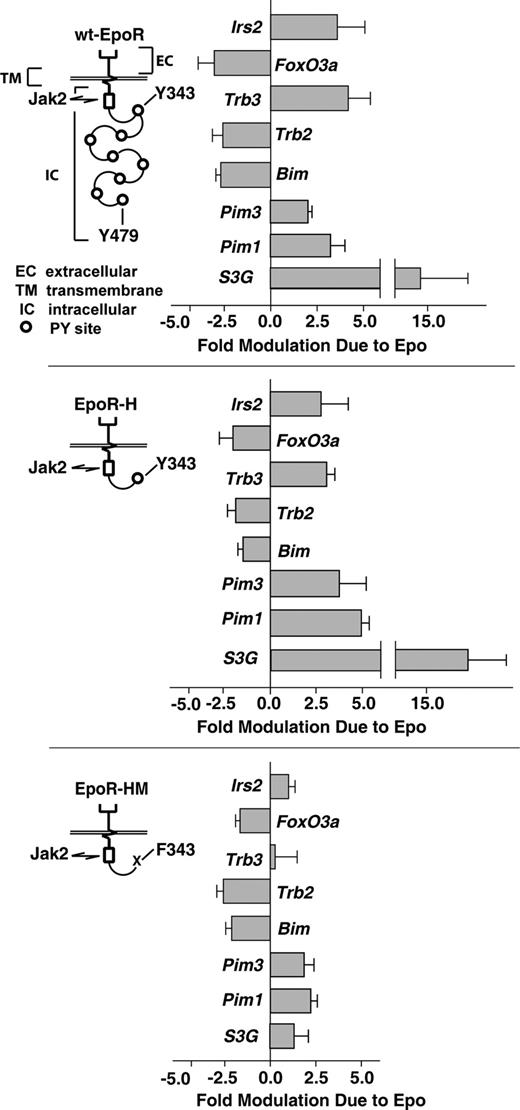
This was investigated using purified KitposCD71highTer119neg erythroblasts as expanded from bone marrow of mice expressing wt-EPOR, EPOR-H, and EPOR-HM alleles. These alleles retain, respectively, all 8 cytoplasmic PY sites; a single PY343 STAT5–binding site; or no PY sites and only juxtamembrane box-1 and -2 motifs for JAK2 binding (Figure 3). As analyzed by RT-QPCR, EPOR-H interestingly retained the capacity to modulate all 8 survival genes that were discovered to be modulated via the wt-EPOR. Furthermore, for Pim1 and Pim3, induction via EPOR-H was somewhat enhanced (Figure 3 top and middle panels). In contrast, the PY-null allele EPOR-HM retained only repressive regulation over FoxO3a, Trb2, and Bim (Figure 3 bottom panel). Specifically, induction of Irs2, Trb3, and S3G via EPOR-HM allele was near background, while Pim1 and Pim3 were induced only approximately 2-fold.
EPO induction of Irs2, Trb3, S3G, Pim1, and PIm3 depends upon EPOR-H PY343 signals, while EPO inhibition of Foxo3a, Trb2, and Bim is EPOR-PY independent. Erythroid progenitor cells were expanded (in SP34-EX medium) from bone marrow preparations for wt-EPOR, EPOR-H, and EPOR-HM mice (n = 3 independent mice per group). For wt-EPOR, EPOR-HM, and EPOR-H EPO receptor alleles, schematics are provided. KitposCD71highTer119neg erythroblasts then were isolated, cultured for 5.5 hours in the absence of hematopoietic cytokines, and exposed to EPO (± 2.5 U/mL). At 90 minutes, RNA was isolated. cDNA was then prepared, and levels of Irs2, Trb3 Trb2, Foxo3a, Bim, Pim1, Pim3, and S3G transcripts were determined by quantitative PCR. Values are mean-fold modulation plus or minus SD.
EPO induction of Irs2, Trb3, S3G, Pim1, and PIm3 depends upon EPOR-H PY343 signals, while EPO inhibition of Foxo3a, Trb2, and Bim is EPOR-PY independent. Erythroid progenitor cells were expanded (in SP34-EX medium) from bone marrow preparations for wt-EPOR, EPOR-H, and EPOR-HM mice (n = 3 independent mice per group). For wt-EPOR, EPOR-HM, and EPOR-H EPO receptor alleles, schematics are provided. KitposCD71highTer119neg erythroblasts then were isolated, cultured for 5.5 hours in the absence of hematopoietic cytokines, and exposed to EPO (± 2.5 U/mL). At 90 minutes, RNA was isolated. cDNA was then prepared, and levels of Irs2, Trb3 Trb2, Foxo3a, Bim, Pim1, Pim3, and S3G transcripts were determined by quantitative PCR. Values are mean-fold modulation plus or minus SD.
An EPOR/PY343/STAT5 signaling axis mediates EPO-dependent GAB2 activation
Investigations next sought to examine roles for select EPO-regulated survival factors at the protein level. Attention first was paid to EPO-regulated PI3K-associated adaptor molecules. For Irs 1-4, transcriptome analyses of KitposCD71highTer119neg erythroblasts per se indicated high-level expression selectively of Irs2 (with Irs1, Irs 3, and Irs 4 expressed at approximately 40-fold lower levels; data not shown). At the protein level, IRS2 also was readily detected in KitposCD71highTer119neg erythroblasts. However, EPO exposure did not lead to detectable modulation of IRS2 phosphorylation.
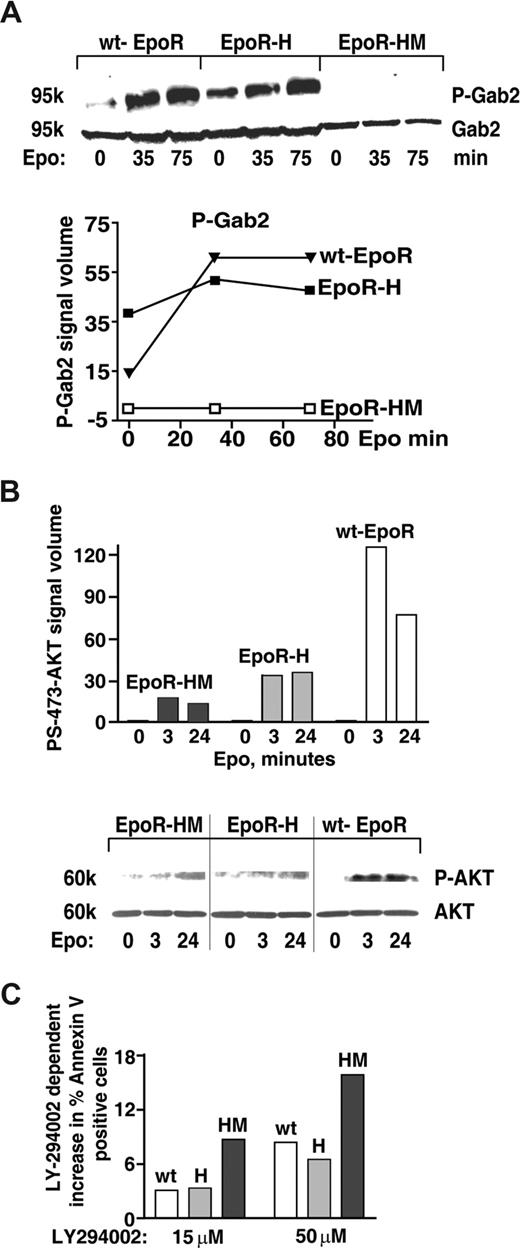
Based on the above-defined activities of EPOR-H, and recent reports of STAT5 interaction with an alternative PI3K docking factor, GAB2,55 Gab transcript expression and possible EPO-induced GAB phosphorylation next were investigated. Among Gabs1-4, Gab2 proved to be expressed at appreciable levels in KitposCD71highTer119neg erythroblasts (transcriptome data not shown). In addition, GAB2's PY452 phosphorylation proved to be rapidly and strongly induced by EPO. Interestingly, this response (GAB2 PY452 phosphorylation) also was efficiently supported in EPOR-H, but not EPOR-HM proerythroblasts (Figure 4A). These analyses were consistent with a prior report of EPO-induced GAB2 phosphorylation,57 and further link this pathway to STAT5 signals. In addition, and following EPO exposure, coimmunoprecipitation of phospho-GAB2 with STAT5 also could be demonstrated in wt-EPOR erythroblasts (data not shown).
An EPOR-PY343 STAT5 axis mediates GAB2 but not AKT activation. (A) KitposCD71highTer119neg erythroblasts were prepared from wt-EPOR, EPOR-H, and EPOR-HM bone marrow cell expansion cultures. Cells were then cultured for 5.5 hours in IMDM containing transferrin (50 μg/mL), insulin (15 ng/mL), and BSA (0.5%). EPO (2 U/mL) was then added and at 0, 35, and 75 minutes lysates were prepared and levels of PY452 GAB2 and total GAB2 were determined by Western blotting. Outcomes were analyzed quantitatively by scanning densitometry (bottom panel). (B) Coupling to AKT is deficient for not only EPOR-HM but also EPOR-H alleles. Bone marrow–derived KitposCD71high erythroblasts were prepared and purified (as described in panel A) from EPOR-HM, EPOR-H, and wt-EPOR mice. Following withdrawal of hematopoietic cytokines (for 5.5 hours), cells were exposed to EPO (2.5 U/mL) and at the indicated intervals lysates were prepared. Levels of phospho-S473-AKT then were determined by Western blotting (and normalized for AKT levels/loading). Note the limited activation of AKT in not only EPOR-HM but also EPOR-H erythroblasts. Vertical lines indicate reassembled segments from a single, uniform en-hanced chemiluminescence (ECL) exposure. (C) Bone marrow–derived KitposCD71highTer119neg erythroblasts expressing an EPOR-H allele are efficiently protected by EPO against apoptosis, while EPOR-HM erythroblasts are not; KitposCD71high wt-EPOR, EPOR-H, and EPOR-HM erythroblasts were expanded and purified. Cells were then treated with LY274002 to inhibit PI3K,56 and were cultured in the presence of EPO at 0.2 U/mL. At 18 hours of culture, frequencies of apoptotic cells were determined by annexin-V staining and flow cytometry. Representative outcomes for LY274002 dosing at 15 and 50 μM are shown.
An EPOR-PY343 STAT5 axis mediates GAB2 but not AKT activation. (A) KitposCD71highTer119neg erythroblasts were prepared from wt-EPOR, EPOR-H, and EPOR-HM bone marrow cell expansion cultures. Cells were then cultured for 5.5 hours in IMDM containing transferrin (50 μg/mL), insulin (15 ng/mL), and BSA (0.5%). EPO (2 U/mL) was then added and at 0, 35, and 75 minutes lysates were prepared and levels of PY452 GAB2 and total GAB2 were determined by Western blotting. Outcomes were analyzed quantitatively by scanning densitometry (bottom panel). (B) Coupling to AKT is deficient for not only EPOR-HM but also EPOR-H alleles. Bone marrow–derived KitposCD71high erythroblasts were prepared and purified (as described in panel A) from EPOR-HM, EPOR-H, and wt-EPOR mice. Following withdrawal of hematopoietic cytokines (for 5.5 hours), cells were exposed to EPO (2.5 U/mL) and at the indicated intervals lysates were prepared. Levels of phospho-S473-AKT then were determined by Western blotting (and normalized for AKT levels/loading). Note the limited activation of AKT in not only EPOR-HM but also EPOR-H erythroblasts. Vertical lines indicate reassembled segments from a single, uniform en-hanced chemiluminescence (ECL) exposure. (C) Bone marrow–derived KitposCD71highTer119neg erythroblasts expressing an EPOR-H allele are efficiently protected by EPO against apoptosis, while EPOR-HM erythroblasts are not; KitposCD71high wt-EPOR, EPOR-H, and EPOR-HM erythroblasts were expanded and purified. Cells were then treated with LY274002 to inhibit PI3K,56 and were cultured in the presence of EPO at 0.2 U/mL. At 18 hours of culture, frequencies of apoptotic cells were determined by annexin-V staining and flow cytometry. Representative outcomes for LY274002 dosing at 15 and 50 μM are shown.
For GAB2, one prime binding partner is PI3-kinase.34 EPOR-PY343–mediated STAT5 engagement of GAB2 therefore was predicted to lead to PI3-kinase and ATK kinase activation. However, as shown in Figure 4B not only EPOR-HM but also EPOR-H erythroblasts proved to only inefficiently support EPO-dependent AKT activation (Figure 4B). As a correlate, EPO-dependent survival effects were analyzed directly (and in parallel) in wt-EPOR, EPOR-H, and EPOR-HM proerythroblasts treated with the PI3K inhibitor LY29400256 (Figure 4C). Outcomes demonstrated compromised survival for EPOR-HM erythroblasts, but near wild-type survival response for EPOR-H cells. EPOR-PY343 survival signals beyond those provided via GAB2 (and PI3-kinase plus AKT) therefore are predicted to exist.
TRB3 and SERPINA-3G act to inhibit erythroid progenitor cell apoptosis due to cytokine withdrawal
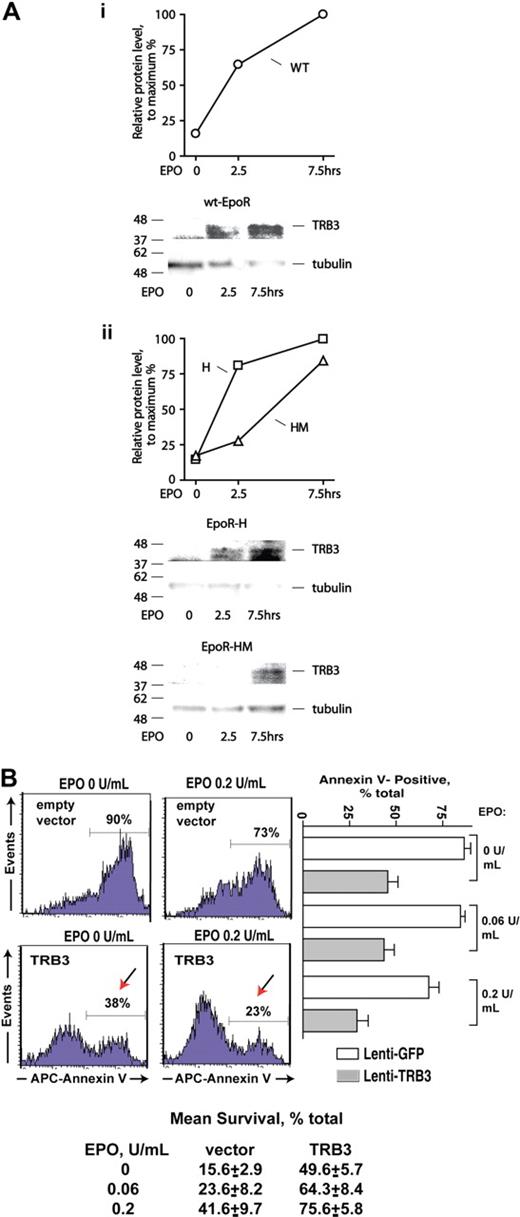
Transcriptome and RT-QPCR analyses pointed to Trb3 and Serpina 3G (S3G) as additional potential EPOR-PY343 (and STAT5)–regulated erythroblast (anti)apoptotic factors (Figures 1,Figure 2–3). TRB3 first was investigated further at the protein and functional levels. In particular, Western blot analyses were performed using purified KitposCD71highTer119neg primary erythroblasts expressing wt-EPOR, EPOR-H, or EPOR-HM alleles. EPO rapidly induced TRB3 expression (Figure 5A). In wt-EPOR, EPOR-H, and EPOR-HM erythroblasts, no induction of TRB3 was observed at 2.5 hours, but did subsequently become detectable at an extended time point (7.5 hours). These results, therefore, corresponded reasonably with transcriptional analyses. Experiments next tested the capacity of TRB3 to promote erythroid progenitor cell survival. Here, an EF1alpha-IRES-GFP lentivirus encoding TRB3 was prepared (together with an empty control vector), and each was used to stably transduce UT7epo cells at limiting MOIs. GFP-positive cells then were isolated by FACS. Possible effects of ectopically expressed TRB3 on survival proved to substantially affect UT7epo cell survival, and at limiting EPO doses significantly inhibited apoptosis. Specifically and for 3 independent analyses, survival advantages of more than 300% over controls were afforded (Figure 5B lower panel). TRB3 therefore appears to act in an EPO signaling axis to enforce erythroid progenitor cell survival (when EPO becomes limiting).
TRB3 induction via EPOR alleles, and TRB3-mediated survival effects in EPO-dependent erythroid progenitor cells. (A) KitposCD71highTer119neg erythroblasts were expanded, and purified from marrow progenitors as prepared from wt-EPOR, EPOR-H, and EPOR-HM mice. Cells then were cultured for 5 hours in 50 μg/mL transferrin, 0.1% BSA, 0.1 mM 2-MF, 15 ng/mL insulin, IMDM and subsequently exposed to EPO (2.5 U/mL). At the intervals indicated, cell lysates were prepared and analyzed for TRB3 expression via Western blotting. Outcomes for the wt-EPOR are illustrated in panel Ai, and for EPOR-HM and EPOR-H alleles in panel Aii. (B) TRB3 inhibits apoptosis due to cytokine withdrawal in EPO-dependent UT7epo cells. Lentiviruses encoding TRB3 or GFP only (empty control vector) were prepared and used to stably transduce UT7epo cells. GFPpos cells were then isolated by FACS, and possible effects of TRB3 on survival were assessed in the presence of EPO at limiting doses. As assayed via staining with APC–annexin-V, significant protection against apoptosis was afforded by TRB3. Representative flow cytometry data are shown (left panels) together with mean TRB3 survival effects (± SE, n = 3). Mean outcomes for 2 independent sets of experiments also are shown (botom panel).
TRB3 induction via EPOR alleles, and TRB3-mediated survival effects in EPO-dependent erythroid progenitor cells. (A) KitposCD71highTer119neg erythroblasts were expanded, and purified from marrow progenitors as prepared from wt-EPOR, EPOR-H, and EPOR-HM mice. Cells then were cultured for 5 hours in 50 μg/mL transferrin, 0.1% BSA, 0.1 mM 2-MF, 15 ng/mL insulin, IMDM and subsequently exposed to EPO (2.5 U/mL). At the intervals indicated, cell lysates were prepared and analyzed for TRB3 expression via Western blotting. Outcomes for the wt-EPOR are illustrated in panel Ai, and for EPOR-HM and EPOR-H alleles in panel Aii. (B) TRB3 inhibits apoptosis due to cytokine withdrawal in EPO-dependent UT7epo cells. Lentiviruses encoding TRB3 or GFP only (empty control vector) were prepared and used to stably transduce UT7epo cells. GFPpos cells were then isolated by FACS, and possible effects of TRB3 on survival were assessed in the presence of EPO at limiting doses. As assayed via staining with APC–annexin-V, significant protection against apoptosis was afforded by TRB3. Representative flow cytometry data are shown (left panels) together with mean TRB3 survival effects (± SE, n = 3). Mean outcomes for 2 independent sets of experiments also are shown (botom panel).
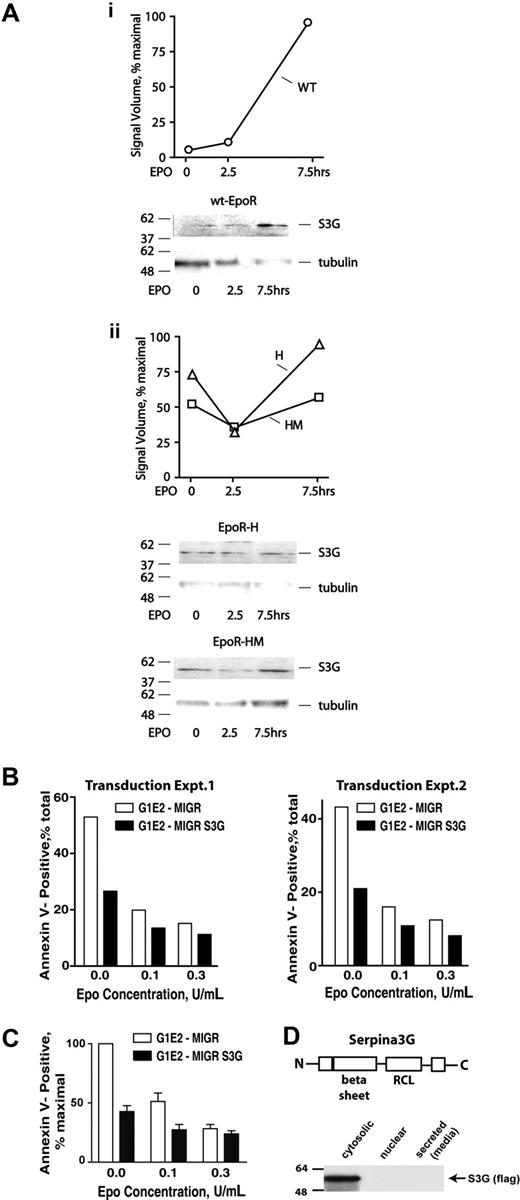
SERPINA-3G (S3G) next was studied, initially via Western blot experiments. In wt-EPOR erythroblasts, EPO clearly induced S3G expression at increased levels (Figure 6Ai). In both EPOR-H and EPOR-HM erythroblasts, however, S3G expression interestingly proved to be sustained upon EPO withdrawal (Figure 6Aii). This outcome was unexpected, and suggests that S3G may be subject to significant posttranscriptional regulation (here, in a potentially compensatory mode for minimal EPOR alleles).
EPO modulation of endogenous S3G expression in wt-EPOR, EPOR-H, and EPOR-HM erythroblasts, and S3G effects on erythroid progenitor cell survival. (A) KitposCD71highTer119neg erythroblasts were isolated from wt-EPOR, EPOR-H, and EPOR-HM bone marrow expansion cultures. Cells then were cultured for 5 hours in 50 μg/mL transferrin, 0.1% BSA, 0.1 mM 2-ME, 15 ng/mL insulin, IMDM. At the time intervals indicated, cell lysates were prepared and analyzed for S3G expression via Western blotting. Outcomes for the wt-EPOR are illustrated in panel Ai, and for EPOR-HM and EPO-H alleles in panel Aii. (B) Epo-dependent G1E/JC4 cells were transduced with a MIGR-S3G retroviral construct, or with an empty MIGR vector as a negative control. For G1E/JC4-S3G and G1E/JC4-MIGR cells in exponential growth phase, EPO was withdrawn for 6 hours and subsequently was provided at the doses indicated. At 24 hours, frequencies of apoptotic cells were assayed by staining with annexin-V and flow cytometry. Transduction sets nos. 1 and 2 represent independently transduced cell populations. (C) Average effects of S3G on G1E/JC4 cell survival for 4 independent analyses also are illustrated. Values are normalized means plus or minus SE. (D) Via Western blotting of cellular fractions and concentrated media, ectopically expressed S3G was observed to localize to a cytosolic fraction. S3G's basic structure, including its serpin domain and reactive center loop (RCL), also is diagrammed.
EPO modulation of endogenous S3G expression in wt-EPOR, EPOR-H, and EPOR-HM erythroblasts, and S3G effects on erythroid progenitor cell survival. (A) KitposCD71highTer119neg erythroblasts were isolated from wt-EPOR, EPOR-H, and EPOR-HM bone marrow expansion cultures. Cells then were cultured for 5 hours in 50 μg/mL transferrin, 0.1% BSA, 0.1 mM 2-ME, 15 ng/mL insulin, IMDM. At the time intervals indicated, cell lysates were prepared and analyzed for S3G expression via Western blotting. Outcomes for the wt-EPOR are illustrated in panel Ai, and for EPOR-HM and EPO-H alleles in panel Aii. (B) Epo-dependent G1E/JC4 cells were transduced with a MIGR-S3G retroviral construct, or with an empty MIGR vector as a negative control. For G1E/JC4-S3G and G1E/JC4-MIGR cells in exponential growth phase, EPO was withdrawn for 6 hours and subsequently was provided at the doses indicated. At 24 hours, frequencies of apoptotic cells were assayed by staining with annexin-V and flow cytometry. Transduction sets nos. 1 and 2 represent independently transduced cell populations. (C) Average effects of S3G on G1E/JC4 cell survival for 4 independent analyses also are illustrated. Values are normalized means plus or minus SE. (D) Via Western blotting of cellular fractions and concentrated media, ectopically expressed S3G was observed to localize to a cytosolic fraction. S3G's basic structure, including its serpin domain and reactive center loop (RCL), also is diagrammed.
Next, EPO-dependent G1E/JC4 cells were implemented as a model for analyses of S3G action. This model was chosen based on a capacity of G1E/JC4 cells to also inducibly activate a GATA1-ER fusion protein, and terminally differentiate.58 (Differentiation, however, did not prove to be affected by S3G; data not shown.) G1E/JC4 cells were transduced with a MIGR-(flag)S3G retrovirus, and empty control vector, and GFPpos lines were isolated from 2 independent transductions. Possible effects of ectopically expressed S3G on survival then were assessed. Exponentially growing G1E/JC4-S3G and control G1E/JC4-MIGR cells were cultured in the presence of EPO at limiting concentrations. At 18 hours of culture, frequencies of apoptotic cells were determined by staining with APC–annexin-V, and via flow cytometry. S3G proved to significantly inhibit apoptosis as incurred due to EPO withdrawal (Figure 6B,C). This effect was observed reproducibly for separate sets of independently transduced G1E/JC4 cells, and in repeated analyses.
Because the majority of serpins are secreted,40,41 cell fractionation experiments also were performed to determine SERPINA-3G's subcellular localization. In these experiments, no detectable amounts of (flag)-SERPINA-3G were observed in concentrated conditioned media, or in nuclear extracts. Instead, (flag)-SERPINA-3G appeared to be localized to a cytosolic compartment (Figure 6D). Possible release from membrane compartments, however, presently cannot be discounted, and subcellular localization may not fully reflect that of endogenous S3G.
Model for EPO-regulated survival circuits in primary KitposCD71highTer119neg erythroblasts
Previously, BCL-XL has been shown via gene disruption experiments to be important for erythroblast survival,31 and Bcl-xl as well as Bcl2 have been proposed to comprise EPO and STAT5 response genes.29,59,60 These considerations raised questions concerning the perhaps overlapping roles of these BCL factors versus the presently defined EPO-regulated survival factors as mediators of EPO's antiapoptotic effects. Experiments therefore were conducted to address this point (and were also prompted by an apparent lack of Bcl-xl or Bcl2 gene regulation by EPO in transcriptome analyses). Here, staged KitposCD71highTer119neg erythroblasts were purified to more than 99% homogeneity, and were cultured in the absence of EPO for 6 hours. Cells were then exposed to EPO (5 U/mL) and at 0, 2.5, and 7.5 hours, BCL-XL and BCL2 levels were analyzed by Western blotting. In parallel, possible modulation of Bclx and Bcl2 transcript levels was assessed by RT-QPCR at 0, 30, 90, and 270 minutes of EPO exposure. As a positive control, Pim1 levels also were analyzed. In these highly EPO-responsive KitposCD71highTer119neg cells, essentially no modulation of BCL-XL or BCL2 was observed (Figure 7A). These results therefore further underline the potential importance of alternate survival factors such as S3G and TRB3 as candidate mediators of EPO's antiapoptotic effects. In Figure 7B, a model is outlined for presently defined EPO-regulated survival circuits in highly EPO-dependent early-stage bone marrow erythroblasts.
EPO-independent activation of BCL-XL and BCL2 expression at discrete stages of (pro)erythroblast development. (A) In primary bone marrow erythroblasts, neither BCL-XL nor BCL2 expression is EPO-modulated. KitposCD71highTer119neg erythroblasts were expanded from wt-EPOR bone marrow preparations. Cells then were cultured for 5 hours in 50 μg/mL transferrin, 0.1% BSA, 15 ng/mL insulin, 0.1 mM 2-ME, IMDM. EPO was then added (2.5 U/mL), and at 0, 2.5, and 7.5 hours cell lysates were prepared. Lysates then were analyzed by Western blotting for levels of BCL-XL, BCL2, PIM1, and beta-tubulin. In parallel, possible EPO modulation of Bcl-x or Bcl2 transcripts also was analyzed, here at 0, 30, 90, and 270 minutes of EPO exposure. Values are mean-fold modulation plus or minus SE. (B) Mapping of EPOR subdomains to EPO-regulated survival factor circuits. Presently defined EPO-modulated transcriptional response circuits are outlined. An EPOR JAK2-only circuit mediates Foxo3a, Bim, and Trb2 repression. In parallel, an EPOR/PY343/STAT5 axis enhances Pim1 and Pim3 expression, and affords EPO induction of Irs2, S3G, and Trb3.
EPO-independent activation of BCL-XL and BCL2 expression at discrete stages of (pro)erythroblast development. (A) In primary bone marrow erythroblasts, neither BCL-XL nor BCL2 expression is EPO-modulated. KitposCD71highTer119neg erythroblasts were expanded from wt-EPOR bone marrow preparations. Cells then were cultured for 5 hours in 50 μg/mL transferrin, 0.1% BSA, 15 ng/mL insulin, 0.1 mM 2-ME, IMDM. EPO was then added (2.5 U/mL), and at 0, 2.5, and 7.5 hours cell lysates were prepared. Lysates then were analyzed by Western blotting for levels of BCL-XL, BCL2, PIM1, and beta-tubulin. In parallel, possible EPO modulation of Bcl-x or Bcl2 transcripts also was analyzed, here at 0, 30, 90, and 270 minutes of EPO exposure. Values are mean-fold modulation plus or minus SE. (B) Mapping of EPOR subdomains to EPO-regulated survival factor circuits. Presently defined EPO-modulated transcriptional response circuits are outlined. An EPOR JAK2-only circuit mediates Foxo3a, Bim, and Trb2 repression. In parallel, an EPOR/PY343/STAT5 axis enhances Pim1 and Pim3 expression, and affords EPO induction of Irs2, S3G, and Trb3.
Discussion
EPO is an essential factor for developing erythroblasts,3,61 and its prime actions are thought to affect progenitor cell survival.4,6-8 Consistent with this concept, EPO also has been demonstrated to exert cytoprotective effects in injured cardiac, renal, brain, and retinal cells.10,11 In a distinct potential cancer cell context, EPO furthermore has been reported to increase mortality in patients with head and neck, and metastatic breast cancers.31,62 Together, these observations have heightened interest in better understanding EPO's specific response pathways. The present study focuses on EPO survival signals, and uses primary bone marrow erythroblasts plus global transcriptome analyses to define new candidate pathway components.
As one core point for discussion, survival circuits modulated via the PY-null allele EPOR-HM first are considered. Despite a conserved evolution among vertebrates,20,63 EPOR cytoplasmic PY-binding motifs for SH2 and PTB domain signal transduction factors appear to be nonessential for steady-state (but not stress) erythropoiesis.18,54 Prior analyses of EPOR-HM signaling have ruled out the potential for compensatory engagement of at least certain EPOR-R PY–coupled signal transduction factors (eg, STAT-5, STAT-3, STAT-1).18,42 Coupling (via PI3K) to AKT also is decreased substantially for this allele (Figure 4). MEK1,2 and ERK1,2 signaling via EPOR-HM, in contrast, is sustained.18 Inhibition of this latter pathway, however, appears to selectively impact on differentiation rather than growth or survival potentials.18 This raises questions as to how EPOR-PY–independent JAK2-only signaling might inhibit programmed cell death. Present analyses indicate that this, in part, may involve EPOR plus JAK2-mediated repression of Bim, and FoxO3a. In particular, EPOR-HM's capacity to enforce the EPO-dependent repression of these inhibitory factors approximates that exerted by the wt-EPOR and EPOR-H. As a BH3-only factor, BIM can destabilize mitochondrial membranes by antagonizing BCL2 and BCL-XL.37,38 FOXO3a similarly can exert proapoptotic effects, in part, by transcriptionally activating FAS-L, TRAIL, and PUMA.50,51,64 Notably, TRAIL and FAS-L each have been implicated as effectors of anemia in the clinical contexts of polycythemia vera and myelodysplastic syndromes.65,66 EPOR-HM repression of FOXO3a and BIM therefore is predicted to lead to enhanced erythroblast survival. EPOR-HM also retained a capacity to repress Trb2 expression. As studied in TF1 cells, TRB2 has been reported to antagonize MCL1 and induce apoptosis.67 Gain-of-function studies in hematopoietic stem cells, however, have implicated myeloleukemogenic roles for TRB2. Additional investigations therefore will be required to better understand the consequences on erythroblast development of TRB2's downmodulation by EPO.
Within EPOR-H, an allele that efficiently supports erythropoiesis during anemia,42 restoration of a PY343 STAT5–binding site restored near wild-type levels of EPO modulation of 5 candidate survival genes Pim1, Pim3, Trb3, Serpina-3G, and Irs2 (Figures 1,Figure 2–3). Via gene disruption experiments, PIM kinases have been shown to be important for maintaining cell survival and normal cell size.39 Pim1 is expressed at high levels in hematopoietic tissues,68 and its disruption also leads to microcytic anemia.69 Mechanistically, PIMs initially were indicated to act convergently with AKT pathways to regulate mTOR.39 Genetic dissections, however, indicate separable yet parallel survival pathways.70 These considerations, together with recent evidence for STAT5 plus PIM kinase–enforced leukemogenic events,71 further support a case that PIM1 and PIM3 likely comprise important EPO (and STAT5) response survival factors for EPO-dependent erythroblasts. For TRB3, its sustained expression was induced multifold by EPO and also depended upon EPOR/PY343/STAT signaling (Figures 1,Figure 2–3,5). Certain prior studies predicted that TRB3 might act in negative feedback loops including inhibitory effects on AKT.48 Uncertainty exists, however, concerning TRB3's molecular action mechanisms.72 In particular, TRB3 recently has been demonstrated to act as an E3 ubiquitin ligase coupling factor with COP1, and to target adipocyte acetyl coenzyme A carboxylase for turnover.47 To provide initial insight into EPO-related roles for TRB3, a lentiviral vector encoding TRB3 presently was prepared, and was used to stably transduce EPO-dependent erythroid progenitor cells. When EPO was limiting, TRB3 significantly enhanced survival (Figure 5). Ways via which TRB3 acts within an EPO/EPOR signaling axis to inhibit cell death are presently uncertain. By speculation, this might involve TRB3 targeting of a key proapoptotic factor in developing erythroblasts. Overall, TRB3's apparent ability to exert differential effects in various tissues may also relate to versatile capacities as an E3 ubiquitin ligase coupler.
For Serpina-3G (S3G), its induced expression likewise proved to depend sharply on EPOR/PY343/STAT5 signals, and was efficiently promoted in primary proerythroblasts by EPOR-H but not by EPOR-HM. Serpins typically are secreted protease inhibitors with broad-range biologic functions. Within a hematopoietic growth factor context, one example involves granulocyte colony-stimulating factor (GCSF) inhibition of SERPINA-1 and SERPINA-373 within a stem cell niche. Specifically, GCSF repression of these secreted serpins relieves an inhibition of proteases, which then act to promote CD34 cell release.73 For SERPINA-3G, one rationale for considering possible cytoprotective roles is a recent case described for promoting helper T-cell expansion.52 These studies of T cells, however, did not involve a cytokine signaling context. Presently, ectopic expression of S3G in EPO-dependent erythroid progenitors is shown to protect against apoptosis due to cytokine withdrawal (Figure 6C). The problem as to how S3G mechanistically affords cytoprotection is a substantially more open question. One hypothesis involves S3G inhibition of a proapoptotic protease. Lysosome-derived cathepsins have been considered as one candidate target,52 but others (including caspases per se) are possible. Certain serpins, in fact have evolved to function via diverse mechanisms including chaperone effects and chromatin binding activities.41,42 Investigations of both SERPINA-3G and TRB3's action mechanisms in EPO-dependent erythroblasts therefore should be of significant future interest. With regard to induction pathways, this might include analyses of putative conserved STAT-binding elements, which (based on TRANSFAC analyses) are predicted to occur in S3G at a region approximately 150-bp upstream of exon 1, and for Trb3 within a −4500-bp upstream region that includes 3 consensus STAT elements (in silico analyses; not shown).
Two PI3-kinase pathway–associated docking factors, Irs2 and GAB2, also are presently shown to be regulated by EPO. For Irs2, activation was at the transcript level. EPO-induced S731 phosphorylation of IRS2, however, was not detected. GAB2, in contrast, proved to be regulated by EPO at the level of S473 phosphorylation, and this furthermore depended sharply upon intactness of an EPOR-PY343 STAT5–binding site. GAB2 also can couple to p85 and GRB2.34,74 This then provides routes for EPOR-PY343 (and STAT5) coupling to AKT and ERK1/2 survival pathways. Silencing of GAB2 plus STAT5, in fact, has recently been shown to inhibit BCR-ABL–dependent survival of chronic myeloid leukemia cells.75
A final point for discussion concerns potential coupling of EPOR signals to BCL-XL expression. Via gene disruption experiments, Bcl-x has been shown to be essential for red cell development.31 In addition, and in several cell line models, Bcl-x (and Bcl2) has been observed to be upmodulated by EPO.59,60 Bcl-x expression further has been argued to be induced by EPO via an EPOR/JAK2/PY343/STAT5 axis.29 In the presently studied primary erythroblast system, however, EPO modulation of BCL-x or BCL2 was not detectable at any significant level. Findings do not rule out possible EPO effects on multistep temporally attenuated pathways to Bcl-x induction or the prospect that EPO might affect BCL-X or BCL2 expression at later developmental stages. For KitposCD71highTer119neg erythroblasts, however, findings are inconsistent with a model involving proposed direct EPOR/PY343/STAT5 induction pathways.29,59 For such early-stage highly EPO-dependent proerythroblasts, the present findings also underline the likely significance of alternate survival factors as supportive of EPO's essential effects within an apparently narrow window of proerythroblast development.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant R01-HL44491, and MMCRI Core Facilities as sponsored via NIH P20-RR018789 and NIH P20-RR015555.
National Institutes of Health
Authorship
Contribution: P.S., A.D., J.F., E.H., O. Bogacheva, O. Bogachev, M.M., S.B., A.P., C.E., and D.M.W. each contributed in major ways to the design and execution of essentially all experiments, and contributed to data analyses and interpretations, figure con-struction, and paper assembly.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Don M. Wojchowski, Director, Stem and Progenitor Cell Biology Program, Maine Medical Center Research Institute, 81 Research Dr, Scarborough, ME 04074; e-mail: wojchd@mmc.org.
References
Author notes
P.S. and A.D. contributed equally to this work.