NOTCH1 mutations are common in T-lineage acute lymphoblastic leukemia (T-ALL). Twin studies and retrospective screening of neonatal blood spots provide evidence that fusion genes and other chromosomal abnormalities associated with pediatric leukemias can originate prenatally. Whether this is also the case for NOTCH1 mutations is unknown. Eleven cases of T-ALL were screened for NOTCH1 mutations and 4 (36%) had mutations in either the heterodimerization (HD) or proline glutamic acid/serine/threonine (PEST) domains. Of these 4, 3 could be amplified by mutation-specific polymerase chain reaction primers. In one of these 3, with the highest sensitivity, NOTCH1 mutation was detected in neonatal blood spots. In this patient, the blood spot was negative for SIL-TAL1 fusion, present concomitant with NOTCH1 mutation, in the diagnostic sample. We conclude that NOTCH1 can be an early or initiating event in T-ALL arising prenatally, to be complemented by a postnatal SIL-TAL1 fusion.
Introduction
There are substantial evidence-based studies of twins with leukemia, retrospective scrutiny of archived neonatal blood spots, and cord blood screening that most pediatric leukemias are initiated in utero.1,2 This conclusion applies to B-cell–precursor acute lymphoblastic leukemia (ALL) with TEL-AML13,4 or hyperdiploidy,5,6 infant pro-B-lineage ALL (B ALL) with MLL-AF4 fusion,7,8 acute myeloid leukemia (AML) with AML1-ETO9 as well as to Down syndrome patients with acute megakaryocytic leukemia (AMKL) and GATA1 mutations.10 In contrast, there are currently little data on the developmental timing of T-lineage acute lymphoblastic leukemia (T-ALL)/non-Hodgkin lymphoma (NHL)
NOTCH1 mutations occur in more than 50% of patients with T-ALL, in concert with other genetic changes.11,12 From these data on clinical samples alone, one cannot deduce the temporal sequence of genetic events to decipher whether NOTCH1 mutation was initiating or secondary. The high prevalence of these mutations provided an opportunity to ask if NOTCH1 mutations occur prenatally. We have screened matched diagnostic, leukemic DNA from patients with T-ALL with their archived neonatal blood spots for concordant NOTCH1 mutations.
Study design
Eleven pediatric patients diagnosed with T-ALL were entered into the study, with ethical approval and with matched blood spots. Release of diagnostic samples (leukemia) and archived neonatal blood spots of patients were both approved by the standing Ethics Committee of the Institute of Child Health and Hospital for Sick Children (Great Ormond Street), and informed consent was obtained in accordance with the Declaration of Helsinki.
Identification of patient-specific mutation in NOTCH1
Genomic regions including the heterodimerization (HD-N and HD-C) and PEST domains of NOTCH1 were amplified by polymerase chain reaction (PCR) using leukemic DNA samples as previously described,11 purified from agarose gel and directly sequenced. Mutations were subcloned into plasmid vectors and confirmed by sequencing.
Detection of SIL-TAL1 fusion in T-ALL patients
Detection of NOTCH1 mutations in blood spots
PCR primers amplifying NOTCH1 mutations in the leukemic samples were designed from the sequence data and optimal PCR conditions were established for each specific primer pair using leukemic DNA samples.
Small pieces (∼1mm2 ) were cut out from each blood spot, mixed with PCR reagents containing Ampdirect Plus (Shimadzu, Kyoto, Japan). and NovaTaq (Novagen, Madison, WI) at 50 μL final volume and amplified.4,9
The NOTCH1 normal genomic sequences for HD-N (for patients MGT01 and MGT09) or PEST domain (for MGT11) were amplified as an internal control for blood spot DNA: HDN-f and HDN-r, 5′-GCTTGATGGGGTGCTTGCGCAG-3′ for amplification of HD-N region and PEST-f, 5′-ACCACCACAGCCGCACCTTG-3′ for PEST region.
Results and discussion
The presence of NOTCH1 mutations was first assessed in the eleven leukemic DNA samples. Heterodimerization (HD) and PEST domains were screened only because they are the domains in which most mutations are found.11,14,15 Four patients (MGT01, 09, 10, and 11) had mutations (confirmed by cloning and sequencing) in either HD (MGT01 and 09) or PEST domains (MGT10 and 11). Three involved insertions of unrelated short sequences and one had a duplication of a relatively long sequence from within NOTCH1 (Table 1). Only one patient sample (MGT01) of the eleven had a SIL-TAL1 fusion. This was a type 1 fusion6 (data not shown).
Details of NOTCH1 mutations in 4 cases of T-ALL
| Patients . | Age at diagnosis . | Mutations in NOTCH1 . | SIL-TAL1 . |
|---|---|---|---|
| MGT01 | 6 y 10 mon | HD; 4894ins (-TCTTACCGAGAAACGAAGACAAG-) | + (Type I) |
| MGT09 | 9 y 1 mon | HD; 4894ins (-TCTTTGTCGCCAAG-) | − |
| MGT10 | 4 y 7 mon | PEST; 7123ins (-GCCCTCCCTGCAGCATGGTAGGTGAGGCCCTCCC-)* | − |
| MGT11 | 5 y 6 mon | PEST; 7403ins (-AGACTGCACTGCA-) | − |
| Patients . | Age at diagnosis . | Mutations in NOTCH1 . | SIL-TAL1 . |
|---|---|---|---|
| MGT01 | 6 y 10 mon | HD; 4894ins (-TCTTACCGAGAAACGAAGACAAG-) | + (Type I) |
| MGT09 | 9 y 1 mon | HD; 4894ins (-TCTTTGTCGCCAAG-) | − |
| MGT10 | 4 y 7 mon | PEST; 7123ins (-GCCCTCCCTGCAGCATGGTAGGTGAGGCCCTCCC-)* | − |
| MGT11 | 5 y 6 mon | PEST; 7403ins (-AGACTGCACTGCA-) | − |
Sequences of patient-specific insertional mutation are underlined.
+ indicates present, −, not present.
dup(7097-7122).
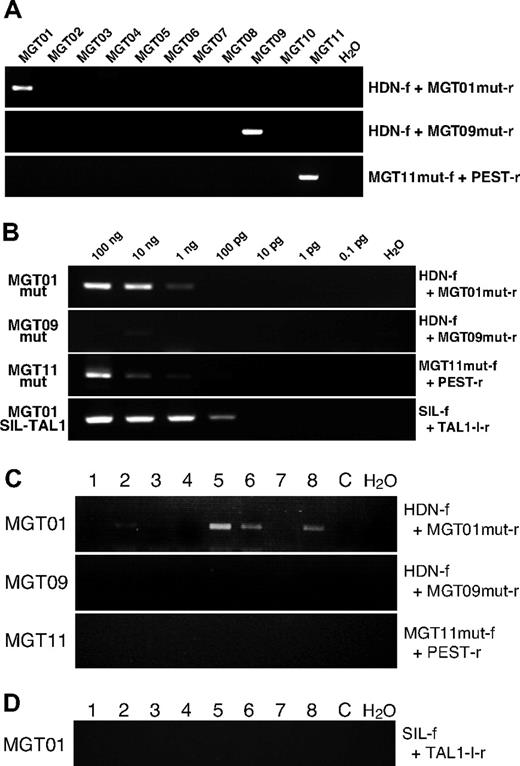
PCR primers were designed based on the sequences of patient-specific mutations and specificity and sensitivity were assayed with diagnostic DNA samples and cloning vectors with mutated sequences by serial dilutions. In one patient sample (MTG10), no primer set worked well, therefore samples from only 3 (MTG01, MTG09 and MTG11) with NOTCH1 mutations were further analyzed. With the patient-specific primer sets, positive PCR products were obtained only in corresponding patient DNA showing specificity of the primers (Figure 1A). Sensitivity of mutation-specific primers was assayed by serially diluting diagnostic DNA and cloned plasmid DNA. Variable and modest sensitivity was obtained. Down to 10−5 pg of mutated DNA could be detected with the primer set for MGT01 and 10−3 pg for MGT09 and MGT11 when plasmids with mutated NOTCH1 were used as a screen (data not shown). Using the diagnostic leukemic DNA as a screen for sensitivity, the threshold for detection was 1 ng for MGT01 (∼100 cells) and 10 ng for MGT11 (∼1000 cells). No specific mutation sequence could be detected in MGT09 up to 100 ng (Figure 1B), which may reflect the fact that the DNA was derived from a bone marrow sample that was substantially hemodiluted.
Detection of NOTCH1 mutation via PCR. (A) Specificity of PCR primers for patient-specific mutations was examined by PCR with diagnostic DNA samples of T-ALL patients (MGT01 to MGT11). Primers used for the PCR are shown with the panels. Two rounds of PCR were necessary to obtain a clear amplification band in patient MGT09, possibly because of a low number of leukemic cells. (B) Sensitivity assay of mutation-specific primers. Serially diluted diagnostic DNA was subjected to PCR amplification with Ampdirect Plus in 40 cycles of PCR reaction. F indicates forward and r, reverse PCR primers for mut, mutation. Sequences of the primers for specific mutation of each patient were as follows: MGT01mut-r, 5′-GTGCGTCACGCTTGGCGACTTTTG-3′ for patient MGT01; MGT09mut-r, 5′-CGTCACGCTTGGGCACAAAG-3′ for MGT09 and MGT11mut-f; 5′-GAGAGCCGAGCCAGGCACACT-3′ for MGT11. MGT01mut-r and MGT09mut-r were used in combination with HDN-f, and MGT11mut-f was used with PEST-r primers. When necessary, 2-round, seminested PCR was conducted with the primers; HDN-f2, 5′-ACTGCGACCAGGGCTGCAACAG-3′ (with MGT01mut-r or MGT09mut-r) and PEST-r2, 5′-GTTGTCCACAGGCGAGGAGTAG-3′ (with MGT11mut-f). (C) Results of representative PCR for patient-specific mutation are shown. Of 8 MGT01 blood spot pieces, 5 showed positive bands (of which 2 were very faint and became clear after seminested PCR amplification (data not shown). All slices from blood spots of MGT09 and MGT11 were negative for their respective NOTCH1 mutation. At least 16 pieces of blood spots, in total, were examined in each patient. C indicates control blood spot without NOTCH1 mutation. (D) Results of SIL-TAL1 amplification on the MGT01 blood spot. Sixteen pieces were examined in total and all were negative, as shown here for 8 slices, even after 2 rounds of PCR amplification.
Detection of NOTCH1 mutation via PCR. (A) Specificity of PCR primers for patient-specific mutations was examined by PCR with diagnostic DNA samples of T-ALL patients (MGT01 to MGT11). Primers used for the PCR are shown with the panels. Two rounds of PCR were necessary to obtain a clear amplification band in patient MGT09, possibly because of a low number of leukemic cells. (B) Sensitivity assay of mutation-specific primers. Serially diluted diagnostic DNA was subjected to PCR amplification with Ampdirect Plus in 40 cycles of PCR reaction. F indicates forward and r, reverse PCR primers for mut, mutation. Sequences of the primers for specific mutation of each patient were as follows: MGT01mut-r, 5′-GTGCGTCACGCTTGGCGACTTTTG-3′ for patient MGT01; MGT09mut-r, 5′-CGTCACGCTTGGGCACAAAG-3′ for MGT09 and MGT11mut-f; 5′-GAGAGCCGAGCCAGGCACACT-3′ for MGT11. MGT01mut-r and MGT09mut-r were used in combination with HDN-f, and MGT11mut-f was used with PEST-r primers. When necessary, 2-round, seminested PCR was conducted with the primers; HDN-f2, 5′-ACTGCGACCAGGGCTGCAACAG-3′ (with MGT01mut-r or MGT09mut-r) and PEST-r2, 5′-GTTGTCCACAGGCGAGGAGTAG-3′ (with MGT11mut-f). (C) Results of representative PCR for patient-specific mutation are shown. Of 8 MGT01 blood spot pieces, 5 showed positive bands (of which 2 were very faint and became clear after seminested PCR amplification (data not shown). All slices from blood spots of MGT09 and MGT11 were negative for their respective NOTCH1 mutation. At least 16 pieces of blood spots, in total, were examined in each patient. C indicates control blood spot without NOTCH1 mutation. (D) Results of SIL-TAL1 amplification on the MGT01 blood spot. Sixteen pieces were examined in total and all were negative, as shown here for 8 slices, even after 2 rounds of PCR amplification.
Excised blood spot segments were placed directly into PCR reactions with Ampdirect Plus to maximize the opportunity to amplify rare sequences4 and at least 16 segments of each spot analyzed. Normal NOTCH1 genomic sequences corresponding to the site of mutation on the region of HD-N (MGT01 and MGT09) and PEST (MGT11) were amplified as PCR controls and all segments (at least 8 segments in each patient) examined showed positive amplification (data not shown), indicating DNA on the Guthrie cards was intact.
Of the 3 patient samples studied, one, MGT01, showed unambiguous amplification of specific mutated sequence on several segments of blood spots (Figure 1C). In total, 20 pieces of the Guthrie card were subjected to PCR amplification and 11 were positive. Several of the positive bands were purified and sequenced and all had the patient-specific HD mutation (Table 1). A total of 20 pieces of control blood spots from the Guthrie cards of nonleukemic children were examined as additional controls with the MGT01 mutation-specific primer set. No amplification bands were observed. Patient MGT01 also had the SIL-TAL1 gene fusion. Sixteen fragments of blood spots for patient MGT01 were analyzed for the presence of the specific SIL-TAL1 fusion sequence but no PCR product was observed after 2 rounds of PCR amplification (70 cycles of PCR in total; Figure 1D).
Patient MGT01's blood spots, positive for NOTCH1 mutation, were the only ones for which we obtained sensitivity comparable with that achieved and required with leukemia fusion genes that register positive in blood spots – approximately 100 cells.2,4,7
Negative results in neonatal blood spot screening are uninterpretable but tend to underestimate the frequency of prenatally-initiated leukemias.2 This is particularly so when, as in the present study with NOTCH1 mutation, levels of sensitivity achievable with subtle sequence mutations, while maintaining specificity, are very modest compared with fusion genes.4,7
No general conclusion can be drawn about the usual timing of this common genetic abnormality in T-ALL, but these data indicate that NOTCH1 mutation can occur prenatally as an early or possibly initiating event in T-cell leukemogenesis. This interpretation accords with the view that NOTCH1 regulates self-renewal properties of stem cells or progenitors16,17 and can initiate T-cell leukemogenesis as a transgene in mice18 or zebra fish.19 In animal models of T-ALL, secondary NOTCH1 mutations have been detected in leukemias initiated by other genetic changes.20,21 As in this particular positive case, MGT01, the concurrent diagnostic SIL-TAL1 fusion sequence was absent from the neonatal blood spot (with a sensitivity threshold of ∼10 cells/mutation copies; Figure 1B), then it is likely that the latter was a secondary, postnatal event complementing the functional impact of prenatal NOTCH1 mutation.
There are only 2 other studies addressing the possible prenatal origins of T-ALL. The first was our previous report of a pair of monozygotic twins with T-NHL/ALL.22 Malignant cells from the pair shared an identical TCRβ rearrangement (including an 11 bp N region) indicative of a single cell clonal origin in utero. Cells from these twins did not have a NOTCH1 mutation (A.M. Ford and M.G., unpublished observation, March 2007). Fasching et al23 reported that 2 patients with T-ALL had detectable TCR gamma clonal rearrangements in their neonatal blood spots, identical to those in the leukemic cells at diagnosis. Collectively, these data argue that pediatric T-ALL can be initiated prenatally.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Ian Hann and Jane Chalker (Great Ormond Street Hospital) for access to clinical samples and Prof Kinuko Mitani for support (M.E. and M.E.-I.).
This work was supported by the Leukaemia Research Fund, United Kingdom.
Authorship
Contribution: M.E.-I. and M.E. carried out all experimental work. H.K. provided clinical samples and diagnostic details plus Guthrie cards. M.G. conceived and planned the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Present address for M.E.-I. and M.E.: Department of Hematology, Dokkyo University School of Medicine, Tochigi, Japan.
Correspondence: Prof Mel Greaves, Section of Haemato-Oncology, Institute of Cancer Research, Brookes Lawley Building, 15 Cotswold Road, Belmont, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: mel.greaves@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal