Abstract
Dendritic cells are key initiators and regulators of the immune response. Dendritic cell commitment and function require orchestrated regulation of transcription. Gata1 is a transcription factor expressed in several hematopoietic lineages. However, Gata1 function has not been explored in the monocytic or dendritic cell compartment. Here, we show that Gata1 is expressed in myeloid and plasmacytoid dendritic cells and that Gata1 ablation affects the survival of dendritic cells. Furthermore, lipopolysaccharide (LPS) stimulation of dendritic cells prompts Gata1 up-regulation, which is accompanied by increased levels of BclX and Ifng. Our findings show that Gata1 is a transcriptional regulator of dendritic cell differentiation and suggest that Gata1 is involved in the dendritic cell and macrophage lineage separation.
Introduction
Dendritic cells (DCs) are key initiators and regulators of the immune response. DCs were originally described by the exclusive capacity to stimulate naive T cells.1 Later studies showed that DCs are also involved in the stimulation of regulatory responses through the propagation of regulatory T cells.2 Two types of DCs have been defined in mice and humans based on differential surface marker expression, origin, or function in the immune response: myeloid DCs (mDCs) and plasmacytoid DCs (pDCs).3 Differential expression of toll-like receptors (TLRs) on mDCs and pDCs defines the recognition of distinct microbial agents (ie, mDCs predominantly express TLR4 recognizing endotoxin and pDCs express TLR3, TLR7, and TLR9).4
DCs are distributed in lymphoid (spleen, lymph nodes, thymus, bone marrow) and nonlymphoid tissues. DCs are renewed from local DC precursors or circulating monocytes, which primarily derive from bone marrow (BM) progenitors.5–8 mDCs and pDCs originate from either common lymphoid progenitors (CLPs) or common myeloid progenitors (CMPs), showing a redundant potential of the hematopoietic system to generate DCs.9 The potential of CLPs or CMPs to differentiate into DCs is terminated upon definitive B-cell or megakaryocyte/erythrocyte commitment, respectively.10 Myeloid-derived DCs share precursors with macrophages (Mfs) until late stages of differentiation.11 The mechanisms that determine DC versus Mf differentiation remain largely unclear, and transcription factors play a central role in their commitment. Reconstitution assays and genetically modified mice facilitated understanding the role of transcription factors such as Gfi1, Id2, Ikaros, Relb, Mafb, and PU.1 in DC versus Mf differentiation.12
Transcription factors Gata1, Gata2, and Gata3 are essential in the hematopoietic system. Gata3 is necessary for the specialization of naive T cells into T-helper 2 (Th2) lymphocytes.13 Gata2 is important for the maintenance of hematopoietic multipotent progenitor cells and remains expressed in mast cells and megakaryocytes.14,15 Gata1 is expressed at basal levels in hematopoietic progenitors16 and regulates the differentiation of the megakaryocyte/erythroid lineage.17 Gata1 is also required for mast-cell and eosinophil (Eo) development.18,19 In contrast, Gata1 is not required for Mf differentiation and, furthermore, forced expression of Gata1 in a myeloid progenitor line prevents normal Mf differentiation in vitro.20,21
Thus far, the expression and role of Gata1 in DC differentiation has not been reported. In this study, we show that Gata1 is expressed in DCs and that Gata1 is required for the survival of DC precursors in vitro and in vivo. In addition, Gata1 is required in both mDCs and pDCs but is irrelevant for the closely related Mfs. Our findings demonstrate that Gata1 is a transcriptional regulator of DC differentiation and suggest that Gata1 is involved in the final DC and Mf separation.
Materials and methods
Animals
Mice bearing a modified Gata1 allele flanked with loxP sites (Gata1-lox mice)22 were crossed with mice expressing a tamoxifen-inducible Cre recombinase under the ROSA 26 promoter (ER-Cre mice).23 As the Gata1 gene is X-linked, we used Gata1-lox ER-Cre males (knockout [KO]), Gata1-lox males (wild type [WT]), and ER-Cre males for in vivo and in vitro tamoxifen treatment in order to obtain pancellular Gata1 recombination. Tx (Tamoxifen Free Base; Sigma, St Louis, MO) was dissolved in 10% ethanol to 90% sunflower oil (Sigma) to a concentration of 5 mg/100 μL by sonication. In 3 independent experiments, 24 KO, 24 WT, and 16 Cre males were fed with a gavage 5 mg/day Tx during 5 consecutive days, followed by 2 days without treatment, and this sequence was repeated 4 times. Mice were killed and different tissues (ie, tail, liver, blood, BM, spleen, and lung) were collected for further analysis. Cre males were indistinguishable from WT males upon Tx treatment. Mice were injected intravenously with 5 μg lipopolysaccharide (LPS, Sigma) and killed 24 hours later for further analysis. Mice were specific pathogen-free and kept with free access to food and water, under the guidelines for animal experimentation approved by the Erasmus University Animal Welfare Committee.
Cell cultures
Standard medium RPMI-1640 plus glutamax-I, 50 μg/mL gentamycine, and 5 × 10−5 M β-mercaptoethanol (Gibco-BRL) was used for the following: interleukin-5 (IL-5) cultures (Eos), 10 × 106/mL BM cells supplemented with 30% fetal calf serum (FCS, Sigma) and 24 ng/mL IL-5 (Pharmingen, BD Biosciences, San Diego, CA); macrophage colony-stimulating factor (M-CSF) cultures (Mfs), 2 × 105/mL BM cells supplemented with 5% FCS and 50 ng/mL recombinant mouse M-CSF (rmM-CSF, R&D Systems, Abingdon, United Kingdom); granulocyte-macrophage CSF (GM-CSF) cultures (mDCs), 2 × 105/mL BM cells supplemented with 5% FCS and 20 ng/mL rmGM-CSF (a gift of Prof K. Thielemans, Vrije Universiteit Brussels, Belgium; when applicable, 100 ng/mL LPS [from Escherichia coli; Sigma] was added at day 8); Flt3-L cultures (mDCs + pDCs), BM progenitors were negatively selected using Dynabeads as indicated in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) and cultures of 106/mL progenitors were supplemented with 10% FCS and 200 ng/mL recombinant human Flt3-ligand (a gift of Amgen, Thousand Oaks, CA), and, when required, DCs generated with Flt3-L were sorted using Dynabeads as indicated in Document S1.
When applicable, cultured cells were treated with 500 nM 4-hydroxy-tamoxifen (OHT; Sigma). The collection time of cultured cells was based on differentiation status of each cell type, as defined by flow cytometry (ie, 10-12 days for Flt3-L cultures and 7-11 days for GM-CSF, IL-5, and M-CSF cultures).
Colony-forming unit–granulocyte, erythroid, monocyte, megakaryocyte (CFU-GEMM) assays were performed according to the manufacturer's instructions (MethoCult GF M3434; Stem Cell Technologies, Vancouver, BC). Murine erythroleukemia (MEL; clone C88) cells were cultured as described.24
ELISA
IL-6 and interferon-γ (IFN-γ) production was measured in the supernatant of cell cultures using enzyme-linked immunosorbent assays (ELISA) according to the manufacturer's protocol (BD Biosciences).
Phenotypic analysis and cell sorting by flow cytometry
Different cell types obtained ex vivo or in vitro were defined using differential expression of surface molecules for flow-cytometry analysis or cell sorting (Figure 1). The list of antibodies used is available in Document S1. For flow-cytometry analysis, aliquots of 2 × 106 cells were incubated with a cocktail of monoclonal antibodies (mAbs). For intracellular Gata1 staining, cells were permeabilized using the Perm/Wash Kit (BD Pharmingen). For sorting of CMPs, lineage-negative cells were first enriched using Dynabeads (Document S1) and subsequently labeled with a mix of mAbs (Figure 1E). Before sorting (FACS-Aria; BD Biosciences, Amsterdam, the Netherlands), cell suspensions were filtered through a 30-μm cell strainer. Purity was 95% to 98%, unless stated otherwise.
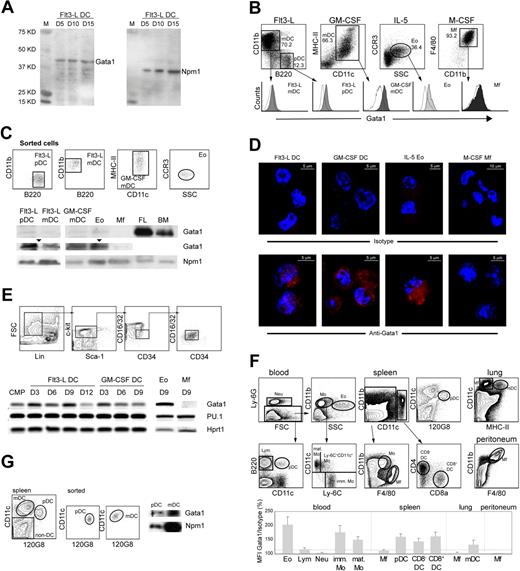
Gata1 is expressed in mouse DCs. (A) Western blot of nuclear extracts of bone marrow cells grown in the presence of Flt3-L, probed with Gata1 antibody. Cells were harvested at the culture days (D) indicated. For loading control, the same blot was reprobed with Nucleophosmin (Npm1) antibody. (B) Histograms of Gata1 expression in pDCs (CD11cmedCD11b−B220+) and mDCs (CD11c+ CD11b+ B220−) from Flt3-L cultures, mDCs from GM-CSF cultures (CD11chiMHCII+), Eos from IL-5 cultures (CCR3+), and Mfs from M-CSF cultures (F4/80hiCD11bhi). Empty histograms show the fluorescence of the isotype control; the signals of the isotype controls depend on cell size and granularity. Filled histograms show Gata1 staining. SSC indicates side scatter. (C) Sorted DCs from Flt3-L (pDCs and mDCs) and GM-CSF (mDCs) cultures express Gata1 as shown by Western-blot analysis of nuclear extracts. Npm1 was used as loading control. Gata1 expression in DCs is lower than in bone marrow (BM) and fetal liver (FL) but clearly visible on a longer exposure (arrowheads). Sorted Eos from IL-5 cultures served as a positive control; Mfs from M-CSF cultures did not express Gata1 at detectable levels. (D) Immunofluorescence staining of cells from Flt3-L, GM-CSF, IL-5, and M-CSF cultures. The top panel shows staining with Alexa595-conjugated antirat antibody (isotype control). The bottom panel shows staining with Gata1 antibody, followed by Alexa595-conjugated antirat antibody. Nuclei are stained with DAPI (blue). Gata1 is located in the cytoplasm and nucleus of DCs and Eos. Images collected using a LSM 510 confocal microscope (Zeiss, Oberkochen, Germany) equipped with 543-nm and 800-nm lasers. A 20×/0.5 NA was used. Pictures were taken in slides. The acquisition software used was AIM 3.2 SP2. Images were analyzed using Imaris software 4.2. (E top) Dot plots representing the sorting strategy to obtain purified CMPs from BM. (Bottom) RT-PCR analysis of Gata1 and PU.1 mRNA levels during Flt3-L–, GM-CSF–, and at the end of IL-5– and M-CSF–stimulated cultures of purified CMPs. D indicates day of culture. (F) Relative expression levels of Gata1 in vivo. Mean fluorescence intensity (MFI) of Gata1-stained samples for each cell type was divided by the MFI value of the isotype control, multiplied by 100. Eo indicates eosinophils; Lym, lymphocytes; Neu, neutrophils; imm Mo, immature monocytes; mat Mo, mature monocytes; and Mf, macrophages. Data represent average relative MFI (± SEM) calculated from 3 independent experiments. (G left) Dot plots showing the sorting strategy of pDCs and mDCs from the spleen. (Right) Sorted spleen pDCs and mDCs express Gata1 as shown by Western-blot analysis of nuclear extracts. Npm1 was used as loading control.
Gata1 is expressed in mouse DCs. (A) Western blot of nuclear extracts of bone marrow cells grown in the presence of Flt3-L, probed with Gata1 antibody. Cells were harvested at the culture days (D) indicated. For loading control, the same blot was reprobed with Nucleophosmin (Npm1) antibody. (B) Histograms of Gata1 expression in pDCs (CD11cmedCD11b−B220+) and mDCs (CD11c+ CD11b+ B220−) from Flt3-L cultures, mDCs from GM-CSF cultures (CD11chiMHCII+), Eos from IL-5 cultures (CCR3+), and Mfs from M-CSF cultures (F4/80hiCD11bhi). Empty histograms show the fluorescence of the isotype control; the signals of the isotype controls depend on cell size and granularity. Filled histograms show Gata1 staining. SSC indicates side scatter. (C) Sorted DCs from Flt3-L (pDCs and mDCs) and GM-CSF (mDCs) cultures express Gata1 as shown by Western-blot analysis of nuclear extracts. Npm1 was used as loading control. Gata1 expression in DCs is lower than in bone marrow (BM) and fetal liver (FL) but clearly visible on a longer exposure (arrowheads). Sorted Eos from IL-5 cultures served as a positive control; Mfs from M-CSF cultures did not express Gata1 at detectable levels. (D) Immunofluorescence staining of cells from Flt3-L, GM-CSF, IL-5, and M-CSF cultures. The top panel shows staining with Alexa595-conjugated antirat antibody (isotype control). The bottom panel shows staining with Gata1 antibody, followed by Alexa595-conjugated antirat antibody. Nuclei are stained with DAPI (blue). Gata1 is located in the cytoplasm and nucleus of DCs and Eos. Images collected using a LSM 510 confocal microscope (Zeiss, Oberkochen, Germany) equipped with 543-nm and 800-nm lasers. A 20×/0.5 NA was used. Pictures were taken in slides. The acquisition software used was AIM 3.2 SP2. Images were analyzed using Imaris software 4.2. (E top) Dot plots representing the sorting strategy to obtain purified CMPs from BM. (Bottom) RT-PCR analysis of Gata1 and PU.1 mRNA levels during Flt3-L–, GM-CSF–, and at the end of IL-5– and M-CSF–stimulated cultures of purified CMPs. D indicates day of culture. (F) Relative expression levels of Gata1 in vivo. Mean fluorescence intensity (MFI) of Gata1-stained samples for each cell type was divided by the MFI value of the isotype control, multiplied by 100. Eo indicates eosinophils; Lym, lymphocytes; Neu, neutrophils; imm Mo, immature monocytes; mat Mo, mature monocytes; and Mf, macrophages. Data represent average relative MFI (± SEM) calculated from 3 independent experiments. (G left) Dot plots showing the sorting strategy of pDCs and mDCs from the spleen. (Right) Sorted spleen pDCs and mDCs express Gata1 as shown by Western-blot analysis of nuclear extracts. Npm1 was used as loading control.
Immunofluorescence
Cultured cells were collected on poly-L-lysine slides (Sigma). Cells were fixed with 4% paraformaldehyde (PFA). Cells were permeabilized with sodium citrate, blocked 2 hours at 4°C with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), and incubated overnight at 4°C with rat anti-Gata1 (N6). Gata1 was visualized using antirat-Alexa596 (1 hour at 4°C). Slides were mounted with DAPI/Vectashield and images collected using a LSM 510 confocal microscope (Zeiss, Oberkochen, Germany) equipped with 543-nm and 800-nm lasers. Images were analyzed using Imaris software 4.2 (Bitplane, Zurich, Switzerland).
Western blot
BM cells were lysed in 2 × Laemmli buffer (whole cell extracts). Nuclear extracts of cultured and splenic DCs and Western-blotting analysis were performed as described.22
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed as described in the Upstate protocol,25 with 4 × 107 cells as starting material for each sample. Antibodies used were Gata1 (N6), PU.1, rat IgG, and rabbit IgG as isotype controls (Santa Cruz Biotechnology, Santa Cruz, CA). Precipitation of the bound chromatin fraction was done with Protein A/G Plus Agarose Beads (Santa Cruz Biotechnology). Input and unbound fractions were kept for quantitative polymerase chain reaction (qPCR) analysis. Primers used are included in Document S1. The annealing temperature used was 60°C. qPCR analysis was performed using SYBR Green I and an iCycler (BioRad, Hercules, CA) or Opticon I (MJ Research, Waltham, MA) system. Enrichment for DNA fragments in the immunoprecipitated fractions was calculated as described.26
PU.1 reporter assays
The PU.1 promoter (NM_011355.1, MGI:98282) expanding 365 bp upstream of the ATG was amplified by PCR from mouse DNA and cloned into pAD5-GFP lentiviral vector (a modified pRRLsin.PPT.CMV.GFP.Wpre27 ), substituting the CMV promoter. The GATA mutant site was generated as described in Document S1. All clones were verified by sequencing.
Recombinant lentiviruses were produced by transient transfection of 293T cells according to standard protocols.28 Supernatant containing recombinant lentiviruses was harvested at days 1, 2, and 3; pooled; and filtered through a 0.45-μm filter before ultracentrifugation (79 090g, 2 hours, 4°C). Ultracentrifuged virus was resuspended in 100 μL sterile PBS. DCs were transduced at day 4 of culture in 6-well plates (6 × 105 cells/well at the start of the culture) with 40 μL of virus suspension and centrifuged at 537.6g for 1 hour at room temperature. LPS was added at day 8 and cells collected for flow-cytometry analysis at day 11 of DC culture. MEL cells were transduced in 6-well plates (1.5 × 106 cells/well) with 10 μL of virus suspension.
Statistical analysis
Statistical analyses were done by a Mann-Whitney statistical test for independent samples, or 2-tailed t test, using the SPSS software package, version 10 (Chicago, IL). Results are presented as the mean (± SEM), unless otherwise indicated.
Results
Gata1 is expressed in mouse dendritic cells
The most commonly used protocols for the generation of DCs in vitro use Flt3-L or GM-CSF as stimulating factors. We found that Gata1 is expressed in Flt3-L–stimulated cultures (Figure 1A). We therefore set out to study Gata1 expression in DCs generated in vitro. Flt3-L stimulates the differentiation of pDCs and mDCs resembling steady-state lymphoid-organ DCs,5,29 whereas GM-CSF generates DCs that closely resemble monocyte-derived inflammatory mDCs.30 We generated Eos (IL-5) and Mfs (M-CSF) as positive and negative controls for Gata1 expression, respectively.18,20
We combined cell-specific labeling (Figure 1B) with intracellular Gata1 staining in flow cytometry. Gated Eos, mDCs, and pDCs (Flt3-L) and mDCs (GM-CSF) were positive for Gata1 expression, whereas Mfs were negative (Figure 1B). Western-blotting analysis of nuclear extracts of sorted cells confirmed Gata1 expression in all DC types and Eos but not in Mfs (Figure 1C). Next, we analyzed the cellular localization of Gata1 protein by performing immunofluorescence staining. Gata1 located in the cytoplasm and nucleus of Flt3-L–, GM-CSF–, and IL-5–cultured cells but was not detectable in M-CSF–cultured cells (Figure 1D). We conclude that Gata1 protein is present in mature mDCs and pDCs generated in vitro.
Gata1 is present throughout in vitro DC differentiation from purified CMPs
We studied Gata1 expression throughout Flt3-L, GM-CSF, IL-5, and M-CSF cultures derived from sorted CMPs (Figure 1E). We also analyzed the expression of the PU.1 transcription factor, previously described as a Gata1 antagonist31 and essential in the monocytic lineage.32
Gata1 was present throughout the course of the Flt3-L and GM-CSF cultures, and also in IL-5 (Eos) cultures, whereas in M-CSF (Mfs) cultures, Gata1 levels were undetectable by the end of the culture period. Expression of PU.1 was present throughout the 4 cultures derived from CMPs. The observation that Gata1 is present in DCs but not in Mfs whereas PU.1 is present in both lineages supports the notion that these factors have a role in cell differentiation/specification.33–35
In vivo–differentiated mouse DCs express Gata1
We performed intracellular flow cytometry staining in freshly isolated cells from spleen, blood, and nonlymphoid organs such as lung and peritoneum. Cells were defined using surface marker expression (Figure 1F). Gata1 signals in lymphocytes, neutrophils, and Mfs were not significantly higher than the thresholds of their isotype controls, implying that Gata1 is not expressed in these cells. In agreement with previous knowledge, Gata1 was present in (Eos). We observed that both immature and mature monocytes in the blood express Gata1. Interestingly, splenic pDCs and mDCs, either CD8− or CD8+, and lung mDCs also express Gata1 (Figure 1F). We next analyzed Gata1 expression by Western blotting in sorted mouse splenic mDCs and pDCs, demonstrating that Gata1 protein is present in both DC subsets in vivo, albeit at low levels (Figure 1G).
Deletion of Gata1 in vivo leads to reduction of the DC precursor pool
Gata1 null embryos die between days 10.5 and 11.5 of gestation,36 precluding studies on the consequences of Gata1 loss in adult mice. To circumvent this limitation, we generated Gata1-lox ER-Cre double-transgenic mice in which a virtually complete loss of Gata1 expression is obtained upon 4 weeks of tamoxifen (Tx) treatment (Figure 2A). Absence of Gata1 in these mice leads to reduction of total cell number in the BM and spleen, depletion of the erythroid compartment, and severe anemia (Saho Tsukamoto, L.G., Mikiko Suzuki, Harumi Yamamoto-Mukai, Kinuko Ohneda, S.P., and Masayuki Yamamoto, manuscript in preparation).
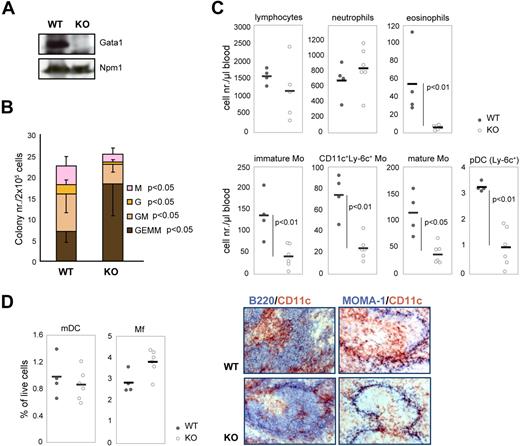
Recombination of Gata1 in vivo affects monocytic/DC lineage development. (A) Western blot of WT and KO BM whole-cell extracts after 4 weeks of Tx treatment. Npm1 was used as loading control. (B) CFU-GEMM assays performed on the BM of WT and KO mice after 4 weeks of Tx treatment. GEMM indicates granulocyte erythroid monocyte megakaryocyte; GM, granulocyte monocyte; G, granulocyte; and M, monocyte. Error bars indicate SD. (C) Dot plots depicting the numbers of different leukocytes (defined by flow cytometry; Figure 1F) in the blood after 4 weeks of Tx treatment. Circles represent cell numbers/μL blood in Tx-treated WT mice (n = 4, gray circles) and in Tx-treated KO mice (n = 6, white circles). Thick horizontal lines represent the average cell numbers/μL blood. Mo indicates monocytes; and pDCs, plasmacytoid dendritic cells. The P values are derived from Mann-Whitney statistical analysis of independent samples. (D; left) Dot plots depicting the frequency of mDCs and Mfs (defined by flow cytometry; Figure 1F,G) in the spleen after 4 weeks of Tx treatment. (Right) Immunohistochemistry of the spleen after 4 weeks of Tx treatment. Sections were stained with antibodies against CD11c (brown; monocytes and mDCs) and B220 (blue; B cells) or MOMA-1 (blue; Mfs). Images collected using a Leica DM-LB microscope (Leica, Wetzlar, Germany). Camera used was a Leica DC-500. A 10×/0.30 NA dry objective was used. Pictures were taken in slides. The acquisition software used was Imaging for Windows (Kodak), belonging to Windows 2000 SP4.
Recombination of Gata1 in vivo affects monocytic/DC lineage development. (A) Western blot of WT and KO BM whole-cell extracts after 4 weeks of Tx treatment. Npm1 was used as loading control. (B) CFU-GEMM assays performed on the BM of WT and KO mice after 4 weeks of Tx treatment. GEMM indicates granulocyte erythroid monocyte megakaryocyte; GM, granulocyte monocyte; G, granulocyte; and M, monocyte. Error bars indicate SD. (C) Dot plots depicting the numbers of different leukocytes (defined by flow cytometry; Figure 1F) in the blood after 4 weeks of Tx treatment. Circles represent cell numbers/μL blood in Tx-treated WT mice (n = 4, gray circles) and in Tx-treated KO mice (n = 6, white circles). Thick horizontal lines represent the average cell numbers/μL blood. Mo indicates monocytes; and pDCs, plasmacytoid dendritic cells. The P values are derived from Mann-Whitney statistical analysis of independent samples. (D; left) Dot plots depicting the frequency of mDCs and Mfs (defined by flow cytometry; Figure 1F,G) in the spleen after 4 weeks of Tx treatment. (Right) Immunohistochemistry of the spleen after 4 weeks of Tx treatment. Sections were stained with antibodies against CD11c (brown; monocytes and mDCs) and B220 (blue; B cells) or MOMA-1 (blue; Mfs). Images collected using a Leica DM-LB microscope (Leica, Wetzlar, Germany). Camera used was a Leica DC-500. A 10×/0.30 NA dry objective was used. Pictures were taken in slides. The acquisition software used was Imaging for Windows (Kodak), belonging to Windows 2000 SP4.
We treated Gata1-lox ER-Cre (KO) and Gata1-lox (WT) males with Tx and analyzed the colony-forming potential of bone marrow myeloid progenitors. Gata1 loss affected the contribution of the different committed progenitors to the BM progenitor pool (Figure 2B). Earlier progenitors (CFU-GEMMs) accumulated, whereas late committed progenitors (CFU-GMs, CFU-Gs, and CFU-Ms) were reduced in KO bone marrow compared with WT bone marrow.
We next analyzed the changes caused by Gata1 deletion in the circulating leukocyte compartment by flow cytometry. After 4 weeks of Tx treatment, the total number of lymphocytes in the blood was slightly reduced in KO mice and neutrophil numbers were slightly increased compared with WT mice (Figure 2C, see Figure 1F for phenotypic definition). In agreement with the literature,18 the number of Eos was severely reduced in KO mice. Importantly, all subtypes of blood monocytes and pDCs were significantly reduced in KO mice (Figure 2C).
Recombination of Gata1 affected the steady-state DC compartment in the spleen, albeit mildly. The number of mDCs was slightly reduced, whereas the number of Mfs increased (Figure 2D). Immunohistochemistry of the spleen also indicated that mDCs in the spleen are affected by Gata1 deletion. Costaining with either B220 (B cells) or MOMA-1 (Mfs) showed that only CD11c+ cells (monocytes and mDCs) were reduced in KO spleens compared with WT spleens (Figure 2D). Collectively, our results suggest that Gata1 loss affects the monocytic/DC lineage in vivo.
Deletion of Gata1 reveals its role in cell survival during DC differentiation in vitro and ex vivo
To investigate the role of Gata1 during DC differentiation, we generated DCs from the BM of Gata1-lox ER-Cre (KO) mice or control Gata1-lox (WT) mice in vitro and induced Gata1 recombination with OHT. Mfs were used as a control not sensitive to Gata1 deletion.20 At day 10, the cell yield from untreated KO and WT cultures was similar. PCR and recombination analysis showed that the recombination efficiency was 50% after 2 days of OHT treatment and almost complete 4 days after treatment (data not shown).
OHT treatment of BM cells starting at day 0 of culture did not affect the viable cell yield in WT compared with untreated control cultures. In contrast, the cell yield in OHT-treated KO DC and Mf cultures was severely reduced relative to untreated controls (Figure 3A), consistent with the notion that myeloid progenitors require functional Gata1 for differentiation.17
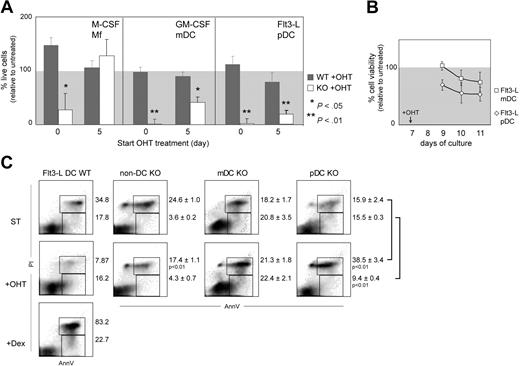
Recombination of Gata1 in vitro affects DC survival. (A) Cell culture efficiency upon tamoxifen (OHT) treatment. Average efficiency (± SEM) from at least 3 independent experiments is plotted. Efficiency is calculated as the ratio between the number of viable cells of treated and nontreated cultures multiplied by 100. The P values are derived from Mann-Whitney statistical analysis of independent samples. (B) mDCs and pDCs were sorted from Flt3-L cultures at day 7 and treated with OHT. Viability was determined by MTT test 2, 3, and 4 days after treatment initiation and calculated as the percentage of the viability of untreated controls. Values are average (± SEM) of 3 independent samples. (C) Analysis of apoptosis in pDCs, mDCs, and non-DCs sorted from the spleen (Figure 1G) and treated ex vivo with OHT for 48 hours. As a positive control for apoptosis, Flt3-L–derived DCs were exposed 24 hours to 1 μM dexamethasone (Flt3-L DCs + Dex). Dot plots are merged from 3 independent experiments; percentages correspond to the average (± SEM). The P values are derived from 2-tailed t tests.
Recombination of Gata1 in vitro affects DC survival. (A) Cell culture efficiency upon tamoxifen (OHT) treatment. Average efficiency (± SEM) from at least 3 independent experiments is plotted. Efficiency is calculated as the ratio between the number of viable cells of treated and nontreated cultures multiplied by 100. The P values are derived from Mann-Whitney statistical analysis of independent samples. (B) mDCs and pDCs were sorted from Flt3-L cultures at day 7 and treated with OHT. Viability was determined by MTT test 2, 3, and 4 days after treatment initiation and calculated as the percentage of the viability of untreated controls. Values are average (± SEM) of 3 independent samples. (C) Analysis of apoptosis in pDCs, mDCs, and non-DCs sorted from the spleen (Figure 1G) and treated ex vivo with OHT for 48 hours. As a positive control for apoptosis, Flt3-L–derived DCs were exposed 24 hours to 1 μM dexamethasone (Flt3-L DCs + Dex). Dot plots are merged from 3 independent experiments; percentages correspond to the average (± SEM). The P values are derived from 2-tailed t tests.
Lineage commitment of Mfs and DCs in vitro occurs around day 5 of culture, as judged by the expression of F4/80 (Mfs) in M-CSF and CD11c (DCs) in GM-CSF and Flt3-L cultures. To study the effects of Gata1 loss in committed DC and Mf cultures, we added OHT to the cultures at day 5. OHT-treated WT cultures displayed normal cell numbers and culture composition compared with untreated WT cultures (Figure 3A). Treatment of KO M-CSF cultures from day 5 onwards also did not affect the cell yield or culture composition. Because these Mfs had completely recombined Gata1, this confirms that committed Mf progenitors do not require Gata1.20 In contrast, the yield of live DCs in OHT-treated KO cultures was severely diminished. The majority of remaining cells in the cultures had not recombined Gata1 (data not shown). Due to Gata1 deletion, the number of live GM-CSF–stimulated mDCs was reduced by 50%. In Flt3-L–stimulated KO cultures, the number of live pDCs was reduced by 80% (Figure 3A) and the number of live mDCs was 23.3% ± 9.8% of the untreated sample. This suggests that both mDCs and pDCs depend on Gata1 irrespectively of the growth factor used for their generation.
Next, we sorted mDCs and pDCs from Flt3-L cultures, treated them with OHT, and measured cell viability for 3 consecutive days, starting after 2 days of OHT treatment to allow Cre recombination (Figure 3B). The viability of both pDCs and mDCs declined upon OHT treatment, indicating that Gata1 loss induces cell death in both DC types generated in vitro. Of note, the viability of pDCs deteriorated faster than that of mDCs.
To test the effect of Gata1 loss on DCs differentiated in vivo, we sorted spleen pDCs and mDCs from WT and KO mice (as defined in Figure 1G) and treated them ex vivo with OHT (Figure 3C). Treatment of WT cells did not affect cell survival compared with untreated control cultures. Two days after OHT treatment, the percentage of early apoptotic (AnnV+PI−) and late apoptotic cells (AnnV+PI+) in a non-DC population (B and T cells) was not increased. In contrast, the frequency of late apoptotic cells (AnnV+PI+) in OHT-treated pDCs was significantly higher compared with untreated cells. For mDCs, only a modest increase in early and late apoptotic cells was observed. Taken together, these data suggest that Gata1 ablation affects pDCs more severely than mDCs in vitro and in vivo and that Gata1 loss in DCs reduces cell survival.
LPS induces Gata1 expression in DCs in vitro and in vivo
The TLR4 agonist LPS induces mDC maturation into highly potent antigen-presenting cells37 and changes in the gene-expression profile.38 We analyzed by quantitative reverse transcriptase–PCR (qRT-PCR) transcriptional changes upon LPS stimulation in GM-CSF–derived mDCs. Effective LPS-induced mDC maturation was confirmed by up-regulation of major histocompatibility complex II (MHC II) and costimulatory molecules (CD80, CD86, and CD40). We analyzed Nfkb1 as a positive control for transcriptional activation by LPS induction in mDCs.39 After 12 hours of LPS stimulation, Nfkb1 was up-regulated approximately 4-fold, indicating an effective response to LPS. After 60 hours, Nfkb1 transcription was reduced compared with the levels observed in the controls (Figure 4A). After 12 hours of LPS stimulation, Gata1 levels declined to 56% of the levels found in untreated mDCs. Interestingly, 60 hours after LPS addition, Gata1 expression was increased approximately 6-fold (Figure 4A).
LPS induces Gata1 expression in DCs in vitro and in vivo. (A) qRT-PCR analysis of gene expression in GM-CSF cultures 12 hours and 60 hours upon LPS stimulation. Data are derived from 4 to 8 independent samples obtained in 2 experiments and analyzed in triplicate. (B) qRT-PCR analysis of gene expression in CD8− mDCs and pDCs sorted from the spleen of LPS-treated (+ LPS) and nontreated (+ PBS) mice. Hprt1, Gapdh, and ubiquitin were used as controls. (C top) qRT-PCR analysis of gene expression in GM-CSF cultures 60 hours upon LPS stimulation and relative to untreated samples. (Middle) qRT-PCR analysis of gene expression in GM-CSF cultures treated with OHT for 36 hours, relative to untreated samples. (Bottom) qRT-PCR analysis of gene expression in LPS-stimulated GM-CSF cultures treated with OHT for 36 hours and relative to LPS-stimulated control samples. Data are derived from 4 to 8 independent samples obtained in 2 experiments and analyzed in triplicate. The average (± SD) of the enrichment (fold increase) relative to nonstimulated samples (set to 1) is depicted. Gapdh and Hprt1 were used as controls. RFE indicates relative fold enrichment. The P values are derived from Mann-Whitney statistical analysis of independent samples. ND indicates not detectable.
LPS induces Gata1 expression in DCs in vitro and in vivo. (A) qRT-PCR analysis of gene expression in GM-CSF cultures 12 hours and 60 hours upon LPS stimulation. Data are derived from 4 to 8 independent samples obtained in 2 experiments and analyzed in triplicate. (B) qRT-PCR analysis of gene expression in CD8− mDCs and pDCs sorted from the spleen of LPS-treated (+ LPS) and nontreated (+ PBS) mice. Hprt1, Gapdh, and ubiquitin were used as controls. (C top) qRT-PCR analysis of gene expression in GM-CSF cultures 60 hours upon LPS stimulation and relative to untreated samples. (Middle) qRT-PCR analysis of gene expression in GM-CSF cultures treated with OHT for 36 hours, relative to untreated samples. (Bottom) qRT-PCR analysis of gene expression in LPS-stimulated GM-CSF cultures treated with OHT for 36 hours and relative to LPS-stimulated control samples. Data are derived from 4 to 8 independent samples obtained in 2 experiments and analyzed in triplicate. The average (± SD) of the enrichment (fold increase) relative to nonstimulated samples (set to 1) is depicted. Gapdh and Hprt1 were used as controls. RFE indicates relative fold enrichment. The P values are derived from Mann-Whitney statistical analysis of independent samples. ND indicates not detectable.
Next, we sorted CD8− mDCs (responsive to LPS through TLR4) and pDCs (nonresponsive to LPS) from the spleen of LPS-treated and nontreated control mice (as defined in Figure 1F) and determined whether LPS treatment in vivo induces similar changes in mDCs. Nfkb1 expression levels were up-regulated in LPS-treated CD8− mDCs, showing that this DC subset responded to LPS stimulation (Figure 4B). As expected, pDCs did not respond to LPS, and neither Nfkb1 nor Gata1 expression levels were changed. In concordance with the in vitro data, we could detect up-regulation of Gata1 in LPS-stimulated mDCs (Figure 4B). These data are consistent with a role for Gata1 in the final maturation/activation of DCs in vivo.
BclX, Ifng, and PU.1 are regulated by Gata1 in mDCs
Based on these observations, we analyzed the expression of a selection of genes in GM-CSF KO and WT cultures upon LPS treatment. We included members of the Bcl family that are required for cell survival and are regulated by Gata1 in erythroid cells.21,40 Furthermore, the balance between Bcl family members is crucial to set the lifespan of DCs after TLR activation.41 To follow changes in apoptotic signals, we measured the expression of caspase 3 (Casp3) and acinus (Acin1).42 In addition, we investigated the expression of interferon gamma (Ifng), a potential Gata1 target gene,43 and the expression of the Gata1 antagonistic transcription factor PU.1.
Coinciding with Gata1 up-regulation 60 hours after LPS stimulation, Ifng, Bcl2, and BclX were also up-regulated compared with control mDCs. In addition, expression of Acin1 and Casp3 was reduced 60 hours after LPS stimulation (Figure 4C). Taken together, Gata1 up-regulation upon LPS treatment correlates with an apparent antiapoptotic response in DCs. The effect of Gata1 deletion on the expression of selected genes was analyzed in KO mDCs 36 hours after OHT treatment initiation, because induced Gata1 ablation led to increasing cell death at later time points. WT mDCs treated with OHT showed no significant changes in the expression of the genes analyzed (data not shown). After 36 hours of OHT treatment, Gata1 expression was reduced to 40% of the level in untreated mDCs, and this reduction was accompanied by a 50% down-regulation of BclX expression (Figure 4C). However, we failed to detect significant changes in Bcl2, Casp3, or Acin1 expression, implying that the latter genes are not directly regulated by Gata1 (Figure 4C). This notion is further supported by the analysis of cells upon LPS stimulation combined with Gata1 deletion. Gata1 levels in LPS-stimulated KO mDCs treated with OHT were reduced to approximately 20% compared with LPS-treated control mDCs. Interestingly, deletion of Gata1 in LPS-treated mDCs correlated with a significant reduction of BclX and Ifng expression compared with LPS-stimulated controls (Figure 4C). These data strongly suggest that BclX and Ifng genes are activated by Gata1 in mDCs.
Expression of PU.1 in WT mDCs was reduced to 0.5% of the nonstimulated value, which coincided with the up-regulation of Gata1 60 hours after LPS stimulation. Furthermore, this down-regulation was much less pronounced in the absence of Gata1 (Figure 4C). These data support the notion of cross-regulation between the 2 factors.34,35
LPS stimulation shifts the balance of PU.1 and Gata1 binding to the PU.1 promoter
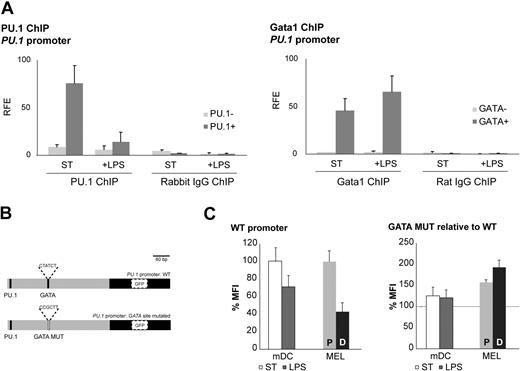
In view of the proposed cross-regulation between Gata1 and PU.134,35 and our finding of coexpression of Gata1 and PU.1 in DC cultures (Figure 1E), we performed Gata1 and PU.1 ChIP assays on the PU.1 promoter. PU.1 is known to transactivate its own promoter,44 and we found that PU.1 binds to the PU.1 promoter in nonstimulated mDCs (Figure 5A). Interestingly, PU.1 binding was decreased in LPS-stimulated cells, correlating with the reduced PU.1 mRNA levels in these cells (Figure 4C). Gata1 ChIP assays showed that Gata1 binds to the PU.1 promoter under both conditions (Figure 5A). Collectively, these data show that the balance between PU.1 and Gata1 binding to the PU.1 promoter shifts in favor of Gata1 upon LPS treatment, suggesting a mechanism for suppression of the PU.1 promoter by Gata1 after LPS treatment of mDCs.
Gata1 regulates PU.1 in DCs. (A) Gata1 and PU.1 ChIP assays were performed with cells from GM-CSF cultures stimulated for 3 days with LPS (+ LPS) or nonstimulated (ST) starting from day 8 of culture. PU.1 ChIP analysis on the PU.1 promoter: amplicon containing PU.1 binding site (PU.1+); amplicon not containing a PU.1 binding site (PU.1−). Gata1 ChIP analysis of the PU.1 promoter: amplicon containing GATA binding site (GATA +); amplicon not containing a GATA binding site (GATA−). Average (± SD) of at least 2 independent experiments analyzed each in triplicate is shown. Corresponding isotype ChIPs (rat IgG or rabbit IgG) are depicted. RFE indicates relative fold enrichment. (B) PU.1 promoter reporter constructs. GATA MUT indicates mutant GATA binding site. (C) GFP expression of the PU.1 promoter reporter constructs in mDCs cultured under standard conditions (ST) and after LPS treatment (LPS) and in proliferating MEL cells (P) and DMSO-induced MEL cells (D). The mean fluorescence intensity (MFI) ± SD obtained from 3 independent experiments is depicted. GFP expression levels with the wild-type promoter are set at 100.
Gata1 regulates PU.1 in DCs. (A) Gata1 and PU.1 ChIP assays were performed with cells from GM-CSF cultures stimulated for 3 days with LPS (+ LPS) or nonstimulated (ST) starting from day 8 of culture. PU.1 ChIP analysis on the PU.1 promoter: amplicon containing PU.1 binding site (PU.1+); amplicon not containing a PU.1 binding site (PU.1−). Gata1 ChIP analysis of the PU.1 promoter: amplicon containing GATA binding site (GATA +); amplicon not containing a GATA binding site (GATA−). Average (± SD) of at least 2 independent experiments analyzed each in triplicate is shown. Corresponding isotype ChIPs (rat IgG or rabbit IgG) are depicted. RFE indicates relative fold enrichment. (B) PU.1 promoter reporter constructs. GATA MUT indicates mutant GATA binding site. (C) GFP expression of the PU.1 promoter reporter constructs in mDCs cultured under standard conditions (ST) and after LPS treatment (LPS) and in proliferating MEL cells (P) and DMSO-induced MEL cells (D). The mean fluorescence intensity (MFI) ± SD obtained from 3 independent experiments is depicted. GFP expression levels with the wild-type promoter are set at 100.
Next, we performed reporter assays with the PU.1 promoter in mDCs. We cloned the −365-bp PU.1 promoter in a lentiviral GFP reporter vector and prepared a version with a mutant GATA binding site (Figure 5B). We transduced mDCs and analyzed GFP expression by flow cytometry in standard conditions and upon LPS stimulation. We used MEL cells as a control cell line. Upon DMSO-induced differentiation of MEL cells, PU.1 and Gata1 mRNA levels display kinetics similar to those observed in LPS-stimulated mDCs (ie, down-regulation of PU.1 and up-regulation of Gata1). GFP expression driven by the wild-type PU.1 promoter was down-regulated in mDCs upon LPS treatment and in MEL cells upon DMSO induction (Figure 5C). Mutation of the GATA binding site in the PU.1 promoter resulted in enhanced expression of GFP in mDCs and MEL cells and the reduction of GFP expression upon LPS treatment of mDCs, and DMSO induction of MEL cells was no longer observed (Figure 5C). Collectively, these data are consistent with the notion that Gata1 represses the PU.1 promoter upon LPS stimulation of mDCs.
Ifng is a direct target of Gata1 in mDCs
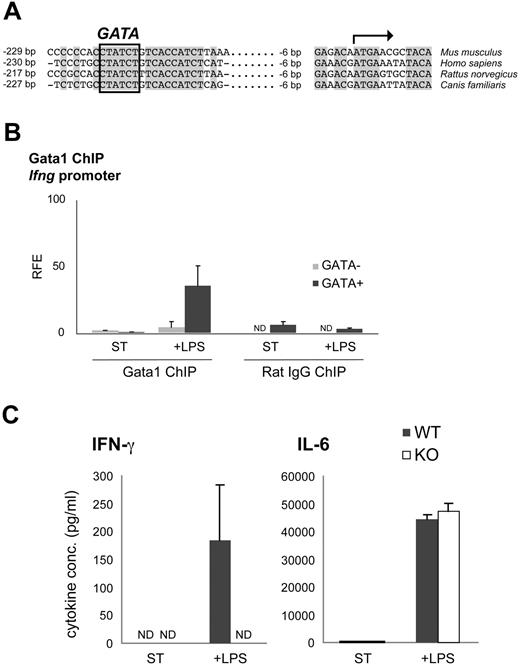
It has been suggested that Ifng could be a potential Gata1 target gene,43 and multiple sequence alignment of the Ifng promoter of human, mouse, rat, and dog reveals a fully conserved GATA site (Figure 6A). To investigate whether Gata1 directly controls the transcriptional activity of Ifng, we performed Gata1 ChIP assays. Gata1 was not detectable at the Ifng promoter in nonstimulated mDCs (Figure 6B). Interestingly, Gata1 associated with the Ifng promoter upon LPS stimulation (Figure 6B), positively correlating with the observed increase in Ifng mRNA levels (Figure 4C). This suggests that Gata1 directly activates Ifng expression in mDCs upon LPS stimulation. To test this, GM-CSF–derived mDCs from Tx-treated KO and WT mice were stimulated for 60 hours with LPS, and cytokine production was analyzed by ELISA. In contrast to WT cultures, KO cultures failed to produce detectable IFN-γ. Production of IL-6 was not affected, demonstrating the specificity of IFN-γ activation by Gata1 (Figure 6C). Collectively, these data strongly suggest that Gata1 directly activates Ifng expression in mDCs upon LPS stimulation.
Gata1 regulates Ifng in LPS-stimulated mDCs. (A) Multiple sequence alignment of the proximal Ifng promoter in human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus) and dog (Canis familiaris). The potential GATA binding site is highlighted. (B) Gata1 ChIP analysis of the Ifng promoter in mDCs derived from GM-CSF cultures stimulated for 3 days with LPS (+ LPS) or grown under standard conditions (ST) starting from day 8 of culture. Amplicon containing GATA binding site (GATA+); amplicon not containing a GATA binding site (GATA−). Average (± SD) of at least 2 independent experiments performed in triplicate is shown. Corresponding isotype ChIPs (rat IgG) are depicted. RFE indicates relative fold enrichment. (C) ELISA on KO and WT GM-CSF culture supernatants stimulated with LPS for 60 hours or grown under standard conditions (ST). Expression of IFN-γ and IL-6 (average ± SEM of 3 independent samples) is shown. ND indicates not detectable.
Gata1 regulates Ifng in LPS-stimulated mDCs. (A) Multiple sequence alignment of the proximal Ifng promoter in human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus) and dog (Canis familiaris). The potential GATA binding site is highlighted. (B) Gata1 ChIP analysis of the Ifng promoter in mDCs derived from GM-CSF cultures stimulated for 3 days with LPS (+ LPS) or grown under standard conditions (ST) starting from day 8 of culture. Amplicon containing GATA binding site (GATA+); amplicon not containing a GATA binding site (GATA−). Average (± SD) of at least 2 independent experiments performed in triplicate is shown. Corresponding isotype ChIPs (rat IgG) are depicted. RFE indicates relative fold enrichment. (C) ELISA on KO and WT GM-CSF culture supernatants stimulated with LPS for 60 hours or grown under standard conditions (ST). Expression of IFN-γ and IL-6 (average ± SEM of 3 independent samples) is shown. ND indicates not detectable.
Discussion
Gata1 has multiple roles in hematopoiesis. Mutations in the amino-terminal zinc finger of Gata1 lead to anemia and macrothrombocytopenia in humans.45 In mice, a targeted deletion in the Gata1 promoter leads to selective loss of the eosinophil lineage.18 Disruption of a hypersensitive site in the Gata1 promoter leads to deregulated megakaryocyte proliferation and impaired production of platelets.46 Erythroid cells that lack Gata1 undergo apoptosis, and Gata1-null mice die at midgestation.20,36 However, Gata1 is thought to be dispensable for the monocyte/macrophage lineage. Ex vivo depletion of Gata1 in common megakaryocyte/erythroid progenitors led to their differentiation switch toward Mfs.20 In addition, overexpression of Gata1 in a murine myeloid cell line prevented their normal differentiation into Mfs and converted them into megakaryocytes.21 Similarly, forced expression of Gata1 in highly purified hematopoietic stem cells induced their commitment exclusively to the megakaryocyte/erythroid lineage.47 A recent study shows that Gata1 expression decreases upon the commitment of granulocyte-monocyte progenitors (GMPs) correlating with increased expression of PU.1, in agreement with the described cross-regulation between the 2 transcription factors.33 However, ectopic activation of Flt3 signaling promotes DC differentiation and Gata1 expression in GMPs while PU.1 is still active,33 implying that Gata1 and PU.1 might not exclude each other in DCs. This prompted us to analyze Gata1 expression and function in DCs and their precursors.
Here, we show that Gata1 is expressed in mouse steady-state mDCs and pDCs in vivo and in vitro, as well as in human DCs (data not shown). We also found expression of Gata1 in circulating inflammatory monocytes, which can be precursors for DCs under inflammatory conditions.6–8 DCs can develop from myeloid and lymphoid progenitors,9 and the presence of Gata1 in all DC types tested implies that Gata1 expression is a DC-specific feature unrelated to their developmental origin. Ablation of Gata1 in vitro in DCs affects their survival in both Flt3-L and GM-CSF cultures. We confirmed these findings by transducing Gata1-lox cells with a lentivirus expressing Cre recombinase (T.B.v.D., unpublished material; data not shown). In contrast to DCs, the closely related Mfs do not express or require Gata1 for their survival. The specific expression and requirement of Gata1 in DCs could be used to resolve the mechanisms of lineage commitment between DCs and macrophages.
We showed that, in addition to the ablation of the erythroid compartment (Saho Tsukamoto, L.G., Mikiko Suzuki, Harumi Yamamoto-Mukai, Kinuko Ohneda, S.P., and Masayuki Yamamoto, manuscript in preparation) and the expected deterioration of the Eo compartment,18 Gata1 deletion in vivo leads to a significant reduction of circulating monocytes and pDCs and a trend in the reduction of splenic mDCs. When we treated sorted splenic DCs with OHT, we found that Gata1 ablation increases the percentage of apoptotic cells in ex vivo cultures, suggesting that splenic steady-state DCs are also sensitive to Gata1 loss.
Gata1 and PU.1 act antagonistically in the hematopoietic compartment.31–33,48 The balance between PU.1 and Gata1 is proposed as a master switch in hematopoietic commitment. However, although Gata1 suppression might be required for commitment to certain lineages, this does not imply that cells are incapable to reactivate the Gata1 transcriptional program when needed. We find coexpression of PU.1 and Gata1 in DCs throughout differentiation. After LPS stimulation, Gata1 levels go up, PU.1 mRNA levels are rapidly down-regulated, and the positive autoregulatory feedback loop of PU.1 is impaired. In addition, we find that Gata1 binds to the PU.1 promoter and this binding is maintained upon LPS stimulation. Thus, the cross-regulation between Gata1 and PU.1 may have important implications in the fine-tuning of DC maturation (Figure 7). Further studies are required to delineate in detail how these transcription factors interact functionally in DCs.
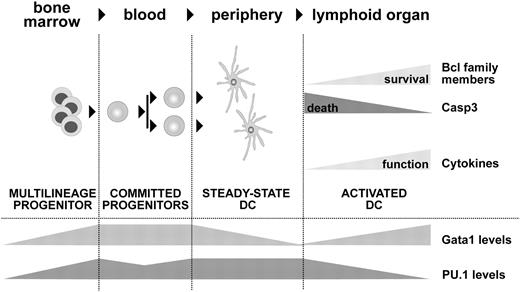
Model of Gata1 function in DCs. Gata1 and PU.1 are coexpressed during DC development. We propose that Gata1 counteracts apoptotic signals at 2 critical developmental stages affecting survival of DC precursors and of TLR-stimulated DCs. Upon TLR4 stimulation, Gata1 levels increase and PU.1 is down-regulated, resulting in an antiapoptotic response as measured by Bcl2 and BclX up-regulation versus Casp3 down-regulation. In addition, this triggers the expression of responsive cytokines such as IFN-γ, thus ensuring optimal activation of the adaptive immune response.
Model of Gata1 function in DCs. Gata1 and PU.1 are coexpressed during DC development. We propose that Gata1 counteracts apoptotic signals at 2 critical developmental stages affecting survival of DC precursors and of TLR-stimulated DCs. Upon TLR4 stimulation, Gata1 levels increase and PU.1 is down-regulated, resulting in an antiapoptotic response as measured by Bcl2 and BclX up-regulation versus Casp3 down-regulation. In addition, this triggers the expression of responsive cytokines such as IFN-γ, thus ensuring optimal activation of the adaptive immune response.
Gata1 has been linked to differentiation, survival, and cell-cycle regulation in different cell types.18,20,21,49–51 Gata1 activates the expression of the Bcl2 and BclX genes in the erythroid lineage.21,40 Bcl2 regulates the lifespan of DCs41 and BclX is required for DC survival in vivo.52 We show that Gata1 up-regulation induced by LPS correlates with Bcl2 and BclX up-regulation. Upon Gata1 depletion with OHT, BclX is down-regulated. This points to BclX as a potential Gata1 target in DCs. We failed to detect direct binding of Gata1 to the BclX promoter by ChIP assays (data not shown), supporting the notion that Gata1 might regulate BclX indirectly.53 Interestingly, we found that LPS-induced up-regulation of Gata1 was accompanied by activation of Ifng expression, revealing this cytokine as a potential novel target for Gata1 in DCs. The Ifng promoter in zebrafish contains potential GATA binding sites,43 suggesting that regulation of interferon expression by Gata1 is conserved through evolution. Sequence alignment of the proximal promoter of Ifng revealed a GATA site fully conserved between mouse and human, and we show here that Gata1 occupies the promoter of Ifng in LPS-stimulated mDCs. This suggests that Gata1 acts as an activator of Ifng expression upon LPS stimulation, in line with published data.54,55 Because production of IFN-γ appears to act in an autocrine fashion in mature DCs,56,57 this suggests a crucial role for Gata1 in the final maturation of DCs (Figure 7). Recent studies showing that GATA binding sites are present in the promoter region of DC-script, a putative DC-specific transcription factor,58 and that ectopic expression of GATA1 in human DCs reduces CCR5 surface expression,59 also support an important role for Gata1 in the DC transcriptional program.
Appropriate reaction of the immune system depends directly on DC function. DCs require an optimal lifespan to pick up antigens, process them, and migrate from the peripheral tissues to the lymph nodes, where they present antigens to mount efficient activation of the adaptive immune response.1,12 Understanding the molecular control of DC development is important for future medical applications of DCs and has gained much attention recently. In this study, we show that Gata1 is expressed in DCs and that it is required for their differentiation, survival, and maturation (Figure 7). Furthermore, the observation that Ifng is a direct Gata1 target gene in DCs encourages further studies to investigate the role of Gata1 in DC function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The Netherlands Organization for Scientific Research (NWO) provided grants for L.G. (ALW 815.02.008), T.N. (Veni 916.66.067), T.B.v.D. (MW 901.08.092), H.H. (Veni 916.56.025), S.P. (MW 901.08.092), and B.N.L. (Vidi 917.46.323). The Dutch Cancer Foundation provided grants for F.G. and S.P. (KWF EUR 2000–2273).
We would like to thank Dr E. Anguita, Dr P. Rodriguez, and Dr J. Strouboulis for advice on the ChIP assays; E. Haak for guidance with the CFU-GEMM assays; Dr H. Braun and P. Papadopoulos for technical support; P. Molenbeek and D. Zondervan for assistance with mouse tamoxifen treatment; and Prof Dr E.A. Dzierzak and Dr K. Ottersbach for reviewing the manuscript.
National Institutes of Health
Authorship
Contribution: L.G. and T.N. designed and performed research, collected and analyzed data, and wrote the paper. T.B.v.D., H.H., N.V., and M.W. performed research. F.G. wrote the paper. S.P. and B.N.L. designed research and wrote the paper.
L.G. and T.N. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sjaak Philipsen, Erasmus MC, Department of Cell Biology, Dr. Molewaterplein 50, 3015GE Rotterdam, The Netherlands; e-mail: j.philipsen@erasmusmc.nl; or Bart N. Lambrecht, Erasmus MC, Department of Pulmonary Medicine, Dr. Molewaterplein 50, 3015GE Rotterdam, The Netherlands; e-mail: b.lambrecht@erasmusmc.nl.