Abstract
Efficient in vivo selection increases survival of gene-corrected hematopoietic stem cells (HSCs) and protects hematopoiesis, even if initial gene transfer efficiency is low. Moreover, selection of a limited number of transduced HSCs lowers the number of cell clones at risk of gene activation by insertional mutagenesis. However, a limited clonal repertoire greatly increases the proliferation stress of each individual clone. Therefore, understanding the impact of in vivo selection on proliferation and lineage differentiation of stem-cell clones is essential for its clinical use. We established minimal cell and drug dosage requirements for selection of P140K mutant O6-methylguanine-DNA-methyltransferase (MGMT P140K)–expressing HSCs and monitored their differentiation potential and clonality under long-term selective stress. Up to 17 administrations of O6-benzylguanine (O6-BG) and 1,3-bis(2-chloroethyl)-1-nitroso-urea (BCNU) did not impair long-term differentiation and proliferation of MGMT P140K–expressing stem-cell clones in mice that underwent serial transplantation and did not lead to clonal exhaustion. Interestingly, not all gene-modified hematopoietic repopulating cell clones were efficiently selectable. Our studies demonstrate that the normal function of murine hematopoietic stem and progenitor cells is not compromised by reduced-intensity long-term in vivo selection, thus underscoring the potential value of MGMT P140K selection for clinical gene therapy.
Introduction
In the last decade, gene therapy has established itself as a promising approach for the treatment of hereditary diseases of the blood-forming system, especially for such patients who cannot be treated by conventional transplantation strategies. Several successful clinical gene therapy studies have demonstrated the curative potential of therapeutic gene delivery by retroviral vectors, most prominently in X-linked severe combined immunodeficiency (SCID-X1) trials,1,2 in adenosine deaminase–deficient (ADA) SCID,3,4 and in X-linked chronic granulomatous disease (X-CGD).5
The occurrence of 3 cases of lymphoproliferative disease in the French SCID-X1 gene therapy trial and subsequent diligent insertional profiling in different models have demonstrated that genotoxicity by insertional mutagenesis plays a greater role in gene-modified progenitor and stem cells than previously anticipated. Insertional activation of the LMO2 oncogene has been demonstrated as a root cause in all 3 cases of malignant lymphoproliferation in the French SCID-X1 trial.6,7 Integration site distribution analysis in leukocyte samples of X-CGD gene therapy patients further demonstrated that insertional activation of genes can also affect the myeloid cell lineage, where vector integrations in MDS1-EVI1, PRDM16, or SETBP1 resulted in a self-limiting expansion of gene-corrected cells.5
Effective selection systems could be used to reduce the risk of insertional mutagenesis, because they make it possible to transplant a lower total number of repopulating cells, and, if selection occurs in vivo, reduce their number further with each selection cycle. Selection of a minority of cell clones reduces the number of insertion sites in engrafted cell clones at risk for oncogene activation. Moreover, efficient in vivo selection of gene-corrected cells would broaden the spectrum of diseases that can be treated by gene therapy to include the majority of congenital diseases in which growth or survival of the cells is not affected to such a great extent by the therapeutic transgene. Under these conditions the genetically corrected cells would not be expected to gain a selective advantage.
Numerous methods of selecting for genetically modified hematopoiesis using the mechanism of chemoresistance gene overexpression have been developed. The multidrug resistance gene (MDR-1),8–12 the dehydrofolate-reductase gene (DHFR),13–16 and the O6-methylguanine-DNA-methyltransferase gene (MGMT)17–19 have been tested extensively in this context. It has been shown that the use of all 3 systems results in an enrichment of gene-corrected blood cells in mice. Nevertheless, in most cases, the enrichment was transient, suggesting that hematopoietic progenitor cells with a restricted proliferative potential were primarily selected. A selection at the stem cell level could unambiguously be demonstrated only with the O6-methylguanine DNA methyltransferase (MGMT) system. MGMT mediates resistance to alkylating agents such as N,N′-bis(2-chloroethyl)-N-nitrosourea (BCNU) and temozolomide by removing methyl and ethyl groups from the O6-position of guanines. MGMT is expressed in different cell types including hematopoietic stem cells but can be inactivated irreversibly by O6-benzylguanine (O6-BG) and other inhibitors. Gene transfer of a mutant form of MGMT (MGMT P140K) that is resistant to O6-BG enables the selection of gene-modified cells by administering BCNU while concomitantly inhibiting wild-type MGMT by applying O6-BG.20 It has been shown previously that MGMT-mediated in vivo selection of hematopoietic stem cells can be achieved in mice and in dogs.19,21–26 Unfortunately, initial studies revealed that MGMT selection using BCNU and O6-BG close to the maximum tolerated dose (MTD) levels involves considerable toxicity, leading to the death of up to 50% of experimental mice and therefore demonstrating the need for dose de-escalation. Moreover, up to the present it had not been determined whether selection and the associated proliferative stress impair the biology of MGMT P140K–expressing stem-cell clones.
Here, we examined the impact of repetitive reduced-intensity BCNU/O6-BG selections on the proliferation and differentiation of long-term hematopoiesis in vivo and monitored the clonality. In order to exaggerate potential effects, BCNU/O6-BG selections were administered up to 17 times for selection in mice that underwent serial transplantations, without detectable evidence of clonal exhaustion or other detrimental effects on the differentiation or longevity of individual stem-cell clones expressing MGMT P140K.
Materials and methods
Mice
C57Bl/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME), and raised and housed at a specific pathogen-free animal facility according to all applicable laws and regulations following approval by the animal care and ethics committee of the University Hospital Freiburg, Germany.
Retroviral gene transfer of MGMT 140PK into murine bone marrow cells
The murine stem-cell virus (MSCV) retrovirus vector coexpressing MGMT P140K and enhanced green fluorescent protein (eGFP) linked by an internal ribosome entry site (IRES) has been described previously.19 Retroviral supernatant was collected from stably transfected E-86 cells in accordance with standard procedures.27 Bone marrow was harvested from femurs and hip bones of male C57Bl/6J mice 48 hours after treatment with 150 μg/g 5-fluorouracil (Sigma, Deisenhofen, Germany). The cells were then transduced as previously described. Briefly, bone marrow (BM) cells were prestimulated for 48 hours at a density of 106 cells/mL in Dulbecco modified Eagle medium (DMEM; Gibco, Paisley, United Kingdom) containing 15% fetal calf serum (FCS; PAN-Biotech, Aidenbach, Germany) supplemented with 100 ng/mL rmFlt-3L, 100 ng/mL rmSCF, and 50 ng/mL rhTPO (all StemCell Technologies, Vancouver, BC). Cells were then harvested, resuspended in retroviral vector–containing medium supplemented with the same growth factors, and transferred into Petri dishes that had been coated with 3 μg/cm2 fibronectin (FN; Sigma) and preloaded with vector-containing medium. After 24 hours cells were harvested, resuspended in fresh vector medium, and transferred into fresh FN-coated and preloaded dishes for another 24 hours.
Transplantation and blood analysis
Bone marrow cells were harvested from the transduction cultures, washed in Hanks balanced salt solution (Sigma) containing 2% FCS (HF), resuspended in 200 μL HF, and injected via tail vein into lethally irradiated (950 cGy) female C57Bl/6J mice (105 cells or 4 × 105 cells per mouse). For serial transplantations, bone marrow from donor femur, tibia, and hip bones was harvested, washed in HF, resuspended in PBS, and transplanted as described above. Four weeks after transplantation and after each administration of the selection drugs (BCNU and O6-BG), eGFP expression in the different peripheral blood cell lineages was analyzed by fluorescence activated cell sorting (FACS Calibur; Becton Dickinson, Heidelberg, Germany). B cells (B220+), monocytes (CD11+), granulocytes (Ly6-G+), erythroid cells (Ter119+), and T cells (CD3+) were stained with fluorochrome-conjugated monoclonal antibodies (Becton Dickinson, Palo Alto, CA). Gates were set to exclude greater than 99.99% of propidium iodide–negative (PI−) cells incubated with irrelevant isotype-matched control antibodies (anti-CD16/32 labeled with the corresponding fluorochromes). Frequencies of repopulating cells were calculated using Poisson statistics and the method of maximum likelihood using L-calc software (StemCell Technologies). Myeloid (CD11+, Ly6+) and lymphoid (B220+, CD3+) cells from peripheral blood cells were separated by fluorescence activated high speed cell sorting (MoFlow; DakoCytomation, Hamburg, Germany).
Drug selection
BCNU (Carmubris; Bristol-Myers Squibb, Munich, Germany) was dissolved in ethanol as recommended and diluted in PBS. O6-BG (Sigma) was dissolved in polyethylene glycol and diluted with prewarmed PBS. Two days after evaluation of the different peripheral blood cell lineages by FACS, either 10 μg/g body weight O6-BG followed by 2.5 μg/g BCNU one hour later or 20 μg/g body weight O6-BG and 5 μg/g BCNU were injected intraperitoneally. Drugs were administered every month throughout the whole experiment. Primary, secondary, and tertiary recipient mice received 7, 6, and 4 rounds of selection, respectively.
In vitro progenitor assays
Murine bone marrow cells were collected by bone marrow aspiration and colony-forming cell (CFC) assays were performed using MethoCult GF M3434 (StemCell Technologies). Single GFP+ colonies were harvested, washed with PBS (Gibco), and DNA isolated using QIAamp MicroKit (Qiagen, Hilden, Germany).
5′-Long terminal repeat integration site analysis
Genomic proviral junction sequences were detected by linear amplification—mediated–polymerase chain reaction (LAM-PCR) as previously described.28 In brief, 5 ng DNA from peripheral blood leukocytes or 1 ng DNA from sorted lymphocytes or granulocytes was used as a template for 2 rounds of a 50-cycle linear PCR with a biotinylated vector-specific primer followed by immobilization on streptavidin-coupled paramagnetic beads and separation using a magnet. This step was followed by hexanucleotide random priming, restriction digestion with TspI, ligation of the linker, and exponential PCR. Two percent of the first PCR product served as a template after an additional magnetic capture purification step for a second (semi-) nested PCR that enabled the visualization of the LAM-PCR amplicons. The LAM-PCR amplicons were separated on a high-resolution Spreadex gel (Elchrom Scientific, Cham, Switzerland). For sequencing (GATC, Konstanz, Germany), the LAM-PCR amplicons were purified (PCR purification kit; Qiagen) and shotgun cloned into the TOPO TA vector (Invitrogen, Carlsbad, CA). Sequences were mapped to the murine genome using the UCSC Blast-like alignment tool (BLAT) genome browser.
Tracking of individual clones
DNA (0.5-2 ng) isolated from peripheral blood samples was preamplified using the Illustra GenomiPhi DNA Amplification Kit (GE Healthcare, Buckinghamshire, United Kingdom) according to instructions of the manufacturer. For subsequent amplification of the unique genomic-proviral junctions, PCR primers were designed. Tracking PCR was performed on 1 to 50 ng preamplified DNA using the unique genomic primers in combination with a vector-specific primer. Specificity of the PCR products was assured by sequencing. Single bands were isolated, purified, and shotgun cloned into a TOPO TA vector (Invitrogen). Sequences were mapped to the murine genome using the UCSC BLAT genome browser.
Statistical analyses
The results are shown as mean values (± SEM) from independent experiments. Differences between groups were assessed using the Student t test. Marked hematopoiesis was defined as selectable if a more than 2-fold increase was observed after repetitive administrations of BCNU/O6-BG. Based on the number of mice with selectable and mice without selectable hematopoiesis within each cell dose group (105 and 4 × 105 cells), the frequency of selectable repopulating cells was calculated by Poisson statistics using the L-calc software (StemCell Technologies).
Results
BCNU/O6-BG selection with reduced intensity and toxicity allows efficient selection of MGMT P140K–expressing hematopoiesis
Murine bone marrow cells were transduced with a retroviral vector encoding the MGMT P140K mutant and eGFP. To reduce the number of insertional events in stem cells, the amount of vector was adjusted to ensure a percentage of transduced cells ranging from 7% to 12% (multiplicities of infection [MOI] 0.4), a transduction rate likely to yield one integration/cell on average. The number of transduced bone marrow cells that reconstituted hematopoiesis long term and could be selected by BCNU/O6-BG administration was established by transplanting 2 cell doses (105 or 4 × 105) into lethally irradiated syngeneic mice. Analysis of the peripheral blood in all animals 4 weeks after transplantation revealed GFP+ white blood cells (WBCs) in 69 of 72 mice (5.3% ± 7.3% in 3 independent experiments). For selection, 2 subgroups were formed in both groups that received injections with different doses of O6-BG and BCNU at monthly intervals (10 μg/g O6-BG followed by 2.5 μg/g BCNU and 20 μg/g O6-BG followed by 5 μg/g BCNU) (Figure 1). Both doses were lower than the previously described LD50 dose of 30 μg/g O6-BG and 10 μg/g BCNU to minimize toxicity. After a total of 7 monthly BCNU/O6-BG administrations 3 of 48 mice had died. One mouse of the control group that had received a transplant with the lower number of cells but not treated with chemotherapy died 15 weeks after transplantation because of leukopenia. Two mice, one receiving the higher cell dose, the other receiving the lower cell dose, died 9 and 29 weeks after transplantation, respectively, in each case 2 to 3 days after selection with the higher dose of BCNU/O6BG. Only 0.2% and 0.04% of the peripheral blood cells of these 2 mice showed GFP expression. A more than 2-fold average increase in gene marking was observed exclusively in those cohorts of mice that received a transplant with 4 × 105 cells and treated with 20 μg/g O6-BG and 5 μg/g BCNU. The average number of GFP-expressing cells increased from 4.7% (± 0.8%) to 36.4% (± 9.8%) GFP+ WBCs after 7 BCNU/O6-BG injections (n = 11, P< .01, Figure 2A and Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). After transplantation of the lower number of bone marrow cells (105), on average 11.6% WBCs expressed GFP for more than 6 months without any systematic increase in response to the BCNU/O6-BG treatment. In all cohorts that were treated with the lower BCNU/O6-BG dose, the average percentage of GFP+ WBCs remained stable for up to 8 months after transplantation. Efficient selection of gene-marked hematopoiesis could be achieved only in mice with more than 3% GFP+ WBCs before the first selection. Whereas GFP+ hematopoiesis in none of 13 mice with a lower than 3% initial proportion of GFP+ peripheral blood cells was selectable, transduced hematopoiesis in 9 of 14 mice with an initial proportion of more than 3% GFP+ blood cells was efficiently selected. Using Poisson statistics, the frequency of selectable hematopoietic units, as defined by a more than 2-fold increase in their relative contribution to total hematopoiesis in response to BCNU/O6-BG, was calculated to be 1 in 5 × 105 transplanted bone marrow cells. According to this calculation, mice in the cohort that received a transplant of the higher cell dose (4 × 105 cells) received on average 0.8 selectable repopulating units per animal. The total number of marked hematopoietic units transplanted was calculated to be 9-fold higher (1 in 5.5 × 104).
Experimental design. Murine bone marrow cells were transduced with retroviral vector coexpressing MGMT P140K and eGFP and transplanted into syngeneic lethally irradiated primary recipient mice. Primary recipients received either 105 or 4 × 105 transduced BM cells. Cohorts of both groups were treated monthly with different dosages of BCNU and O6BG as indicated.
Experimental design. Murine bone marrow cells were transduced with retroviral vector coexpressing MGMT P140K and eGFP and transplanted into syngeneic lethally irradiated primary recipient mice. Primary recipients received either 105 or 4 × 105 transduced BM cells. Cohorts of both groups were treated monthly with different dosages of BCNU and O6BG as indicated.
Efficient selection of MGMT P140K–expressing long-term hematopoiesis by BCNU/O6BG. The percentage of peripheral blood cells that expressed GFP was determined by FACS. Mice were treated monthly with BCNU/O6BG. Control mice underwent transplantation but did not receive the subsequent chemotherapy. (A) Efficient selection of MGMT P140K–expressing hematopoiesis in primary transplant recipient mice was observed only in those cohorts of mice that received 4 × 105 cells (squares) and monthly administrations of 20 μg/g O6BG and 5 μg/g BCNU (black symbols) (*P < .05). Triangles indicate 105 cells transplanted; squares, 4 × 105 cells transplanted; white, no O6BG/BCNU treatment; gray, 10 μg/g O6BG/2.5 μg/g BCNU; and black, 20 μg/g O6BG/5 μg/g BCNU; (n = 12 for each group). The higher dose of BCNU and O6BG allowed selection of marked hematopoiesis even after (B) secondary (n = 10, P < .05) and (C) tertiary (n = 11, P < .05) transplantation.
Efficient selection of MGMT P140K–expressing long-term hematopoiesis by BCNU/O6BG. The percentage of peripheral blood cells that expressed GFP was determined by FACS. Mice were treated monthly with BCNU/O6BG. Control mice underwent transplantation but did not receive the subsequent chemotherapy. (A) Efficient selection of MGMT P140K–expressing hematopoiesis in primary transplant recipient mice was observed only in those cohorts of mice that received 4 × 105 cells (squares) and monthly administrations of 20 μg/g O6BG and 5 μg/g BCNU (black symbols) (*P < .05). Triangles indicate 105 cells transplanted; squares, 4 × 105 cells transplanted; white, no O6BG/BCNU treatment; gray, 10 μg/g O6BG/2.5 μg/g BCNU; and black, 20 μg/g O6BG/5 μg/g BCNU; (n = 12 for each group). The higher dose of BCNU and O6BG allowed selection of marked hematopoiesis even after (B) secondary (n = 10, P < .05) and (C) tertiary (n = 11, P < .05) transplantation.
Unperturbed myeloid differentiation potential of selected long-term hematopoiesis
To test whether long-term selection and the associated proliferative stress impair long-term differentiation and proliferation of MGMT P140K–expressing stem-cell clones, we analyzed the expression of lineage markers in the GFP+ compartment of WBCs in BCNU/O6-BG–treated and untreated mice. The GFP+ cell population contributed to erythroid, monocytic, granulocytic, T-lymphoid, or B-lymphoid lineages regardless of whether they were treated (Figure 3A). The proportion of T cells increased in the selected mice compared with unselected mice (P < .01). A corresponding 2.8-fold decrease in the TER119+ (P < .01) and a 4.9-fold decrease in the MAC1+ (P < .01) transduced cell population in selected animals after 7 rounds of drug treatment indicate that the increase in T lymphocytes occurred at the expense of erythroid and monocytic cells. To exaggerate potential toxic effects, bone marrow from treated and untreated mice was serially transplanted into secondary and tertiary lethally irradiated syngeneic mice (2 recipient mice per donor mouse). Either no or 20 μg/g O6-BG and 5 μg/g BCNU were administered at monthly intervals (6 times in secondary mice and 4 times in tertiary mice). As in the primary recipient mice, BCNU/O6-BG treatment allowed an efficient selection of the marked hematopoiesis (Figure 2B,C and Tables 1 and 2). The mean percentage of marked WBCs increased in secondary transplant recipient mice from 29.9% ± 7.2% to 65.1% ± 8.7% (n = 10, P < .05) (Figure 2B) and in tertiary transplant recipient mice from 18.9% ± 4.4% to 42.5% ± 9.6% (n = 11, P < .05) (Figure 2C). In vivo selection did not affect the in vivo differentiation of MGMT P140K–expressing long-term hematopoiesis in secondary recipient mice, indicating that the reduced erythroid and monocytic differentiation observed in the primary transplant recipient mice was restricted to primary engrafted mice only. Even after up to 17 BCNU/O6-BG administrations, no change in the ability to generate blood cells of different lineages was observed (Figure 3B,C). In tertiary transplant recipients, a small increase of CD3+ marked blood cells but no other lineage could be detected (P = .044) after 4 rounds of BCNU/O6BG treatment compared with the controls.
Multilineage differentiation of BCNU/O6BG-selected MGMT P140K–expressing long-term hematopoiesis. FACS analysis of peripheral blood cells from unselected control mice and mice that received a transplant of 4 × 105 cells before (4 weeks after transplantation) and after repetitive monthly injections of 5 μg/g BCNU and 20 μg/g O6BG (8, 12, 16, 20, 24, 28, and 32 weeks after transplantation, indicated by successive bars) was performed 23 to 25 days after each cycle of BCNU/O6BG treatment. The GFP+ hematopoiesis contributed to erythroid (TER119), monocytic (MAC1), granulocytic (GR1), B-lymphoid (B220), and T-lymphoid (CD3) lineages in unselected control animals as well as in BCNU/O6BG-treated mice even after serial transplantation. In primary recipient mice (A), the marked hematopoiesis produced fewer erythroid cells and monocytic cells accompanied by a relative increase in the proportion of the transduced T lymphocytes (*P < .01). In secondary recipients (B), no difference in the lineage contribution of the transduced hematopoiesis was observed with or without BCNU/O6BG treatment (n = 10). An increase in the proportion of marked T-lymphoid cells was detected again after tertiary transplantation in BCNU/O6BG-treated recipients (n = 11, *P < .05), whereas all other cell lineages remained unaffected (C). Standard error of the mean (± SEM) is shown for 11 to 12 individual mice per bar.
Multilineage differentiation of BCNU/O6BG-selected MGMT P140K–expressing long-term hematopoiesis. FACS analysis of peripheral blood cells from unselected control mice and mice that received a transplant of 4 × 105 cells before (4 weeks after transplantation) and after repetitive monthly injections of 5 μg/g BCNU and 20 μg/g O6BG (8, 12, 16, 20, 24, 28, and 32 weeks after transplantation, indicated by successive bars) was performed 23 to 25 days after each cycle of BCNU/O6BG treatment. The GFP+ hematopoiesis contributed to erythroid (TER119), monocytic (MAC1), granulocytic (GR1), B-lymphoid (B220), and T-lymphoid (CD3) lineages in unselected control animals as well as in BCNU/O6BG-treated mice even after serial transplantation. In primary recipient mice (A), the marked hematopoiesis produced fewer erythroid cells and monocytic cells accompanied by a relative increase in the proportion of the transduced T lymphocytes (*P < .01). In secondary recipients (B), no difference in the lineage contribution of the transduced hematopoiesis was observed with or without BCNU/O6BG treatment (n = 10). An increase in the proportion of marked T-lymphoid cells was detected again after tertiary transplantation in BCNU/O6BG-treated recipients (n = 11, *P < .05), whereas all other cell lineages remained unaffected (C). Standard error of the mean (± SEM) is shown for 11 to 12 individual mice per bar.
Mean white blood cell counts of animals that underwent secondary transplantations
| . | Weeks after BMT . | ||||||
|---|---|---|---|---|---|---|---|
| 4 . | 8 . | 12 . | 16 . | 20 . | 24 . | 28 . | |
| WBC count, × 106 | 3.02 | 5.28 | 5.11 | 3.39 | n.d. | 3.39 | 4.46 |
| SEM | 0.3 | 0.4 | 0.5 | 0.5 | — | 0.5 | 0.5 |
| WBC count, × 106 | 3.56 | 5.06 | 4.38 | 4.00 | n.d. | 4.24 | 4.55 |
| SEM | 0.4 | 0.7 | 0.8 | 0.4 | — | 0.5 | 0.5 |
| . | Weeks after BMT . | ||||||
|---|---|---|---|---|---|---|---|
| 4 . | 8 . | 12 . | 16 . | 20 . | 24 . | 28 . | |
| WBC count, × 106 | 3.02 | 5.28 | 5.11 | 3.39 | n.d. | 3.39 | 4.46 |
| SEM | 0.3 | 0.4 | 0.5 | 0.5 | — | 0.5 | 0.5 |
| WBC count, × 106 | 3.56 | 5.06 | 4.38 | 4.00 | n.d. | 4.24 | 4.55 |
| SEM | 0.4 | 0.7 | 0.8 | 0.4 | — | 0.5 | 0.5 |
WBC counts and plus or minus SEM in the peripheral blood of mice at different time points after transplantation of 4 × 105 cells are shown. The mice were either treated with 20 μg/g O6BG and 5 μg/g BCNU (top 2 rows) or received no further treatment (bottom 2 rows). WBC counts equal × 109/L.
— indicates not available; n.d., not done.
Mean white blood cell counts of animals that underwent tertiary transplantations
| . | Weeks after BMT . | ||||
|---|---|---|---|---|---|
| 4 . | 8 . | 12 . | 16 . | 20 . | |
| WBC count, × 106 | 2.14 | 3.04 | 3.08 | 3.41 | 2.71 |
| SEM | 0.2 | 0.4 | 0.5 | 0.4 | 0.3 |
| WBC count, × 106 | 3.35 | 4.08 | 4.36 | 4.84 | 3.30 |
| SEM | 0.4 | 0.5 | 0.5 | 0.3 | 0.4 |
| . | Weeks after BMT . | ||||
|---|---|---|---|---|---|
| 4 . | 8 . | 12 . | 16 . | 20 . | |
| WBC count, × 106 | 2.14 | 3.04 | 3.08 | 3.41 | 2.71 |
| SEM | 0.2 | 0.4 | 0.5 | 0.4 | 0.3 |
| WBC count, × 106 | 3.35 | 4.08 | 4.36 | 4.84 | 3.30 |
| SEM | 0.4 | 0.5 | 0.5 | 0.3 | 0.4 |
WBC counts and plus or minus SEM in the peripheral blood of mice at different time points after transplantation of 4 × 105 cells are shown. The mice were either treated with 20 μg/g O6BG and 5 μg/g BCNU (top 2 rows) or received no further treatment (bottom 2 rows). WBC counts equal × 109/L.
Stable clonality under BCNU/O6-BG selection
Depletion of stem-cell clones with low-level expression of the MGMT P140K transgene or clonal exhaustion due to the constant proliferative stress caused by long-term selection could lead to pauperization of the clonality of long-term hematopoiesis. We used LAM-PCR and sequence analysis to enumerate marked hematopoietic clones in the peripheral blood of primary, secondary, and tertiary recipient mice with and without selection. At all time points after transplantation, multiple (5 to > 20) insertion sites could be visualized (Figure 4). Neither the number of marked clones nor their longevity differed in selected and unselected mice. Most of the clones that were detected in primary recipient mice could also be visualized in secondary and tertiary recipients (Figures 5, 6). Even after tertiary transplantation and repetitive BCNU/O6-BG administration, no evidence for stem-cell exhaustion was observed. Sequencing of shotgun-cloned genome to vector fusion sequences revealed 17 unique integration sites. Using the UCSC BLAT genome browser, 14 of these integrations could be mapped to known genes (Table 3). One integration was located within the Hox B cluster 4317 bp downstream of Hox B5 and 8302 bp upstream of the Hox B4 gene whose overexpression has been shown to promote hematopoietic stem-cell self-renewal.29–31 One insertion was located 11 812 bp upstream of the Tpt1 gene that encodes for a highly conserved protein known to be involved in cell growth, cell-cycle progression, malignant transformation, and protection of cells against various stress conditions and apoptosis.32 Additional integrations occurred 3012 bp upstream of Gif (gastric intrinsic factor), a well-known vitamin B12 binding protein and 843 bp upstream of Arid5b (AT-rich interactive domain 5B), which plays a role in the regulation of development and/or tissue-specific gene expression.33 Another integration occurred 9566 bp upstream of Ncor2 (nuclear receptor corepressor 2) and 88 473 bp downstream of Scarb1 (scavenger receptor class B, member 1). The Scarb1 locus encodes for a multiligand HDL receptor,34 the Ncor2 locus for a silencing mediator of retinoid and thyroid hormone receptor SMRT that belongs to the Notch signaling pathway.35,36 The Ncor2/Scarb1 locus is a common integration site involved in the development of brain tumors and B-cell malignancy after retroviral insertion in mice.37 One insertion was located in intron 3 of the transmembrane protein encoding gene 6330512M04Rik and another insertion in intron 2 of Creg2. Creg2 encodes for a secreted glycoprotein that enhances the differentiation of pluripotent stem cells.38 One insertion was mapped in exon 3 of the Fmnl3 gene (formin-like 3) that encodes for a protein involved in the Rho-signaling pathway.39 Additional sequences were located 9567 bp upstream of BC043301, encoding for a nucleid acid and metal ion binding protein, and 25 085 bp downstream of 2610301B20Rik. We found 2 integrations in the vicinity of solute carrier family members, 547 bp upstream of Slc25a1, encoding for a mitochondrial deoxynucleotide carrier,40 and 1376811 bp upstream of Slc10a2, which is involved in bile salt uptake in distal ileum.41 One integration occurred 1173 bp in intron 1 of the gene Centd2 whose product shows Rho-GAP– and PIP3-dependent ARF-GAP activity.42 Another integration was located 1266555 bp upstream of Cntnap5a, expressing for a protein involved in cell adhesion (Table 3).
The number of clones contributing to marked hematopoiesis is not affected by BCNU/O6BG treatment. Representative LAM-PCR analysis of peripheral blood of mice at different time points after transplantation of 4 × 105 cells. Peripheral blood samples were analyzed before (4 weeks after transplantation) and with (A) or without (B) repetitive monthly BCNU/O6BG treatments (8, 12, 16, 20, 24, 28, and 32 weeks after transplantation). Multiple insertion sites were detected at all time points irrespective of whether the mice were treated with BCNU/O6BG. DNA (5 ng) was extracted from peripheral blood samples at each time point. M indicates a 100-bp DNA ladder; -c, negative control (5 ng DNA extracted from untransduced C57BL/6J bone marrow cells); and IC, internal control band.
The number of clones contributing to marked hematopoiesis is not affected by BCNU/O6BG treatment. Representative LAM-PCR analysis of peripheral blood of mice at different time points after transplantation of 4 × 105 cells. Peripheral blood samples were analyzed before (4 weeks after transplantation) and with (A) or without (B) repetitive monthly BCNU/O6BG treatments (8, 12, 16, 20, 24, 28, and 32 weeks after transplantation). Multiple insertion sites were detected at all time points irrespective of whether the mice were treated with BCNU/O6BG. DNA (5 ng) was extracted from peripheral blood samples at each time point. M indicates a 100-bp DNA ladder; -c, negative control (5 ng DNA extracted from untransduced C57BL/6J bone marrow cells); and IC, internal control band.
Clonality of serially transplanted long-term hematopoiesis. Representative LAM-PCR analysis of peripheral blood. Multiple clones contributed to the long-term hematopoiesis of the primary (A), secondary (B), and tertiary recipient mouse (C), and the number of these clones was not reduced by BCNU/O6BG administration. M indicates 100-bp DNA ladder; -c, negative control (5 ng DNA extracted from untransduced C57BL/6J bone marrow cells); and IC, internal control band.
Clonality of serially transplanted long-term hematopoiesis. Representative LAM-PCR analysis of peripheral blood. Multiple clones contributed to the long-term hematopoiesis of the primary (A), secondary (B), and tertiary recipient mouse (C), and the number of these clones was not reduced by BCNU/O6BG administration. M indicates 100-bp DNA ladder; -c, negative control (5 ng DNA extracted from untransduced C57BL/6J bone marrow cells); and IC, internal control band.
Tracking analysis of individual clones. The activity of individual clones over time in mice that underwent primary (1°), secondary (pair 2°a, 2°b), and tertiary (pair 3°a, 3°b) transplantation is shown. All mice that underwent tertiary transplantation received bone marrow from the corresponding secondary mouse 2°b not 2°a. Clone numbers refer to listed clones in Table 1. Dark squares indicate time points where the clone was detectable either by LAM-PCR and sequencing or by clone-specific tracking PCR. n.d. indicates not done.
Tracking analysis of individual clones. The activity of individual clones over time in mice that underwent primary (1°), secondary (pair 2°a, 2°b), and tertiary (pair 3°a, 3°b) transplantation is shown. All mice that underwent tertiary transplantation received bone marrow from the corresponding secondary mouse 2°b not 2°a. Clone numbers refer to listed clones in Table 1. Dark squares indicate time points where the clone was detectable either by LAM-PCR and sequencing or by clone-specific tracking PCR. n.d. indicates not done.
Insertion sites detected by LAM-PCR in peripheral blood samples of mice that underwent serial transplantation
| Clone no. . | Ref seq gene . | Description of gene product . | Locus link . | Chromosome (orientation) . | Integration locus . | Genomic locus . |
|---|---|---|---|---|---|---|
| BCNU/O6BG | ||||||
| 1 | Gif | Gastric intrinsic factor; vitamin B12 binding protein | 14603 | 19 (−) | 11833504 | 3012 down |
| 2 | Ncor2 | Nuclear receptor corepressor 2; Notch signaling pathway | 20602 | 5 (+) | 125477598 | 9566 up |
| 3 | Arid5b | AT rich interactive domain 5B; essential for accumulation of lipid store | 71371 | 10 (−) | 67674926 | 843 up |
| 4 | HoxB5/HoxB4 | Homeobox B5 and B4; transcription factors | 15412/15413 | 11 (+) | 96126527 | 8302 up/4317 down |
| 5 | Creg2 | Cellular repressor of E1A-stimulated genes 2; putative secreted glycoprotein | 263764 | 1 (−) | 39581009 | 14719 In2 |
| 6 | Tpt1 | Translationally controlled tumor protein; cell growth, cell cycle progression | 22070 | 14 (−) | 74567674 | 11812 up |
| Control | ||||||
| 7 | 6330512M04Rik | Hypothetical protein LOC320802 | 320802 | 7 (−) | 142149500 | 32149 In3 |
| 8 | Fmnl3 | Formin-like 3; actin-binding, Rho GTPase binding | 22379 | 15 (+) | 99163868 | 34632 Ex3 |
| 9 | Centd2 | Centaurin, delta 2; Rho-GAP/PIP3-dependent ARF-GAP activity | 69710 | 7 (−) | 101253116 | 1173 In1 |
| 10 | BC043301 | cDNA sequence BC043301; nucleic acid and metal ion binding | 210104 | 7 (+) | 43423114 | 9567 up |
| 11 | Slc25a1 | Solute carrier family 25; mitochondrial deoxynucleotide carrier | 13358 | 16 (−) | 17842278 | 547 up |
| 12 | 2610301B20Rik | Hypothetical protein LOC67157 | 67157 | 4 (−) | 10851653 | 25085 down |
| 13 | Cntnap5a | Contactin associated protein-like 5A; cell adhesion | 636808 | 1 (+) | 116246129 | 1266555 up |
| 14 | Slc10a2 | Solute carrier family 10, member2; bile salt uptake in distal ileum | 20494 | 8 (+) | 6482015 | 1376811 up |
| Clone no. . | Ref seq gene . | Description of gene product . | Locus link . | Chromosome (orientation) . | Integration locus . | Genomic locus . |
|---|---|---|---|---|---|---|
| BCNU/O6BG | ||||||
| 1 | Gif | Gastric intrinsic factor; vitamin B12 binding protein | 14603 | 19 (−) | 11833504 | 3012 down |
| 2 | Ncor2 | Nuclear receptor corepressor 2; Notch signaling pathway | 20602 | 5 (+) | 125477598 | 9566 up |
| 3 | Arid5b | AT rich interactive domain 5B; essential for accumulation of lipid store | 71371 | 10 (−) | 67674926 | 843 up |
| 4 | HoxB5/HoxB4 | Homeobox B5 and B4; transcription factors | 15412/15413 | 11 (+) | 96126527 | 8302 up/4317 down |
| 5 | Creg2 | Cellular repressor of E1A-stimulated genes 2; putative secreted glycoprotein | 263764 | 1 (−) | 39581009 | 14719 In2 |
| 6 | Tpt1 | Translationally controlled tumor protein; cell growth, cell cycle progression | 22070 | 14 (−) | 74567674 | 11812 up |
| Control | ||||||
| 7 | 6330512M04Rik | Hypothetical protein LOC320802 | 320802 | 7 (−) | 142149500 | 32149 In3 |
| 8 | Fmnl3 | Formin-like 3; actin-binding, Rho GTPase binding | 22379 | 15 (+) | 99163868 | 34632 Ex3 |
| 9 | Centd2 | Centaurin, delta 2; Rho-GAP/PIP3-dependent ARF-GAP activity | 69710 | 7 (−) | 101253116 | 1173 In1 |
| 10 | BC043301 | cDNA sequence BC043301; nucleic acid and metal ion binding | 210104 | 7 (+) | 43423114 | 9567 up |
| 11 | Slc25a1 | Solute carrier family 25; mitochondrial deoxynucleotide carrier | 13358 | 16 (−) | 17842278 | 547 up |
| 12 | 2610301B20Rik | Hypothetical protein LOC67157 | 67157 | 4 (−) | 10851653 | 25085 down |
| 13 | Cntnap5a | Contactin associated protein-like 5A; cell adhesion | 636808 | 1 (+) | 116246129 | 1266555 up |
| 14 | Slc10a2 | Solute carrier family 10, member2; bile salt uptake in distal ileum | 20494 | 8 (+) | 6482015 | 1376811 up |
All samples analyzed were derived from cloned and sequenced LAM-PCR amplicons as shown in Figures 4,5. The sequenced LAM-PCR products were mapped on the murine genome database, and the site of insertion was identified using the University of California, Santa Cruz BLAT search tools and mouse database from February 2006.
down indicates downstream of gene; up, upstream of gene; In, intron; and Ex, exon.
To analyze the contribution of individual clones to hematopoiesis over time, we performed comprehensive tracking analyses of individual marked clones in BCNU/O6BG-treated mice and control mice that underwent serial transplantation by PCR using allele-specific primers. In line with previous reports demonstrating considerable heterogeneity of the transplantable stem-cell compartment,43 some heterogeneity in the activity of individual clones over time was observed irrespective of whether the mice were treated (Figure 6).
Only a subfraction of all marked stem-cell clones is selected
Given the low number of selectable repopulating units calculated by Poisson statistics, the amplification of multiple vector integrations sites by LAM-PCR in all mice was surprising. To exclude persistence of marked long-lived mature lymphocytes as a potential explanation for the observed discrepancy, we analyzed sorted lymphoid (B220+ or CD3+) and myeloid cells (CD11+ or Ly6+) from 6 mice by LAM-PCR. Approximately 90% of the integration sites amplified from myeloid cells could also be visualized in the lymphoid fraction, demonstrating that multipotent progenitor or stem cells were in fact present. In the lymphoid fraction, only 10% to 20% more integration sites were amplified than in the myeloid fraction, suggesting that any contribution of transplanted long-lived mature lymphoid cells was small or absent. To exclude multiple vector integrations in single progenitor or stem cells, myeloid colonies from bone marrow cells of 6 mice were analyzed. Single colonies were picked from semisolid assays, DNA was isolated, and LAM-PCR amplification of the integration sites was undertaken. In DNA from 4 of 16 colonies, 2 to 4 vector copies were detected by LAM-PCR. In 12 of 16 colonies, only one vector integration per colony could be amplified, confirming that at least predominant single copy vector integration was obtained with the MOI of 0.4. The LAM-PCR amplicons were shotgun cloned and sequenced. Different integration site sequences were detected in colonies from mice that received a transplant of bone marrow cells from different donor mice. No integration sites were observed that were detectable exclusively in colonies but not in the peripheral blood. Taken together, in line with our observation of a marked but not selectable hematopoiesis in mice that received transplants with the lower transduced cell dose, these findings suggest that a stable fraction of clones but not all transduced clones contributed to the efficient selection.
Stem-cell renewal produces clonal identity and similar hematopoietic reconstitution in split marked stem-cell transplants of secondary recipients in pairs
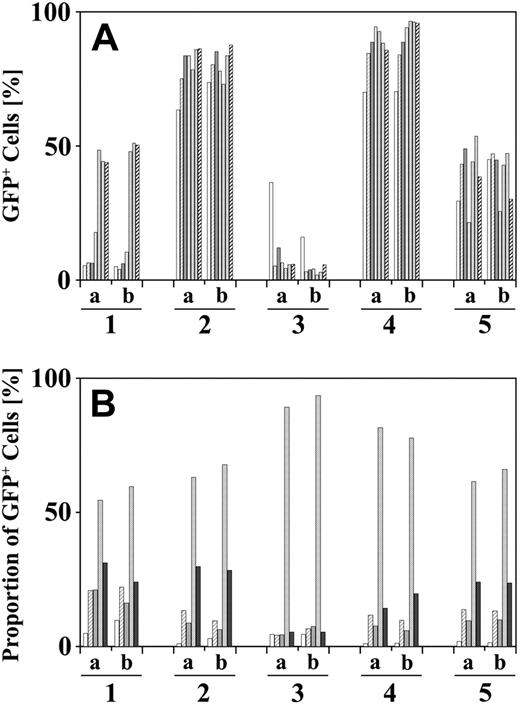
In serial transplantation, bone marrow from each individual mouse that underwent transplantation was split and transplanted into pairs of secondary recipient mice. LAM-PCR revealed a remarkably identical clonal repertoire in both recipient mice. Clone-specific tracking PCR confirmed self-renewal of long-term stem cells (Figure 6). Very similar engraftment kinetics and lineage differentiation were detected in both mice regardless of whether the pairs of mice were treated with the selection drugs (Figure 7). In contrast, as expected, clonal repertoires, kinetics, and differentiation were completely different among mice that received a transplant of cells from different donors.
Similar reconstitution of split bone marrow from individual mice in pairs of secondary transplant recipients. Peripheral blood samples from pairs of secondary mice that received a transplant of bone marrow from identical donors were analyzed by FACS at monthly intervals. Kinetics (A) and differentiation (B) of GFP+ blood cells were similar in pairs of secondary mice receiving bone marrow from the same donor. In panel A, time points in months are indicated by successive bars. In panel B, successive bars indicate the proportion of erythroid progenitors, granulocytes, macrophages, B lymphocytes, and T lymphocytes of the GFP+ blood cells 12 weeks after transplantation. Representative pairs of secondary mice with (mice 1 a,b, 2 a,b, 4 a,b) or without BCNU/O6BG treatment (mice 3 a,b, 5 a,b) are shown.
Similar reconstitution of split bone marrow from individual mice in pairs of secondary transplant recipients. Peripheral blood samples from pairs of secondary mice that received a transplant of bone marrow from identical donors were analyzed by FACS at monthly intervals. Kinetics (A) and differentiation (B) of GFP+ blood cells were similar in pairs of secondary mice receiving bone marrow from the same donor. In panel A, time points in months are indicated by successive bars. In panel B, successive bars indicate the proportion of erythroid progenitors, granulocytes, macrophages, B lymphocytes, and T lymphocytes of the GFP+ blood cells 12 weeks after transplantation. Representative pairs of secondary mice with (mice 1 a,b, 2 a,b, 4 a,b) or without BCNU/O6BG treatment (mice 3 a,b, 5 a,b) are shown.
Discussion
Safe clinical application of drug-resistance gene transfer for genetically modifying hematopoiesis in patients requires a thorough preclinical evaluation of potential toxicities in animal models. Among the different approaches for stem-cell protection and selection available, constitutive expression of MGMT P140K by gene transfer allows a comparatively very efficient selection of gene-modified murine and canine long-term hematopoietic stem cells.19,44 In vivo administration of BCNU and O6-BG in nonobese diabetic (NOD)/SCID mice also selects MGMT P140K–transduced human hematopoiesis.21,25,45,46 As with every selection strategy, potential adverse effects on the biology of long-term stem cells in vivo might be a concern, including the preferential selection of certain blood cell lineages against others, and a progressive reduction of the clonal stem-cell repertoire. In this study, our data demonstrate that selection based on retroviral expression of the MGMT P140K mutant impairs neither the ability of stem cells to generate all blood cell lineages nor their proliferation. Validity of these results was controlled by several measures in our experimental design. We aimed at maximizing biologic side effects including selective lineage bias by an extended drug application schedule that included up to 17 administrations of chemotherapy and 3 serial transplantations, extending the overall observation time up to 20 months. Even under these stringent conditions, we did not detect any evidence of stem-cell exhaustion. In line with this observation, serial transplantation experiments previously demonstrated that murine hematopoietic stem cells expand at each passage approximately 10- to 20-fold without any detectable decline in their activity or their capability to reconstitute all blood cell lineages during 4 transplantations.47,48 In a more recent study, Allsopp et al challenged the concept of an unlimited lifespan of hematopoietic stem cells in serial transplantation. They observed a dramatic drop in the numbers of stem-cells that were defined by their phenotype after 4 passages of serial transplantation.49 This decline was associated with a shortening of telomeres, the linear repetitive DNA structures situated at the ends of chromosomes. Proliferative stress might cause such telomere shortening even at younger age. Shorter telomeres have been shown in leukocytes of patients after hematopoietic stem-cell transplantation compared with their donors,50–54 but the accelerated telomere shortening detectable in peripheral blood monocytes was restricted to the first year after hematopoietic stem-cell transplantation. A subsequent reduction of telomere reserve by approximately 20% probably is functionally insignificant.55 The lack of any evidence for a dysfunction of normal human hematopoietic stem cells after transplantation due to telomere shortening in the literature indicates that stem cells have a large telomere reserve. Mouse stem cells might be especially robust against exhaustion due to the longer telomeres in mice compared with humans. This species difference may need to be taken into account when extrapolating our data to humans.
The influence of retroviral gene transfer on clonal fate can be significant. We have recently shown that the enhancer function of long terminal repeats (LTRs) of retroviral vectors can drive the expression of genes adjacent to the insertion locus. LTR-driven expression of transcription factors can lead to clonal proliferation and hematologic malignancy in mice and humans.5,7,31,56 In fact, insertional mutagenesis by wild-type retroviruses and, recently, retrovirus vectors has been used as a tool to identify genes that can cause hematopoietic progenitor immortalization.57,58 It is therefore conceivable that insertional gene activation might have contributed to the selection of gene-marked hematopoiesis. In the present study, control mouse cohorts that received transplants of gene-marked bone marrow cells from the same transduction cultures, but were not treated with chemotherapy, did not exhibit any evidence of an autonomous proliferation of gene-corrected cells, such as an average increase in the proportion of gene-marked cells. Nor did we observe any clonal dominance or vector-driven hematologic malignancies during long-term follow-up. In contrast to previous studies reporting a high incidence of integration-driven clonal dominance in mice,31 our experiments were specifically designed to decrease the likelihood of such events. We transplanted a low number of 4 × 105 bone marrow cells from which only 8% to 12% carried predominantly one copy of the vector. Nevertheless, our sequence analysis of animals that underwent serial transplantations demonstrates a selective engraftment and survival of clones carrying vector insertions near or in genes that are potentially relevant for the regulation of hematopoietic stem cells. These genes are involved in important cell regulatory processes such as cell growth, cell development, or cell differentiation. Still, even additional proliferative stress of the gene-marked stem cells due to selection, a potential risk factor,59,60 did not reduce the number of active stem-cell clones or result in malignant transformation in our studies. Our results suggest that integration events near or in relevant regulatory genes do not necessarily alter the biology of stem-cell clones under selection or lead to fatal consequences.
The striking similarity in clonal composition, kinetics, and differentiation of the hematopoiesis in secondary recipient mice that received a transplant of cells from the same primary donor compared with the clonal diversity after transplantation of bone marrow cells from different donors has interesting implications for the murine stem cell compartment. A surprisingly high degree of clonal activity in the biology of long-term stem cells seems to be intrinsically determined since it is equally passed on to offspring stem cell generations that reconstitute hematopoiesis in secondary and even tertiary recipient mice. Moreover, our results suggest that extensive self-renewal of stem cells in the primary recipients in vivo must have occurred that resulted in a net symmetric refilling of the stem cell compartment.
MGMT P140K selection offers the selective advantage needed for successful treatment of diseases in which the transgene does not itself establish a selective advantage. We show that a reduced-intensity MGMT P140K selection does not alter hematopoietic stem-cell biology and may increase the safety of clinical gene therapy. Our study supports the view that the likelihood of side effects caused by gene transfer into hematopoietic stem cellsdepends on the dose of gene-modified cells as well as vector copy numbers per cell and can therefore be decreased by limiting their numbers and subsequent selection.61 These data can serve as a proof of principle that selection is possible well below the maximally tolerated dose of the alkylator. Clinical studies will be needed to establish the minimal dose requirements for obtaining genetically modified human long-term stem cells that convey a sustained therapeutic effect after selection.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants Ka 976/4–1 from the Deutsche Forschungsgemeinschaft, D1 KV9527/7 from the German Minister for Education and Research, and the Dr. Mildred Scheel Stiftung fur Krebsforschung, Bonn, Germany to C.K. and H. G. and NIH DK074310 to D.A.W.
We thank Marie Follow and Klaus Geiger for cell sorting. We also gratefully acknowledge helpful discussions and support from Dr Christoph Peters and Dr Roland Mertelsmann.
National Institutes of Health
Authorship
Contribution: C.R.B. designed and performed research, analyzed data, and wrote the paper; I.H.P. performed research; M.S. designed research and analyzed data; S.F. performed research; D.A.W. designed research and provided reagents; C.K. designed research and wrote the paper; H.G. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christof von Kalle, The National Center for Tumor Diseases (NCT), German Cancer Research Center, Im Neuenheimer Feld 350; e-mail: christof.kalle@nct-heidelberg.de.