Abstract
Insertional oncogenesis is a possible consequence of the integration of gamma-retroviral (RV) or lentiviral (LV) vectors into the human genome. RV common insertion sites (CISs) have been identified in hematopoietic malignancies and in the nonmalignant progeny of transduced hematopoietic stem/progenitor cells (HSCs), possibly as a consequence of clonal selection in vivo. We have mapped a large number of RV and LV integrations in human CD34+ HSCs, transduced in vitro and analyzed without selection. Recurrent insertion sites (hot spots) account for more than 21% of the RV integration events, while they are significantly less frequent in the case of LV vectors. RV but not LV hot spots are highly enriched in proto-oncogenes, cancer-associated CISs, and growth-controlling genes, indicating that at least part of the biases observed in the HSC progeny in vivo are characteristics of RV integration, already present in nontransplanted cells. Genes involved in hematopoietic and immune system development are targeted at high frequency and enriched in hot spots, suggesting that the CD34+ gene expression program is instrumental in directing RV integration. The lower propensity of LV vectors for integrating in potentially dangerous regions of the human genome may be a factor determining a better safety profile for gene therapy applications.
Introduction
Gene therapy of genetic blood disorders requires stable genetic modification of hematopoietic stem cells. Gene transfer vectors derived from murine gamma-retroviruses, such as the Moloney murine leukemia virus (MLV), have been used for more than a decade to transduce human bone marrow (BM)–derived or mobilized hematopoietic stem/progenitor cells (HSCs) in a clinical context. Retroviral vector–mediated gene transfer has recently achieved therapeutic efficacy, allowing correction of life-threatening diseases such as severe combined immunodeficiencies (SCIDs)1–3 or chronic granulomatous disease (CGD).4 MLV-derived vectors, however, have also raised significant safety concerns for the genotoxic risk potentially associated with their uncontrolled integration into the human genome.5–7 Indeed, insertional activation of a T-cell proto-oncogene has been correlated with the occurrence of lymphoproliferative disorders in 3 patients treated with retrovirally transduced hematopoietic cells for X-linked SCID (X-SCID).1 Recent studies have shown that gamma-retroviral vectors integrate preferentially within transcribed genes and around promoters and CpG islands,8 where insertion of the viral long terminal repeat (LTR) transcriptional enhancer has a high probability to interfere with gene regulation.9 Nevertheless, no adverse event related to viral insertion was reported in other clinical trials for X-SCID,3 adenosine deaminase-deficient SCID (ADA-SCID),2 CGD,4 or graft-versus-host disease (GVHD),9 suggesting the existence of specific risk factors that are incompletely understood.10
Analysis of MLV integration patterns in natural or experimentally induced leukemias/lymphomas showed the existence of insertion sites recurrently associated with a malignant phenotype. These “common insertion sites” (CISs) include proto-oncogenes or other genes associated with cell growth and proliferation, the activation or deregulation of which has a causal relationship with the establishment and/or progression of neoplasia.11 Some of these sites, such as the EVI1-MDS1 locus, have been identified at relatively high frequency also in the nonmalignant progeny of transduced hematopoietic cells in mice,12 nonhuman primates,13 and humans,4 indicating that insertion into certain genes may cause clonal amplification of transduced progenitors in vivo. From these studies, however, it is not clear whether clonal dominance is entirely the result of in vivo selection, or is favored by the existence of highly preferred regions of retroviral integration that make clonal amplification more likely to occur. This issue is highly relevant in understanding the different outcomes of different gene therapy clinical trials, in assessing the relative safety of using MLV-derived vectors in specific clinical applications, and in comparing the safety profile of alternative vectors (eg, HIV-derived lentiviral vectors) or vector designs.
We report an analysis of gamma-retroviral (RV) and lentiviral (LV) vector integration hot spots from large collections of integration sites obtained from human cord blood (CB)– and BM–derived CD34+ HSCs transduced in vitro and analyzed without selection. Hot spots account for more than 20% of the MLV integration sites, while they are significantly less frequent in the case of HIV-derived vectors. Integration sites associated with clonal dominance and neoplasia in both mice and humans, including LMO2, are hot spots of gamma-RV but not LV integration in human hematopoietic cells.
Materials and methods
Retroviral vectors
CB–derived CD34+ cells were transduced with the previously described LGSΔN and LGSΔN-ΔCAAT RV vectors,14 driving the expression of green fluorescent protein (GFP) under an intact or a U3-deleted MLV LTR, and of ΔLNGFR under an internal SV40 promoter. CB-derived CD34+ cells were also transduced with the self-inactivating (SIN) pRRLsin-18.pptCMV-GFPwpre LV vector15 containing a U3-deleted HIV-1 LTR and a cytomegalovirus (CMV)–driven GFP cassette,16 or with the pHR2pptCMV-GFPwpre or the pHR2pptGSΔN LV vectors, retaining HIV-1 wild-type LTRs and driving the expression of GFP or ΔLNGFR under internal CMV or SV40 promoters. To generate the pHR2pptCMVGFPwpre construct, a pptCMVGFPwpre fragment from the pRRLsin-18.pptCMVGFPwpre vector was cloned into ClaI-EcoRI sites of pHR2MD-NGFR.17 To obtain the pHR2pptGSΔN LV construct, the pHR2pptCMVGFPwpre vector was digested with BamHI/EcoRI and ligated to a GFP-SV40ΔLNGFR cassette. BM–derived CD34+ cells were transduced with previously described RV vectors expressing either the ADA (GIADA12 ) or the γc receptor3 cDNA.
RV vector supernatants were produced by transient transfection of the amphotropic Phoenix packaging cell line. Infectious particle titer was determined on K562 cells. Vesicular stomatitis virus–G protein (VSV-G) pseudotyped LV particles were prepared by transient cotransfection of 293T cells, collected and concentrated as already described,17 and titrated on 293T cells. Transduction efficiency was evaluated by flow cytometry. Amphotropic or GaLV-pseudotyped ADA and γc receptor RV vectors were titered as previously described.2,3
Transduction of human CD34+ cells
CD34+ HSCs were purified from CB Ficoll fractions by magnetic sorting (MiniMACS; Miltenyi, Auburn, CA) and prestimulated for 24 to 48 hours in serum-free Iscove modified Dulbecco medium (IMDM) supplemented with 20% BIT (Stem Cell Technologies; Vancouver, BC, Canada), 20 ng/mL human thrombopoietin (TPO), 100 ng/mL Flt3-ligand (PeproTech, Rocky Hill, NJ), 20 ng/mL IL-6, and 100 ng/mL stem-cell factor (SCR; R&D Systems, Minneapolis, MN). RV transduction was performed by spinoculation (3 rounds at 1500 rpm for 45 minutes) in the presence of 4 μg/mL polybrene. LV transduction was performed by overnight incubation at a MOI of 200 in the presence of 4 μg/mL polybrene. Transduction efficiency was evaluated by analysis of enhanced GFP (EGFP) and/or ΔLNGFR expression by flow cytometry using a mouse anti–human NGFR antibody (Becton Dickinson, San Jose, CA).
BM- or peripheral blood (PB)–derived CD34+ cells were purified from healthy donors or patients with SCID by magnetic sorting, prestimulated for 24 hours in IMDM containing human serum or serum-free X-Vivo-10 medium and a cytokine cocktail (Flt3-ligand, SCF, TPO, IL-3), and transduced by 3 cycle-exposure to the GIADA1 or the γc receptor RV vector supernatant as previously described.2,3
Cloning and analysis of RV insertion sites
Integration sites were cloned by linker-mediated polymerase chain reaction (LM-PCR) or linear amplification–mediated PCR (LAM-PCR), as described.18,19 Briefly, genomic DNA was extracted from 0.5 to 5 × 106 infected cells and digested with MseI and a second enzyme to prevent amplification of internal 5′ LTR fragments (PstI for RV vectors and SacI/NarI for LV vectors). An MseI double-stranded linker was then ligated, and LM-PCR was performed with nested primers specific for the linker and the 3′ LTR (MLV: 5′-GACTTGTGGTCTCGCTGTTCCTTGG-3′ and 5′- GGTCTCCTCTGAGTGATTGACTACC-3′; and HIV: 5′- AGTGCTTCAAGTAGTGTGTGCC-3′ and 5′- GTCTGTTGTGTGACTCTGGTAAC-3′). PCR products were shotgun-cloned (TOPO TA cloning kit; Invitrogen, Carlsbad, CA) into libraries of integration junctions, which were then sequenced to saturation. A valid integration contained the MLV or HIV nested primer, the entire MLV or HIV genome up to a CA dinucleotide, and the linker nested primer. Sequences between the 3′ LTR and the linker primers were mapped onto the human genome by the University of California Santa Cruz (UCSC) BLAT alignment tool (http://genome.ucsc.edu), accessed May 2004. Random genomic sequences originated by LM-PCR (genomic MseI-MseI, PstI-MseI, NarI-MseI, or SacI-MseI fragments) were used as controls. Sequences featuring a unique best hit with 95% or greater identity to the human genome were considered genuine integration sites, and classified as intergenic when occurring at an arbitrarily chosen distance of more than 30 kb from any “known gene” (UCSC definition), perigenic when 30 kb or less upstream or downstream of a known gene, and intragenic when within the transcribed portion of at least 1 known gene. In case of multiple transcript variants, the most represented and/or the longest isoform was chosen. Gene density analysis was performed using the UCSC Table Browser tool (http://genome.ucsc.edu). For each integration, the number of known genes (a single isoform in case of multiple variants) contained in a range of 1 Mb around the insertion site was calculated. For all pairwise comparisons, we applied a 2-sample test for equality of proportions with continuity correction using the Rweb 1.03 statistical analysis package (http://math.montana.edu/Rweb/).
Gene expression profiling
The expression profile of CD34+ cells was determined by microarray analysis. RNA was isolated from 1 to 2 × 106 CB- and BM-derived CD34+ cells stimulated with cytokines according to the same protocols used for RV (CB- and BM-derived cells) or LV (CB-derived cells) vector transduction, transcribed into biotinylated cRNA, hybridized to Affymetrix HG-U133A Gene Chip arrays (Santa Clara, CA) and analyzed as previously described.9 To correlate retroviral integration and gene activity, expression values from the CD34+ cell microarrays were divided into 4 classes (ie, absent, low below the 25th percentile in a normalized distribution, intermediate between the 25th and the 75th percentiles, and high above the 75th percentile).
Functional clustering analysis
Functional cluster analysis of genes targeted by retroviral integrations and from control sequences was performed using the DAVID 2.1 Functional Annotation Tool21,22 (http://david.abcc.ncifcrf.gov). In the DAVID annotation system, a Fisher exact test corrected for multiple comparisons (DAVID's EASE score21 ) is adopted to measure the level of gene enrichment in Gene Ontology (GO) annotation terms with respect to a background population, and GO categories considered overrepresented when yielding an EASE score lower than 0.05. A list of 417 cancer-associated CISs was obtained from the Mouse Retrovirus Tagged Cancer Gene Database,23 where murine genes were replaced with human homologs. Genes were analyzed also by the network-based Ingenuity pathways analysis tool (Ingenuity Systems, www.ingenuity.com). Gene identifiers were uploaded into the application, and mapped to their corresponding Focus Gene in the Ingenuity Pathways Knowledge Base. Networks were algorithmically generated based on the direct or indirect interaction between Focus Genes. The functional analysis of each network identified the biological functions and/or diseases that were most significant to the genes in the network. (Fisher exact test). A list of 596 human proto-oncogenes was compiled from the University of New South Wales (UNSW) Embryology DNA-Tumor Suppressor and Oncogene Database24 and the Tumor Gene Database.25
Results
Retroviral integration preferences in human CD34+ HSCs
Human CD34+ HSCs were purified from umbilical CB pools, BM from patients with ADA-SCID and X-SCID, or PB from a healthy donor. CB CD34+ cells were transduced with MLV-derived gamma-RV or HIV-derived LV vectors carrying a GFP reporter gene and either a wild-type or a U3-deleted (SIN) LTR. BM CD34+ cells were transduced with MLV-derived RV vectors expressing either ADA2 or γc receptor3 from a wild-type LTR. PB CD34+ cells were transduced with the vector expressing γc receptor.3 Transduction efficiency ranged from 15% (SIN-RV) to more than 90% (SIN-LV) depending on the vector and target cell type, and remained stable throughout the culture period. DNA was obtained 1 to 12 days after infection, from cells that underwent 1 (all BM and PB samples) to 5 to 6 (all CB samples) cell doublings in culture. Vector-genome junctions were cloned and sequenced by a LM-PCR or LAM-PCR approach adapted to the different vector types, and mapped onto the human genome. Cumulatively, we mapped 1030 RV and 849 LV integrations in CB- or BM-derived CD34+ cells. A total of 595 RV integrations were obtained from CB cells transduced with wild-type (395) or SIN (200) LTR vectors expressing ΔLNGFR from an internal promoter, and 435 RV integrations were obtained from BM cells transduced with wild-type LTR vectors expressing ADA (190) or γc receptor (245). All LV integrations were obtained from CB cells transduced with wild-type (404) or SIN (445) LTR vectors expressing GFP or ΔLNGFR from an internal promoter.
Among RV integrations, 172 (16.7%) were in an intergenic position, 566 (55.0%) were within the transcribed portion of at least 1 gene, and 292 (28.3%) were at a distance of 30 kb or less upstream or downstream of 1 or more genes (Table 1; the complete list of sequences is available at GenBank with the accession number ER916114–ER918350). Among LV integrations, 148 (17.4%) were in an intergenic position, 609 (71.7%) were in an intragenic position, and 92 (10.9%) were in a perigenic position. Conversely, a collection of 798 control sequences randomly cloned by LM-PCR contained 369 (46.2%) intergenic, 308 (38.6%) intragenic, and 121 (15.2%) perigenic sequences. Compared with controls, RV vectors showed a preference for intragenic (2-sample test for equality of proportions with continuity correction; P < .001) and perigenic (P < .001) integration, while LV vectors showed a much higher preference for intragenic positions (P < .001).
RV integration site distribution in human CD34+ HSCs
| . | Intergenic, % . | Intragenic, % . | Perigenic, % . | Total hits . | Within 10 kb from TSS, % . | Total vector/gene interactions* . |
|---|---|---|---|---|---|---|
| CD34+ cells | ||||||
| RV vectors | 16.7 | 55.0 | 28.3 | 1030 | 29.3 | 1517 |
| LV vectors | 17.4 | 71.7 | 10.9 | 849 | 16.1 | 1241 |
| Control sequences | 46.2 | 38.6 | 15.2 | 798 | 9.1 | 902 |
| RV hot spots | 16.0 | 56.6 | 27.4 | 219 | 22.2 | 302 |
| LV hot spots | 8.6 | 81.4 | 10.0 | 70 | 13.2 | 2114 |
| Control hot spots | 36.4 | 59.1 | 4.5 | 22 | 13.0 | 23 |
| HeLa cells | ||||||
| RV vectors | 26.4 | 48.1 | 25.5 | 869 | 26.1 | 1219 |
| RV hot spots | 16.5 | 53.2 | 30.3 | 109 | 27.3 | 165 |
| . | Intergenic, % . | Intragenic, % . | Perigenic, % . | Total hits . | Within 10 kb from TSS, % . | Total vector/gene interactions* . |
|---|---|---|---|---|---|---|
| CD34+ cells | ||||||
| RV vectors | 16.7 | 55.0 | 28.3 | 1030 | 29.3 | 1517 |
| LV vectors | 17.4 | 71.7 | 10.9 | 849 | 16.1 | 1241 |
| Control sequences | 46.2 | 38.6 | 15.2 | 798 | 9.1 | 902 |
| RV hot spots | 16.0 | 56.6 | 27.4 | 219 | 22.2 | 302 |
| LV hot spots | 8.6 | 81.4 | 10.0 | 70 | 13.2 | 2114 |
| Control hot spots | 36.4 | 59.1 | 4.5 | 22 | 13.0 | 23 |
| HeLa cells | ||||||
| RV vectors | 26.4 | 48.1 | 25.5 | 869 | 26.1 | 1219 |
| RV hot spots | 16.5 | 53.2 | 30.3 | 109 | 27.3 | 165 |
Distribution of RV and LV integration sites unambiguously mapped in unselected CB- and BM-derived CD34+ HSCs, and RV integrations in HeLa cells from a previously published collection.27 . Integrations (total hits) were distributed as inside (intragenic), outside (intergenic), or at a distance of less than 30 kb upstream or dowstream (perigenic) from known genes (UCSC annotation). Insertions at a distance of ±10 kb from transcription start sites (TSSs) are indicated as percentage of the total vector/gene interactions. Control sequences were obtained from a randomly cloned library of Pst1/MseI-restricted, LM-PCR–amplified human CD34+ cell DNA.
Total number of genes within 30 kb from individual hits + intergenic hits.
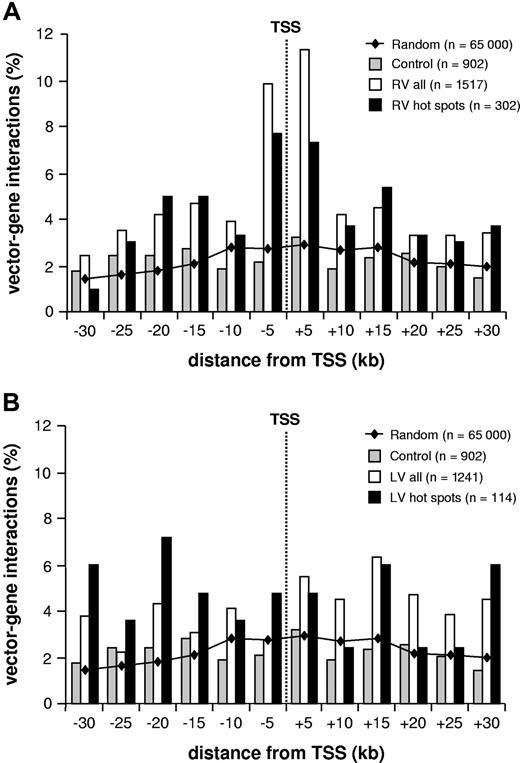
The position of the integrated proviruses with respect to known genes is shown in Figure 1, which considers the total number of vector-gene interactions in an interval of 30 kb around each insertion site (1517 and 1241 for RV and LV vectors, respectively). Compared with randomly cloned or computer-generated26 control sequences, a significant clustering around transcription start sites was observed for RV but not LV vectors. Overall, 29.3% of the total RV vector-gene interactions were within 10 kb from the + 1 position of known genes, compared with 16.1% for LV vectors (P < .001; Table 1; Figure 1). The RV general integration preferences were similar in CD34+ and HeLa cells, as indicated by the analysis of 869 insertions from a previously published collection27 (Table 1).
RV integrations and transcription start sites. Distribution of gamma-RV (A) and LV (B) integration sites in human CD34+ cells within an interval of 30 kb upstream or downstream from the transcription start site (TSS) of known genes (UCSC definition, considering only 1 isoform/gene). The bars show the percentage of distribution in each 5-kb interval of retroviral insertions (□), insertion hot spots (■), and control sequences (▩). The line shows the distribution of 65 000 computer-generated random insertion sites.26 n values indicate vector-gene interactions (ie, the total number of genes within 30 kb from individual insertions plus the intergenic insertions).
RV integrations and transcription start sites. Distribution of gamma-RV (A) and LV (B) integration sites in human CD34+ cells within an interval of 30 kb upstream or downstream from the transcription start site (TSS) of known genes (UCSC definition, considering only 1 isoform/gene). The bars show the percentage of distribution in each 5-kb interval of retroviral insertions (□), insertion hot spots (■), and control sequences (▩). The line shows the distribution of 65 000 computer-generated random insertion sites.26 n values indicate vector-gene interactions (ie, the total number of genes within 30 kb from individual insertions plus the intergenic insertions).
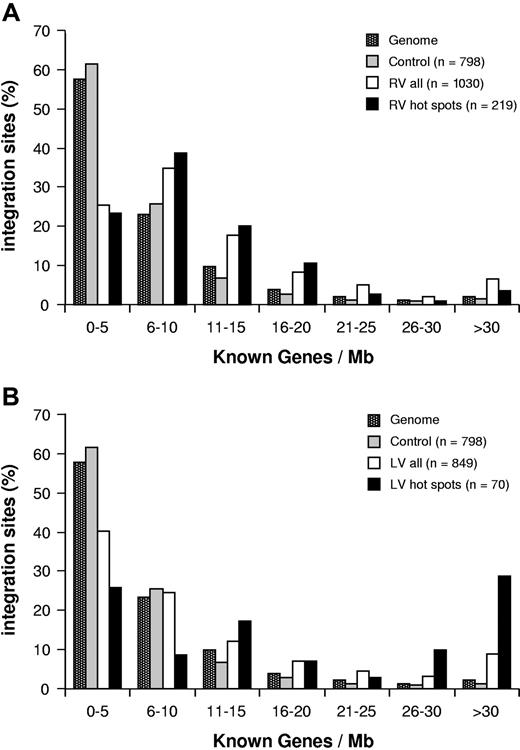
In CD34+ cells, RV integrations showed a significant preference for gene-dense regions: more than 60% of proviruses were found in genomic regions containing 6 to 20 genes/Mb, with a peak of 35% at a density of 6 to 10 genes/Mb, while more than 60% of control sequences mapped to regions with a gene density of less than 5 genes/Mb (P < .001; Figure 2A). On the contrary, LV integrations followed a distribution within regions of different gene density more similar to that of the control sequences and of the human genome, and different from that of RV (P < .001; Figure 2B).
Retroviral integration and gene density. Integration sites (□) and integration hot spots (■) of RV (A) and LV (B) vectors in CD34+ cells are plotted according to the number of known genes contained in a range of 1 Mb around each insertion site, in intervals of 5 genes/Mb. The distribution of control sequences is indicated by ■. ■ represents the frequency of 1-Mb segments in the human genome for each gene density interval. n values indicate the number of independent hits in each group.
Retroviral integration and gene density. Integration sites (□) and integration hot spots (■) of RV (A) and LV (B) vectors in CD34+ cells are plotted according to the number of known genes contained in a range of 1 Mb around each insertion site, in intervals of 5 genes/Mb. The distribution of control sequences is indicated by ■. ■ represents the frequency of 1-Mb segments in the human genome for each gene density interval. n values indicate the number of independent hits in each group.
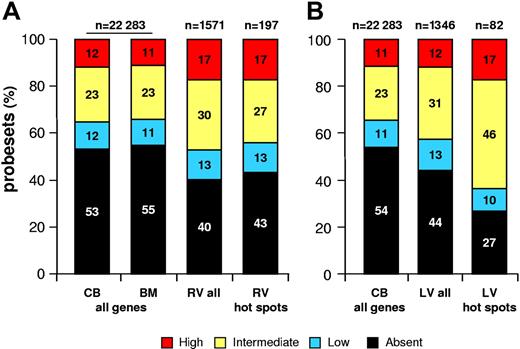
To correlate vector integration with gene activity, we determined the expression profile of more than 16 000 genes by microarray analysis in CB- and BM-derived CD34+ cells activated in culture in the same conditions used for RV and LV transduction. As shown in Figure 3, approximately 60% of 1571 probesets representing 866 genes hit by a RV vector detected a transcript in activated CD34+ cells; among them, 13% were classified as lowly abundant, 30% were classified as intermediately abundant, and 17% were classified as highly abundant, compared with a 45% to 47% “present” call on the whole microarrays (percentages were slightly different between CB- and BM-derived cells) and a 11% to 12%, 23%, and 11% to 12% breakdown in the 3 abundance classes. With the exception of the lowest expression class, all differences were statistically significant (P < .001), indicating that RV vectors integrate preferentially into genes active in CD34+ cells at the time of transduction, and particularly in the fraction of genes expressed at a higher level. A similar correlation with gene activity was observed, as already reported in T cells,28 for genes hit by LV vectors: approximately 56% of 1346 probesets representing 757 hit genes detected a transcript in activated CD34+ cells, with a 13%, 31%, and 12% breakdown in the 3 abundance classes, respectively. Compared with the whole microarray, the fraction of probesets with a present call was significantly higher (56% vs 46%; P < .001), but the difference was accounted for essentially by the intermediately abundant transcripts (31% vs 23%; P < .001; Figure 3), indicating that LV vectors tend to integrate into active genes in CD34+ cells but have no specific preference for genes expressed at high levels when compared with RV vectors (P < .001).
Correlation between retroviral integration and gene activity in CD34+ cells. The bars show the percentage of distribution of expression values from Affymetrix HG-U133A microarrays of cytokine-stimulated CD34+ cells. To correlate retroviral integration and gene activity, expression values from the CD34+ cell microarrays were divided into 4 classes: absent (black), low (below the 25th percentile in a normalized distribution; blue), intermediate (between the 25th and the 75th percentiles; yellow) and high (above the 75th percentile; red). (A) The first 2 bars (all genes) show the distribution of the more than 16 000 genes on the microarray of CB- or BM-derived CD34+ cells activated in the same conditions used for transduction with RV vectors; the other 2 bars represent the expression values of genes targeted by all RV integrations (RV all) or by integration hot spots (RV hot spots), derived from a weighted mean of the CB and BM microarray values. (B) The first bar (all genes) shows the distribution of the more than 16 000 genes on the microarray of CB-derived CD34+ cells activated in the same conditions used for transduction with LV vectors; the other 2 bars represent the expression values of genes targeted by all LV integrations (LV all) or by integration hot spots (LV hot spots). The n values indicate the number of probesets analyzed for each group of genes.
Correlation between retroviral integration and gene activity in CD34+ cells. The bars show the percentage of distribution of expression values from Affymetrix HG-U133A microarrays of cytokine-stimulated CD34+ cells. To correlate retroviral integration and gene activity, expression values from the CD34+ cell microarrays were divided into 4 classes: absent (black), low (below the 25th percentile in a normalized distribution; blue), intermediate (between the 25th and the 75th percentiles; yellow) and high (above the 75th percentile; red). (A) The first 2 bars (all genes) show the distribution of the more than 16 000 genes on the microarray of CB- or BM-derived CD34+ cells activated in the same conditions used for transduction with RV vectors; the other 2 bars represent the expression values of genes targeted by all RV integrations (RV all) or by integration hot spots (RV hot spots), derived from a weighted mean of the CB and BM microarray values. (B) The first bar (all genes) shows the distribution of the more than 16 000 genes on the microarray of CB-derived CD34+ cells activated in the same conditions used for transduction with LV vectors; the other 2 bars represent the expression values of genes targeted by all LV integrations (LV all) or by integration hot spots (LV hot spots). The n values indicate the number of probesets analyzed for each group of genes.
Genes regulating cell growth and proliferation are preferred targets of retroviral integration
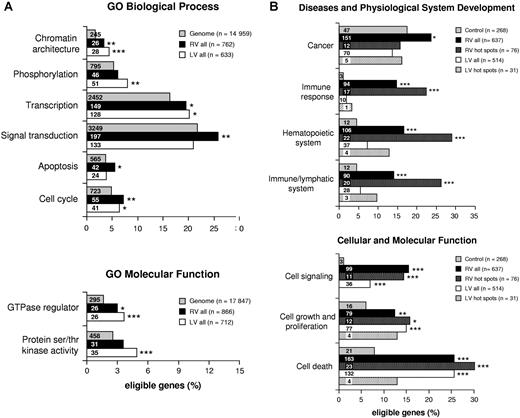
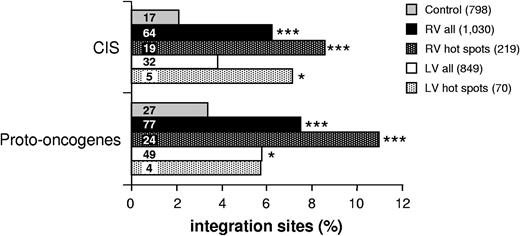
A functional classification by the GO criteria29 of genes hit by RV and LV vectors in CD34+ cells (Tables S1, S2, available on the Blood website; see the Supplemental Materials link at the top of the online article) showed statistically significant biases toward several gene categories (Figure 4A). In particular, genes involved in the establishment and/or maintenance of chromatin architecture, signal transduction, and cell cycle were significantly more represented in the collection of genes hit by RV integrations compared with their expected frequency in the human genome (EASE score < .005). Genes involved in chromatin remodeling and phosphorylation also were hit at a higher-than-expected frequency by LV vectors (EASE score < .0005 and < 0.005, respectively), particularly those with serine/threonine kinase and GTPase activity (EASE score < .0005). Two additional categories (ie, transcription and apoptosis) were overrepresented in genes hit by RV and/or LV vectors, although at less significant levels (EASE score < .05). A different analysis, carried out by the Ingenuity network-based pathway analysis software, indicated that genes involved in cell signaling, cell growth/proliferation, cell death, cancer, and hematopoietic system development were significantly overrepresented in the collection of RV and/or LV integrations with respect to genes annotated in the Ingenuity Pathways Knowledge Base software (.005 < P < .05). These categories were therefore chosen to carry out a direct frequency comparison between RV and LV target genes and our control gene list (Tables S1Table S2. List of all known genes (UCSC definition) hit by lentiviral (LV) proviruses, inside or at a distance of ≤30 kb upstream or downstream, in human CD34+ HSCs (PDF, 86.4 KB)–S3). Genes involved in cell signaling, growth/proliferation, and death were overrepresented in both RV and LV integrations with respect to control sequences (P < .001; Figure 4B), while genes involved in hematopoietic and immune system development, immune response and cancer were significantly overrepresented only in RV integrations (P < .001; Figure 4B). The comparison was then extended to genes specifically annotated in cancer-related databases (see “Materials and methods, Functional clustering analysis” for definitions and data source). RV integrations hit 77 proto-oncogenes and 64 cancer-associated CISs, corresponding to 7.5% and 6.2%, respectively, of the 1030 integrations (Figure 5). Both categories were significantly overrepresented (P < .001) compared with control sequences (27 proto-oncogenes and 17 CISs out of 798 sequences). On the contrary, LV integrations hit 49 proto-oncogenes and 32 CISs out of 849 integrations (Figure 5), a borderline significant difference compared with controls (P = .03 and .07, respectively). Interestingly, HeLa cell integrations show overrepresentation of proto-oncogenes but not CISs (data not shown). This finding is not surprising considering that CIS have been mostly defined in hematopoietic malignancies.
Genes regulating cell growth and proliferation are preferential targets of retroviral integration. (A) GO analysis of integration target genes in CD34+ cells. Genes identified as targets for RV (■) and LV (□) integration were analyzed for significant functional clusters with the DAVID 2.1 software. Functional categories are derived from the GO–Biological Process (establishment and/or maintenance of chromatin architecture, phosphorylation, transcription, signal transduction, apoptosis, cell cycle) and the GO–Molecular Function (GTPase regulator activity, protein serine/threonine kinase activity) classifications. Bars indicate the number of integration target genes annotated within the given category out of n genes eligible for each analysis. Asterisks denote the significance level of overrepresentation of any given category with respect to the human genome (▩), used as background population (***EASE score < .0005; **EASE score < .005; *EASE score < .05). The number of gene identifiers annotated within each functional category is indicated in the bars. (B) Functional clustering analysis comparing integration target and control gene lists. Function/disease categories were those significantly overrepresented in at least 1 integration target gene list (.005 < P < .05) using the Ingenuity Pathways Knowledge Base as background population and the Ingenuity analysis software. Bars represent the percentage of integration target genes belonging to each category among n genes eligible for the analysis. Asterisks denote the probability that differences observed between integration data sets (RV, LV, RV hot spots, and LV hot spots) and the control data set are due to chance alone (2-sample test for equality of proportions with continuity correction; ***P < .001; **P < .005; *P < .05). The number of genes annotated within each category is indicated in the bars.
Genes regulating cell growth and proliferation are preferential targets of retroviral integration. (A) GO analysis of integration target genes in CD34+ cells. Genes identified as targets for RV (■) and LV (□) integration were analyzed for significant functional clusters with the DAVID 2.1 software. Functional categories are derived from the GO–Biological Process (establishment and/or maintenance of chromatin architecture, phosphorylation, transcription, signal transduction, apoptosis, cell cycle) and the GO–Molecular Function (GTPase regulator activity, protein serine/threonine kinase activity) classifications. Bars indicate the number of integration target genes annotated within the given category out of n genes eligible for each analysis. Asterisks denote the significance level of overrepresentation of any given category with respect to the human genome (▩), used as background population (***EASE score < .0005; **EASE score < .005; *EASE score < .05). The number of gene identifiers annotated within each functional category is indicated in the bars. (B) Functional clustering analysis comparing integration target and control gene lists. Function/disease categories were those significantly overrepresented in at least 1 integration target gene list (.005 < P < .05) using the Ingenuity Pathways Knowledge Base as background population and the Ingenuity analysis software. Bars represent the percentage of integration target genes belonging to each category among n genes eligible for the analysis. Asterisks denote the probability that differences observed between integration data sets (RV, LV, RV hot spots, and LV hot spots) and the control data set are due to chance alone (2-sample test for equality of proportions with continuity correction; ***P < .001; **P < .005; *P < .05). The number of genes annotated within each category is indicated in the bars.
CISs and proto-oncogenes are overrepresented in RV integrations and integration hot spots. Comparative analysis of the frequency of genes annotated in the CIS and proto-oncogene databases (see “Materials and methods, Functional clustering analysis” for definitions and data source) between integration target and control gene lists. Bars represent the percentage of RV and LV integrations, RV and LV integration hot spots, and control sequences targeting at least 1 proto-oncogene or CIS. The n values indicate the number of independent hits in each group. Asterisks denote the level of enrichment with respect to control data set (2-sample test for equality of proportions with continuity correction; ***P < .001; *P < .05).
CISs and proto-oncogenes are overrepresented in RV integrations and integration hot spots. Comparative analysis of the frequency of genes annotated in the CIS and proto-oncogene databases (see “Materials and methods, Functional clustering analysis” for definitions and data source) between integration target and control gene lists. Bars represent the percentage of RV and LV integrations, RV and LV integration hot spots, and control sequences targeting at least 1 proto-oncogene or CIS. The n values indicate the number of independent hits in each group. Asterisks denote the level of enrichment with respect to control data set (2-sample test for equality of proportions with continuity correction; ***P < .001; *P < .05).
Overall, these analyses show that both RV and LV vectors have a general tendency to integrate into genes involved in the regulation of cell growth and proliferation, and that RV integrations have a specific bias for genes associated with oncogenic transformation. An Ingenuity network analysis confirmed these biases and showed, in addition, that a significant number of genes hit by RV integrations are functionally linked in gene networks involved in apoptosis (Table S4; Figure 6A), signal transduction, transcriptional regulation, and cancer (Table S4; Figure 6B).
Genes hit by retroviral integration are functionally linked in gene networks. Representative networks originated by Ingenuity analysis of RV target genes (Table S4 for a complete list). Both networks are made of 35 target genes, with an Ingenuity score of 42 or higher. The color code indicates the most significant biological functions associated to each network (P < .001). (A) RV network 1; (B) RV network 4 (networks are identified in Table S4).
Genes hit by retroviral integration are functionally linked in gene networks. Representative networks originated by Ingenuity analysis of RV target genes (Table S4 for a complete list). Both networks are made of 35 target genes, with an Ingenuity score of 42 or higher. The color code indicates the most significant biological functions associated to each network (P < .001). (A) RV network 1; (B) RV network 4 (networks are identified in Table S4).
RV but not LV vectors show a high frequency of integration hot spots
The RV and LV insertion site collections were analyzed for the presence of integrations at recurrent sites (hot spots), using essentially the same criteria previously applied to the definition of cancer-associated CISs (at least 2 independent insertions in less than 30 kb, 3 in less than 50 kb, and 4 in less than 100 kb11,20 ). Overall, 219 (21.3%) of 1030 RV insertion sites met these criteria, identifying 97 hot spots in the genome of CD34+ cells (Table S5). A total of 109 (12.5%) of 869 integrations met the same criteria in HeLa cells, defining 52 hot spots (data not shown). LV vectors showed a significantly lower propensity to integrate at recurrent sites, with only 70 (8.2%) of 849 integrations meeting the definition criteria, and identifying 33 hot spots (Table S5). Comparing the 3 collections, 1 hot spot appeared to be a recurrent site for both RV (4 hits) and LV (3 hits) integration (Chromosome 17q23.2: 55188652–55285672), while 3 hot spots were found in common between CD34+ and HeLa cells (data not shown). It is worth noting that 22 (2.8%) of 798 control sequences also met the hot spot definition criteria (Table S5), defining a background level of false positivity in the LM-PCR analysis. The different subgroups of RV integrations contributed to the hot spot list proportionally to their size, with no apparent bias related to the type of transduced cell (CB, BM, or PB), the vector used for transduction (wt-LTR or SIN-LTR), or the number of cell doublings undergone in culture before harvesting (Table S6). In particular, nonexpanded cell populations (BM- and PB-derived), which collectively contributed less than half of the 1030 total RV integrations, contributed with at least 1 integration to 56 (58%) of the 97 RV hot spots (Table S5).
The position of RV hot spot integrations with respects to known genes reflected the RV general integration preferences, with intergenic, perigenic, and gene-dense regions overrepresented to the same extent observed in the entire collection of RV integrations, and clustering around transcription start sites (TSSs) only slightly decreased (P = .015; Table 1; Figures 1A, 2A). On the contrary, LV hot spots showed a higher frequency of integration in intragenic (81.4% vs 71.7%) and gene-dense (65.7% vs 35.6% in the more than 11 genes/Mb range) regions (Table 1; Figure 2B). Similarly, RV hot spots occurred in the same proportion of expressed genes observed for all RV integrations (Figure 3A), while LV hot spots contained a significantly higher proportion of expressed genes (73.2% vs 55.9%; P = .003; Figure 3B).
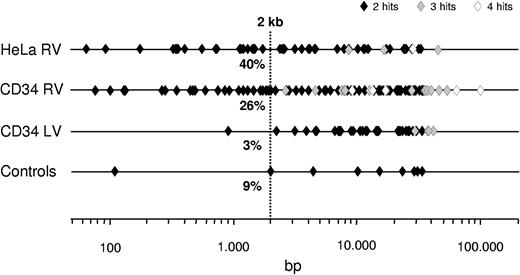
Interestingly, the maximum distance between independent integrations defining a hot spot was significantly lower for RV vectors compared with LV vectors and control sequences with hot spot characteristics. Overall, 52% and 67% of the RV hot spots in CD34+ and HeLa cells span less than 10 kb, including those containing 3 or 4 independent integrations, compared with 36% and 27% for LV and control sequences, respectively (Figure 7). One-fourth (26.0%) of the RV hot spots in CD34+ cells and almost one-half (40.4%) of those in HeLa cells contained 2 independent integrations in less than 2 kb, compared with only 3% of the LV hot spots.
Schematic representation of the maximum distance between individual hits within RV and LV hot spots. Symbols represent single hot spots originated from 2 (♦), 3 (♦), or 4 (◇) hits in the genome of CD34+ HSCs (1030 RV and 849 LV integrations) and HeLa cells (869 RV integrations), plotted according to the maximum distance between individual integrations (in base pairs, log scale). Also shown are “false positive” hot spots generated by applying the definition criteria to a library of LM-PCR–amplified random sequences of human CD34+ DNA (798 sequences). A total of 26.0% of the 97 RV hot spots in CD34+ cells and almost one-half (40.4%) of the 52 RV hot spots in HeLa cells contained 2 independent integrations in less than 2 kb, compared with only 1 of the 33 LV hot spots.
Schematic representation of the maximum distance between individual hits within RV and LV hot spots. Symbols represent single hot spots originated from 2 (♦), 3 (♦), or 4 (◇) hits in the genome of CD34+ HSCs (1030 RV and 849 LV integrations) and HeLa cells (869 RV integrations), plotted according to the maximum distance between individual integrations (in base pairs, log scale). Also shown are “false positive” hot spots generated by applying the definition criteria to a library of LM-PCR–amplified random sequences of human CD34+ DNA (798 sequences). A total of 26.0% of the 97 RV hot spots in CD34+ cells and almost one-half (40.4%) of the 52 RV hot spots in HeLa cells contained 2 independent integrations in less than 2 kb, compared with only 1 of the 33 LV hot spots.
Proto-oncogenes and cancer-associated CISs are hot spots of RV but not LV integration
The list of RV integration hot spots in CD34+ cells includes proto-oncogenes (eg, LYL1, MYB), cancer-associated CISs (eg, FLI1, EVI2A, EVI2B, NF1), and genes involved in chromosomal translocations in hematopoietic malignancies (eg, LMO2, MKL1, ETV6) (Table 2), all of them occurring at frequencies significantly higher than expected (P < .001) and higher than in the overall list of RV integrations (Figure 5). Interestingly, nonexpanded cell populations contributed with at least 1 integration to 9 (53%) of the 17 hot spots containing a proto-oncogene or a cancer-associated CIS (Table 2), again indicating the absence of biases related to the number of cell doublings in culture. On the contrary, LV hot spots showed little enrichment for proto-oncogenes or CISs, although in this case, low numbers make comparisons poorly significant (Figure 5). Furthermore, RV but not LV hot spots included a very high proportion of genes belonging to the intracellular signaling cascade category (25.3%), which were significantly overrepresented using either the human genome or the total RV integrations as a background population in a GO analysis (EASE score, 1.2 × 10−6 and 2.2 × 10−4, respectively), despite their relatively small number (ie, 22). Interestingly, genes involved in hematopoietic and immune system development and in immune response by Ingenuity pathway analysis were further and significantly enriched in RV hot spots with respect to the entire list of RV integrations (P < .1; Figure 4B).
RV and LV hot spots containing at least 1 proto-oncogene and/or cancer-associated CIS
| Chromosome . | Range, bp . | No. hits . | Gene symbol . | Origin (no. hits) . |
|---|---|---|---|---|
| RV hot spots | ||||
| 14q24.3 | 13 882 | 4 | C14orf43, PNMA1* | CB-RV (2) CB-SIN-RV (2) |
| 11p13 | 48 661 | 3 | AF116668, LMO2* | BM-ADA (1) CB-RV (1) CB-SIN-RV (1) |
| 17q11.2 | 7 827 | 3 | EVI2A,*EVI2B, *NF1,*OMG | BM-X-SCID (1) CB-RV (2) |
| 10q25.2 | 1 920 | 2 | ADD3* | BM-ADA (1) CB-SIN-RV (1) |
| 11q23.2 | 22 851 | 2 | ZBTB16* | BM-ADA (1) CB-RV (1) |
| 11q24.3 | 14 147 | 2 | FLI1* | BM-ADA (1) CB-RV (1) |
| 12p13.2 | 7 360 | 2 | ETV6* | CB-RV (2) |
| 16p13.11 | 18 559 | 2 | ABCC1* | BM-ADA (1) CB-RV (1) |
| 19p13.13 | 137 | 2 | BTBD14B, LYL1,*NFIX,*TRMT1 | BM-X-SCID (1) CB-RV (1) |
| 20p12.3 | 136 | 2 | PLCB1* | CB-SIN-RV (2) |
| 20q13.12 | 19 100 | 2 | C20orf121, PKIG,*SERINC3* | CB-RV (2) |
| 22q13.1 | 29 588 | 2 | AB051446, MKL1,*RUTBC3* | BM-X-SCID (1) CB-SIN-RV (1) |
| 2p11.2 | 779 | 2 | CAPG,*LOC284948, RBED1 | PB-ND (2) |
| 2p21 | 975 | 2 | AK025445, MGC40574, THADA,*ZFP36L2* | CB-RV (2) |
| 4p14 | 11 999 | 2 | N4BP2, RHOH* | CB-RV (2) |
| 6q23.3 | 9 422 | 2 | MYB* | CB-RV (2) |
| 6p24.3 | 1 991 | 2 | RREB1* | CB-RV (1) CB-SIN-RV (1) |
| LV hot spots | ||||
| 9q34.3 | 31 043 | 3 | AK130247, C9orf163, INPP5E, NOTCH1,*PMPCA, DCCAG3 | CB-SIN-LV (2) CB-LV (1) |
| 2p21 | 22 106 | 2 | THADA* | CB-SIN-LV (1) CB-LV (1) |
| 20p12.3 | 24 132 | 2 | PLCB1* | CB-LV (2) |
| 17p13.3 | 25 818 | 2 | RUTBC1, SMG6,*SRR, TSR1 | CB-LV (2) |
| Controls | ||||
| 6q25.1 | 4 561 | 2 | ESR1* | — |
| Chromosome . | Range, bp . | No. hits . | Gene symbol . | Origin (no. hits) . |
|---|---|---|---|---|
| RV hot spots | ||||
| 14q24.3 | 13 882 | 4 | C14orf43, PNMA1* | CB-RV (2) CB-SIN-RV (2) |
| 11p13 | 48 661 | 3 | AF116668, LMO2* | BM-ADA (1) CB-RV (1) CB-SIN-RV (1) |
| 17q11.2 | 7 827 | 3 | EVI2A,*EVI2B, *NF1,*OMG | BM-X-SCID (1) CB-RV (2) |
| 10q25.2 | 1 920 | 2 | ADD3* | BM-ADA (1) CB-SIN-RV (1) |
| 11q23.2 | 22 851 | 2 | ZBTB16* | BM-ADA (1) CB-RV (1) |
| 11q24.3 | 14 147 | 2 | FLI1* | BM-ADA (1) CB-RV (1) |
| 12p13.2 | 7 360 | 2 | ETV6* | CB-RV (2) |
| 16p13.11 | 18 559 | 2 | ABCC1* | BM-ADA (1) CB-RV (1) |
| 19p13.13 | 137 | 2 | BTBD14B, LYL1,*NFIX,*TRMT1 | BM-X-SCID (1) CB-RV (1) |
| 20p12.3 | 136 | 2 | PLCB1* | CB-SIN-RV (2) |
| 20q13.12 | 19 100 | 2 | C20orf121, PKIG,*SERINC3* | CB-RV (2) |
| 22q13.1 | 29 588 | 2 | AB051446, MKL1,*RUTBC3* | BM-X-SCID (1) CB-SIN-RV (1) |
| 2p11.2 | 779 | 2 | CAPG,*LOC284948, RBED1 | PB-ND (2) |
| 2p21 | 975 | 2 | AK025445, MGC40574, THADA,*ZFP36L2* | CB-RV (2) |
| 4p14 | 11 999 | 2 | N4BP2, RHOH* | CB-RV (2) |
| 6q23.3 | 9 422 | 2 | MYB* | CB-RV (2) |
| 6p24.3 | 1 991 | 2 | RREB1* | CB-RV (1) CB-SIN-RV (1) |
| LV hot spots | ||||
| 9q34.3 | 31 043 | 3 | AK130247, C9orf163, INPP5E, NOTCH1,*PMPCA, DCCAG3 | CB-SIN-LV (2) CB-LV (1) |
| 2p21 | 22 106 | 2 | THADA* | CB-SIN-LV (1) CB-LV (1) |
| 20p12.3 | 24 132 | 2 | PLCB1* | CB-LV (2) |
| 17p13.3 | 25 818 | 2 | RUTBC1, SMG6,*SRR, TSR1 | CB-LV (2) |
| Controls | ||||
| 6q25.1 | 4 561 | 2 | ESR1* | — |
Range indicates the maximum distance between hits contained in each hot spot. For the complete list of hot spots, see Table S5.
CB-RV indicates CB-derived CD34+ cells transduced with wt-LTR RV; CB-SIN-RV, CB-derived CD34+ cells transduced with SIN-LTR RV; BM-ADA, BM-derived CD34+ cells from patients with ADA-SCID transduced with wt-LTR RV; BM-X-SCID, BM-derived CD34+ cells from patients with X-SCID transduced with wt-LTR RV; PB-ND, PB-derived CD34+ cells from healthy donor transduced with wt-LTR RV; CB-LV, CB-derived CD34+ cells transduced with wt-LTR LV; and CB-SIN-LV, CB-derived CD34+ cells transduced with SIN-LTR LV; —indicates not applicable.
Indicates proto-oncogene or CIS.
Discussion
RV integration preferences have significant consequences on the potential genotoxicity of different families of vectors used to transfer genes into HSCs. The probability of dominant activation of potentially cancer-causing genes (eg, those involved in the control of stem-cell self-renewal, growth, and differentiation) may in fact differ significantly between RV and LV vectors simply based on the different frequency by which they may target those genes. Here we report a detailed analysis of the RV and LV integration preferences in human CB-, PB- and BM-derived CD34+ HSCs transduced in the same conditions used in clinical applications and analyzed without selection. The general integration preferences of the 2 vector families were similar to those previously described for other mammalian hematopoietic or nonhematopoietic cells (reviewed in Bushman et al8 ), and showed on average a 2-fold higher probability for RV vectors to target gene-dense regions, highly active genes, and promoter-proximal regions. However, RV but not LV integration occurs at high frequency (> 20%) at genomic locations (hot spots) that are significantly enriched in proto-oncogenes and genes involved in the control of cell proliferation.
A high frequency of hot spots, defined by a statistical criterion previously applied to define cancer-associated CISs,11,20 appears to be a hallmark of RV integration in human CD34+ HSCs. We found that more than one-fifth of the RV integrations meet the definition criteria, a frequency more than 7-fold higher than expected from the analysis of a randomly cloned collection of human DNA sequences, and almost 3-fold higher than that found in a collection of LV integrations of comparable size. The average extension of RV hot spots (ie, the maximum distance between all insertions within each spot) was well within the definition criteria, and significantly smaller than that of LV hot spots, spanning less than 10 kb in half of the cases and less than 2 kb in one-fourth of the cases. RV integration appears therefore to have high preference for restricted genomic locations, which may exhibit specific chromatin conformations or features that favor tethering of the preintegration complexes (PICs) with higher probability. These features do not include gene density, proximity to promoters, or gene expression per se, since hot spots integrations show exactly the same preferences observed in the entire collection of RV integrations. Interestingly, we observed that the frequency of hot spots increased progressively during the study, following the increase of the sample size in an almost linear fashion. This may suggest that by analyzing a much higher number of sequences, all RV integrations could be clustered in a defined subset of genomic regions, all having the appropriate features recognized by the PICs. Unfortunately, the molecular bases of the interactions between RV PICs and the mammalian chromatin are poorly understood, and it is difficult to correlate our finding with any specific mechanism. The situation was completely different in the case of LV hot spots, the frequency of which increased only slightly with the increase in the sample size and appeared to plateau. More importantly, insertions in LV hot spots showed strikingly different characteristics with respect to the general LV integration preferences, and were greatly enriched in gene-dense regions and expressed genes. These data suggest that LV integration may happen in a much wider portion of the HSC genome, and that hot spots are generated at low frequency by locations that are more favorable than others to PIC interaction, and are apparently those with a high density of expressed genes. This explanation is consistent with the available evidence that LV PICs are tethered to the human genome by a widely distributed chromatin component loosely associated with gene activity, such as chromatin-remodeling30 or DNA-repair31 complexes, high mobility group (HMG)32 and polycomb group proteins,33 and LEDGF.34,35
Previous studies carried out in patients4 as well as in animal models12,13,36 have indicated that integrations in cancer-associated CISs and growth-controlling genes are enriched in the progeny of RV-transduced, repopulating HSCs. The major conclusion of these studies was that certain viral insertions lead to clonal selection of stem/progenitor cells in vivo. However, the pretransplantation frequency of these insertion events was never accurately measured in the relevant cell population. Our analysis indicates that a bias toward integration into or around certain category of genes (ie, those involved in signal transduction, cell cycle, chromatin remodeling, and transcription) is already present in nontransplanted hematopoietic progenitors, and particularly in integration hot spots. In particular, proto-oncogenes and cancer-associated CISs are enriched at 3- to 5-fold the expected frequency in RV hot spots, indicating a specific preference for genomic locations containing these categories of genes. These include proto-oncogenes expressed in CD34+ hematopoietic progenitors and involved in hematopoietic cell neoplasia, such as LMO2 and EVI2-NF1, targeted at a frequency of approximately 1:350; LYL1 and MYB, targeted at a frequency of approximately 1:500; and others (Table 2). Importantly, there was no significant difference in the number of integrations contributing to oncogene-containing hot spots between nonexpanded (BM- and PB-derived) or moderately expanded (all CB-derived) cell populations, arguing against the likelyhood of clonal outgrowth generated in culture by insertional activation of growth-promoting genes.
A network-based pathway analysis indicates that a significant number of genes targeted by RV integration are functionally linked in transcription-, signal transduction–, apoptosis-, and tumorigenesis-related networks. Interestingly, genes involved in hematopoietic and immune system development are targeted at uniquely high frequency by RV integrations, and further enriched in RV hot spots, suggesting that the gene expression program of a cycling hematopoietic cell is at least in part instrumental in directing RV PICs in certain regions of the genome. Consistent with this hypothesis, almost none of the genes present in CD34+ cells hot spots are found in hot spots from HeLa cells, which most likely operate different regulatory networks. Kustikova et al36 reached similar conclusions in compiling their “insertional dominance database” from the progeny of serially transplanted HSCs in mice, although they explain the observed overrepresentation of certain gene categories and functional networks with in vivo selection rather than with intrinsic properties of the RV integration machinery. Indeed, 18% to 34% of the genes present in the mouse database, depending on the stringency of the comparison, are present also in our list, arguing against an exclusive role for in vivo selection in determining most of the frequency biases. A notable exception is the EVI1-MDS1 locus, which we found only once in nontransplanted cells, although it was found at exceedingly high frequencies in vivo in mice,12,36 nonhuman primates,13 and, at least in 1 case, humans.4 Insertional activation of the EVI1-MDS1 locus should therefore be considered a factor favoring clonal amplification and/or selection in vivo independently from the frequency by which it is targeted by RV integration before transplantation. It should be noted, however, that our data come from a population of hematopoietic progenitors in which the proportion of repopulating stem cells is admittedly low, leaving the possibility that stem cell–specific hot spots went undetected. Unfortunately, an integration analysis in pretransplantation, long-term repopulating stem cells is currently impossible, and it is therefore difficult to come to definitive conclusions as to what proportion of the biases detected in the stem cell progeny in vivo is due to vector preferences, and what proportion is due to in vivo selection. We favor a predominant role of vector-specific factors, also based on our experience with patients with ADA-SCID in whom pretransplantation and posttransplantation integration preferences showed essentially overlapping patterns (A. Aiuti, B.C., A.R., F.M. et al, manuscript submitted).
In conclusion, this study shows previously unrecognized features of RV and LV integration into human HSCs that may have an impact in assessing the prospective genotoxic risk of using either vector system for human gene therapy applications. In particular, the frequency and characteristics of integration hot spots may be substantial factors in determining a differential safety profile for RV and LV vectors of comparable design and content.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from Telethon Italy (GGP06101 and TIGET), the European Commission (VI FP, CONSERT), and Fondazione Cariplo.
Authorship
Contribution: C.C., A.R., and F.M. designed the research and wrote the paper; C.C, G.F., D.S., A. Antonelli, A.M., S.H., and B.C. performed research and analyzed data; M.S., C.v.K., A.T., A. Aiuti, and G.F. contributed vital reagents and data sets.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 287, 41100 Modena, Italy, Phone: +39-059-2055392, Fax: +39-059-2055410; e-mail: fulvio.mavilio@unimore.it.