Abstract
Overexpression of wild-type MN1 is a negative prognostic factor in patients with acute myeloid leukemia (AML) with normal cytogenetics. We evaluated whether MN1 plays a functional role in leukemogenesis. We demonstrate using retroviral gene transfer and bone marrow (BM) transplantation that MN1 overexpression rapidly induces lethal AML in mice. Insertional mutagenesis and chromosomal instability were ruled out as secondary aberrations. MN1 increased resistance to all-trans retinoic acid (ATRA)–induced cell-cycle arrest and differentiation by more than 3000-fold in vitro. The differentiation block could be released by fusion of a transcriptional activator (VP16) to MN1 without affecting the ability to immortalize BM cells, suggesting that MN1 blocks differentiation by transcriptional repression. We then evaluated whether MN1 expression levels in patients with AML (excluding M3-AML) correlated with resistance to ATRA treatment in elderly patients uniformly treated within treatment protocol AMLHD98-B. Strikingly, patients with low MN1 expression who received ATRA had a significantly prolonged event-free (P = .008) and overall (P = .04) survival compared with patients with either low MN1 expression and no ATRA, or high MN1 expression with or without ATRA. MN1 is a unique oncogene in hematopoiesis that both promotes proliferation/self-renewal and blocks differentiation, and may become useful as a predictive marker in AML treatment.
Introduction
Recently, we have found that MN1 overexpression is a prognostic marker in patients with acute myeloid leukemia (AML) with normal karyotype1 characterized by an intermediate prognosis. Patients with high MN1 expression had a significantly worse prognosis compared with patients with low MN1 expression. High MN1 expression has as well been associated with other AML characteristics like inv(16) or overexpression of EVI-1.2,3 This suggested to us that MN1 may play a functional role in the pathogenesis of AML. MN1 is a transcription factor first identified as a fusion partner of TEL in t(12;22) AML.4 Oncogenic potential has been established for the MN1/TEL fusion protein but not for MN1 alone.5,6 MN1 does not contain any of the known transcription factor domains, but several transcriptional activation domains have been described.7 Interestingly, MN1 locates to retinoic acid response elements (RAREs), and has been implicated as a transcription cofactor of the retinoic acid receptor/retinoic X receptor (RAR/RXR) complex.7
AML is a heterogeneous clonal disorder marked by accumulation of undifferentiated myeloid blasts. In our current understanding, 2 distinct genetic events are necessary to consign properties of (1) self renewal and (2) impaired differentiation to leukemic cells, known as the 2-hit model of leukemogenesis.8 In the last 15 years, concepts have been developed to overcome the impaired differentiation in AML using differentiation-inducing agents like granulocyte-colony stimulating factor or all-trans retinoic acid (ATRA) in addition to conventional chemotherapy. In acute promyelocytic leukemia (APL), a specific form of AML characterized by the fusion proteins PML-RARα or PLZF-RARα, addition of ATRA has revolutionized the treatment, as most of the patients are cured.9 Retinoic acid and its receptor RARα regulate myeloid differentiation. In the absence of its ligand, RARα recruits transcriptional corepressors to its target genes and inhibits transcription.10 Binding of ATRA induces conformational changes in the receptor, which causes dissociation of corepressors and promotes recruitment of coactivators, allowing transcription of target genes.11 In APL, the gene PML is fused to RARA, making it resistant to physiologic concentrations of ATRA.12,13 Previous evidence suggests that RARα target genes remain repressed by the fusion protein, and myeloid differentiation is blocked.14 However, at pharmacologic concentrations of ATRA, RARα is able to recruit the activator complex, and APL cells can terminally differentiate.12,15 In a variant form of APL, RARA is fused to PLZF, and these fusions render RARα insensitive even to pharmacologic levels of ATRA.16,17 However, in non-APL AML, a therapeutic benefit of differentiation-inducing agents has not been clearly established,18-23 and leukemic cells are considered resistant to these agents. We investigated whether MN1 plays a role in RAR signaling and in myeloid differentiation induced by ATRA in hematopoietic cells.
We found that MN1 is a potent and presumably sufficient oncogene in murine hematopoiesis by promoting self-renewal and blocking myeloid differentiation. Strikingly, MN1 increased resistance to ATRA-induced differentiation by more than 3000-fold. Resistance to ATRA could be overcome by fusing a transcriptional activator to MN1, suggesting that MN1 blocks differentiation by repression of gene transcription. Moreover, high MN1 expression in patients with AML was associated with ATRA resistance, whereas patients with low MN1 expression survived significantly longer when ATRA was added to standard chemotherapy.
Patients, materials, and methods
Retroviral vectors and vector production
The 4-kb full-length cDNA of human MN1 was subcloned from pMN50 (kindly provided by Dr Zwarthoff, Rotterdam, the Netherlands) into the NotI site of expression vector pSF9124 upstream of the internal ribosomal entry site (IRES) and the enhanced GFP green fluorescent protein gene. As a control, the pSF91 vector carrying only the IRES-enhanced GFP cassette was used. The correct sequence of MN1 was verified (corresponding to GenBank entry X82209.2). The MN1-VP16 fusion gene was generated by subcloning the region encoding the activation domain of VP16, obtained by PCR amplification of pVP16 (Stratagene, La Jolla, CA), and ligating it in-frame to the C-terminus of MN1 in the pSF91-MN1 vector. Constructs were validated by sequencing, and correct expression and transmission were confirmed by Western blot and Southern blot analysis. Generation of recombinant ecotropic retrovirus-producing GP + E86 cells was performed as previously described.25
Mice and retroviral infection of primary bone marrow cells
Parental strain mice were bred and maintained as approved by the University of British Columbia Animal Care Committee. Primary mouse bone marrow (BM) cells were transduced as previously described.26 Briefly, BM cells were harvested from mice treated for 4 days with 150 mg 5-fluorouracil/kg (Faulding, Underdaler, Australia) and stimulated for 48 hours in Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 10 ng/mL human interleukin-6 (hIL-6), 6 ng/mL murine IL-3 (mIL3), and 100 ng/mL murine stem-cell factor (mSCF; all from StemCell Technologies, Vancouver, BC, Canada). The cells were infected by cocultivation with irradiated (4000 rad) GP + E86 viral producer cells in the presence of 5 μg/mL protamine sulfate (Sigma, Oakville, Ontario, Canada). Donors of primary BM cells were C57BL/6J Ly5.1-Pep3b mice, and recipients were C57Bl mice (Ly5.2+/Ly5.2+).
BM transplantation and monitoring of recipients
Purified GFP+ cells (1 × 106/mouse) were injected into the tail vein of lethally irradiated recipient mice that were exposed to a single dose of 810 rad total-body irradiation accompanied by a life-sparing dose of 2.5 × 105 C57Bl BM cells. Viability of mice was monitored daily. At time of death spleens were weighed, and white blood cells (after red blood cell [RBC] lysis in 3% acetic acid) and RBCs were counted using a hemocytometer. Lineage distribution was determined by fluorescence-activated cell-sorter (FACS) analysis (FACSCalibur; Becton Dickinson, Mississauga, ON, Canada) as previously described.26 Monoclonal antibodies used were phycoerythrin (PE)-labeled Gr-1, Mac-1, B220, CD4/CD8, Ter119, c-kit, and Sca-1 (Pharmingen, San Diego, CA); or Cy5-labeled IgE (kindly provided by Dr Krystal, Vancouver, BC, Canada). For comparability, the proportion of PE+ cells was determined by setting the PE+ gate on unstained cells so that 1% of these cells were included. Morphologic and histologic analysis of peripheral blood (PB), BM, and spleen cells was performed as previously described.26 Images were visualized using a Nikon Eclipse 80i microscope (Nikon, Mississauga, ON, Canada), and a 100 ×/1.4 numerical aperture objective, with Zeiss Immersol medium. A Nikon Coolpix 4500 camera (Nikon) and Canon ZoomBrowser EX 2.0 software (Canon, Mississauga, ON, Canada) were used to capture images.
In vitro assays
Cell proliferation was assessed in 15% FBS containing DMEM supplemented with 10 ng/mL hIL-6, 6 ng/mL mIL-3, and 100 ng/mL mSCF. All culture media and growth factors were obtained from StemCell Technologies. Cells were maintained at a cell density below 1 × 106/mL and were counted with the Vi-Cell XR Cell Viability Analyzer (Beckman Coulter, Fullerton, CA). In vitro cytotoxicity assays and in vitro differentiation assays were performed as described.1,27 Cells were plated at a density of 4 × 104 cells/mL and incubated under light-protective conditions. ATRA (Sigma) was dissolved in DMSO (Sigma), and added to the culture medium at the specified concentrations as 1/1000th of the final volume. For prolonged culture in the presence of ATRA, cells were propagated in fresh medium and ATRA or DMSO was re-added every 72 hours.
Clonogenic progenitor assay
Colony-forming cells (CFCs) were assayed in methylcellulose (Methocult M3434; StemCell Technologies) as described previously.25 For each plating, 4000 viable cells/well were plated in duplicate. Colonies were evaluated microscopically 10 to 11 days after plating by using standard criteria.
RT-PCR
RNA was extracted and reverse transcribed as previously described.1 Conventional or quantitative reverse transcription-polymerase chain reaction (RT-PCR) was done as previously described using either the iCycler (Bio-Rad, Hercules, CA; for conventional RT-PCR, the LightCycler (Roche Diagnostics, Mannheim, Germany; used for quantification of MN1 expression in patient samples), or the 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA; used for gene expression studies in murine BM cells).1,28 Relative expression was determined with the second derivative maximum method using the LightCycler Relative Quantification Software as described1 or the 2−ΔΔCT method,29 and the housekeeping Abl1 gene transcript was used to normalize the results. Primers were manufactured by Invitrogen (Burlington, ON, Canada). Primer sequences (5′ to 3′) are provided in Table S3, available on the Blood website (see the Supplemental Materials link at the top of the online article).
Immunoblotting and antibodies
Western blot analysis was performed as previously described.30 Primary antibodies used were goat polyclonal anti-MN1 (catalog no. sc-27349; Santa Cruz Biotechnology, Santa Cruz, CA) and mouse monoclonal anti-GAPDH (catalog no. TRK5G4-6C5; Research Diagnostics, Concord, MA). Secondary horseradish peroxidase-conjugated antibodies used were bovine antigoat (catalog no. sc-2352; Santa Cruz Biotechnology) and donkey antimouse (catalog no. A9044; Sigma).
Southern blot analysis
Genomic DNA was isolated using DNAzol reagent as recommended by the manufacturer (Invitrogen).30 Proviral integrity was confirmed by digesting genomic DNA with the long-terminal repeat (LTR)-specific NheI restriction endonuclease (New England Biolabs, Ipswich, MA), and probed with a GFP-specific probe (8.1 kb). The clonality of leukemic cells was assessed by digestion with the proviral-unique XbaI restriction endonuclease (New England Biolabs), and probed with a GFP-specific probe.
Retroviral integration site analysis
Retroviral integration site analysis was performed as described in detail elsewhere.31 Genomic DNA (1 μg) from leukemic MN1 mice was digested with AseI (New England Biolabs), and the fragments were ligated overnight at room temperature to a double-stranded bubble linker. Nested PCR was performed on one tenth of the ligation products. PCR-A used a linker-specific primer (Vectorette primer 224 5′-CGAATCGTAACCGTTCGTACGAGAATCGCT-3′) and a primer specific for the SF91 vector but not amplifying mouse DNA (SF91-A 5′- GTCGTCACCTGTGCAGAATTGCGAACC -3′). A 1-μL aliquot of PCR-A reaction product (1/25th) was used as a template for the second nested PCR (PCR-B) using nested linker primer B (5′-TACGAGAATCGCTGTCCTCTCCTT-3′) and primer SF91-B (5′-GAACCATGGATTCCACCGTGA-3′). Individual bands of more than 714 bp were excised, subcloned into the PCR4 vector, digested with KpnI and AseI (New England Biolabs), and sequenced, if positive clones were detected. Integration sites were then identified by aligning captured genomic mouse DNA with the mouse genome.
Chromosome preparation and spectral karyotyping
Cytogenetic characterization using spectral karyotyping (SKY) was carried out as described.32 Briefly, metaphase spreads were hybridized with the SKY probe mixture for mouse chromosomes (Applied Spectral Imaging [ASI], Migdal HaEmek, Israel). Signal detection was carried out according to the manufacturer's instructions (ASI). Chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI) and mounted with Vectashield (Vector Laboratories, Burlingame, CA). For image acquisition and analysis of chromosomes, the SpectraCube system coupled to an epifluorescence microscope with a charge-coupled device (CCD) camera and SkyView software (ASI) was used. A total of 10 to 15 metaphases per cell line were analyzed.
FACS analysis of hematopoietic progenitor cell populations
MN1 and MN1VP16 cells were used approximately 30 days after transduction of 5-FU-treated murine BM cells for FACS analysis of hematopoietic progenitor cell populations. Cells were stained as described1 using the following antibodies: PE-Cy5-conjugated antimouse Gr1 (clone Ly6G-6C; BioLegend, San Diego, CA), PE-Cy5-conjugated antimouse Ter119 (clone Ter119; BioLegend), PE-Cy5-conjugated antimouse B220 (clone RA3-6B2; Pharmingen) PE-Cy5-conjugated antimouse CD3 (clone 145-2C11; Pharmingen) PE-Cy5-conjugated antimouse CD4 (clone GK1.5; Pharmingen) PE-Cy5-conjugated antimouse CD8 (clone 53-6.7; Pharmingen), PE-Cy5-conjugated antimouse IL-7R (clone A7R34; eBioscience, San Diego, CA) PE-Cy5-conjugated antimouse Sca-1 (clone D7; eBioscience), APC-conjugated antimouse c-kit (clone 2B8; Pharmingen), PE-conjugated antimouse CD16/32 (clone 2.4G2, FcγR III/II; Pharmingen), biotin-conjugated antimouse CD34 (clone RAM34; eBioscience), and PE-TexasRed-conjugated streptavidin (Pharmingen). DAPI was used to exclude dead cells. The proportion of common myeloid progenitor (CMP)-like cells (GFP+, IL-7R−, Sca1−, Lin−, ckit+, CD34+, Fc RIII/IIlo), granulocyte-macrophage progenitor (GMP)-like cells (GFP+, IL-7R−, Sca1−, Lin−, ckit+, CD34+, Fc RIII/II+), and megakaryocyte-erythroid progenitor (MEP)-like cells (GFP+, IL-7R−, Sca1−, Lin−, ckit+CD34−, Fc RIII/IIlo) from total cells was compared between MN1 and MN1VP16 cells.
Patients and treatment
This study was performed in accordance with the Declaration of Helsinki and with the approval of the review board of the Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany. Before entering the study, patients gave written informed consent. A total of 83 patients aged 60 years or older who were newly diagnosed with AML were included in this analysis. All patients were treated in the treatement trial AML HD98-B. The only exclusion criterion in the present study for patients treated in this protocol was lack of patient material. MN1 expression levels in these patients were measured as described.1 Details of the treatment protocol can be found elsewhere.23
Statistical analysis
Statistical analysis was performed as previously described.1 In brief, AML samples were dichotomized at the median MN1 expression value (normalized copy number, MN1 transcripts per ABL transcripts, 0.0676; range, 0-2.59; fold-change in expression between highest and lowest measurable value, 1356-fold), resulting in 2 expression groups: a low MN1 group with MN1 values below the median value, and a high MN1 group with MN1 values above the median value. Each of these 2 groups was subdivided according to treatment with or without ATRA. Pairwise comparisons between patient characteristics or in vitro data were performed by Student t test for continuous variables and by Fisher exact test for categorical variables. The Kaplan-Meier method and log-rank test were used to estimate the distribution of event-free survival (EFS) and overall survival (OS) and to compare differences between survival curves, respectively. The 2-sided level of significance was set at P values less than .05. Dichotomization of patients at the 25% or 75% quartile instead of the median did not improve separation of ATRA-sensitive from ATRA-resistant patients and therefore is not reported in detail. Multivariate analysis was not performed due to the low patient number per group. The statistical analyses were performed with the statistical software package SPSS 15.0 (SPSS Science, Chicago, IL).
Results
MN1 immortalizes murine bone marrow cells
The human coding sequence of MN1 was retrovirally overexpressed in murine hematopoietic BM cells (human MN1 is 86% identical to mouse predicted Mn1) driven by the spleen-focus forming virus promoter (Figure S1). Intact proviral integration and protein expression were confirmed by Southern and Western blot, respectively (data not shown; Figure 1A; Figure S2 for GFP expression). MN1-transduced BM cells rapidly outgrew nontransduced cells (all cells were GFP+ at 17 days after transduction; Figure S3), giving rise to polyclonal cell lines, which have been maintained for more than 200 days in culture. During this period, cells remained cytokine dependent. Whereas BM cells transduced with empty vector gave rise to slow-growing, IgE-expressing mast cells, MN1-transduced cells were IgE− (data not shown). Cocultured, MN1-transduced, or nontransduced BM cells did not show differences in the rate of apoptosis (AnnexinV+) 2 days after the end of infection (13.5% ± 3.2% vs 15.2% ± 1.3%, respectively; mean ± SD of 3 independent experiments). Immunophenotypically MN1 cells are characterized by high c-kit expression (99%), low Gr-1, Mac-1, and Sca-1 expressions (1%-3%), and absence of lymphoid markers. In CFC assays for myelo-erythroid differentiation, MN1 cells could be serially replated (not continued after the seventh replating) in contrast to control cells (Figure 1B).
MN1 immortalizes murine BM cells and induces AML in mice. (A) Western blot using an antihuman MN1 and antimouse Gapdh antibody in lysates of control (ctl)- and MN1-transduced BM cells. (B) Total number of GMPs in serial replating. Cumulative yield is shown for an initial plating of 4000 transduced BM cells (mean ± SD of 3 [MN1] or 2 [control] independent experiments). (C) Survival curves for mice that received transplants of MN1-transduced (n = 18, 5 independent experiments) or GFP control-transduced (n = 7, 3 independent experiments) BM cells. (D) Representative Wright-Giemsa–stained cytospin preparation of BM cells from a leukemic mouse (MN1). (E,F) Representative hematoxylin-eosin–stained tissue sections from spleen (E) and liver (F) of leukemic mice (MN1). See “Materials and methods” for image acquisition information. (G) Immunophenotype of leukemic BM cells at time of death gated on GFP+ cells (mean ± SD of 9 analyzed mice). (H) Southern blot analysis using 10 μg genomic DNA of bulk BM from leukemic MN1 mice (4 mice each of 2 independent BM transductions; panels B,C). The cDNA for eGFP was used for probing.
MN1 immortalizes murine BM cells and induces AML in mice. (A) Western blot using an antihuman MN1 and antimouse Gapdh antibody in lysates of control (ctl)- and MN1-transduced BM cells. (B) Total number of GMPs in serial replating. Cumulative yield is shown for an initial plating of 4000 transduced BM cells (mean ± SD of 3 [MN1] or 2 [control] independent experiments). (C) Survival curves for mice that received transplants of MN1-transduced (n = 18, 5 independent experiments) or GFP control-transduced (n = 7, 3 independent experiments) BM cells. (D) Representative Wright-Giemsa–stained cytospin preparation of BM cells from a leukemic mouse (MN1). (E,F) Representative hematoxylin-eosin–stained tissue sections from spleen (E) and liver (F) of leukemic mice (MN1). See “Materials and methods” for image acquisition information. (G) Immunophenotype of leukemic BM cells at time of death gated on GFP+ cells (mean ± SD of 9 analyzed mice). (H) Southern blot analysis using 10 μg genomic DNA of bulk BM from leukemic MN1 mice (4 mice each of 2 independent BM transductions; panels B,C). The cDNA for eGFP was used for probing.
Overexpression of wild-type MN1 induces AML in mice
To test the function of MN1 overexpression in hematopoiesis, 1 × 106 freshly transduced GFP+ BM cells (MN1 or control) were transplanted into lethally irradiated mice along with 2.5 × 105 recipient-type BM cells. Mice that received transplants of MN1-expressing cells rapidly died, with a median survival of only 35 days (n = 18; 5 independent experiments using 2 independent viral-producer cell lines), whereas mice that received transplants of control-transduced cells remained healthy after 170 days (n = 7, 3 independent experiments, P < .001; Fig 1C). According to the Bethesda criteria for hematopoietic neoplasms in mice,33 we diagnosed AML in all mice based on infiltration of BM (Figure 1D; 97% GFP+) and spleen (Figure 1E; 92% GFP+) by myeloid cells, spread to the liver (Figure 1F), splenomegaly, high WBC and low RBC counts (Table 1), hemorrhage of the gastrointestinal system and soft tissue apparent upon dissection of leukemic mice, more than 20% of leukemic myeloblastic cells in the BM, and immunophenotypic characteristics of immature myeloid cells (Figures 1G, S4). Leukemias were transplantable to secondary recipients with the same short latency as in the primary mice (Figure S5). Interestingly, 17% of RBCs were GFP+ at the time of leukemic death (range, 0.5%-83%). Due to the short latency to leukemic death, we evaluated the possibility that MN1 induces the leukemic phenotype by itself without the need for a collaborating hit. Accordingly, in high-resolution SKY analysis from leukemic BM we did not find recurrent secondary aberrations (n = 10 mice; Table S1). In addition, the clonality of leukemias was assessed by Southern blot analysis. A monoclonal pattern would have been expected if insertional mutagenesis had provided a collaborating aberration, leading to clonal dominance. However, analysis of DNA isolated from BM of leukemic mice revealed an oligoclonal integration pattern with 6 different patterns from 8 analyzed mice (Figure 1H). Integration analysis of expanded clonal CFCs revealed a range of integration patterns with no more than 2 integrations per clone (data not shown), consistent with oligoclonal disease. Cloning of the insertion sites did not reveal an insertion close to or within known oncogenes or tumor suppressor genes (Table S2). The median MN1 expression in leukemic mice was 1623-fold higher than the median MN1 expression in 142 patients with AML reported previously.1 Collectively, these data suggest that overexpression of MN1 in hematopoietic cells is sufficient to induce AML in mice.
Hematologic parameters of leukemic mall mice at time of death and of normal controls
| . | MN1 . | Control . |
|---|---|---|
| RBC, ×109/L, mean (range); n = 10 | 0.25 (0.8-6.0) | 9.23 (7.8-10.4)* |
| WBC, ×106/L, mean (range); n = 10 | 51.2 (5.6-96.7) | 7.7 (2.8-13.3)* |
| Spleen weight, mean g (range); n = 11 | 0.49 (0.26-0.8) | 0.06 (0.04-0.07) |
| . | MN1 . | Control . |
|---|---|---|
| RBC, ×109/L, mean (range); n = 10 | 0.25 (0.8-6.0) | 9.23 (7.8-10.4)* |
| WBC, ×106/L, mean (range); n = 10 | 51.2 (5.6-96.7) | 7.7 (2.8-13.3)* |
| Spleen weight, mean g (range); n = 11 | 0.49 (0.26-0.8) | 0.06 (0.04-0.07) |
RBC counts, WBC counts, and spleen of leukemic MN1 mice at time of death, and of normal controls.
From hematology mouse phenome database on strain C57BL/6J, Jackson Laboratory, Bar Harbor, ME (www.jax.org).
MN1 antagonizes biologic effects of ATRA in vitro
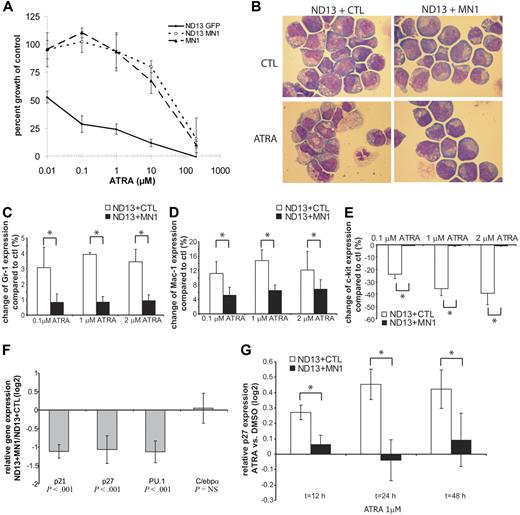
Van Wely et al described that MN1 binds to RAREs and synergizes with ATRA to activate the transcriptional activity of the Moloney sarcoma virus LTR (MSV-LTR).7 We investigated the biologic effect of ATRA and MN1 on proliferation and differentiation of a preleukemic BM cell line immortalized by the NUP98-HOXD13 (ND13) fusion protein,26 which is sensitive to the differentiation-inducing and proliferation-inhibiting effects of ATRA. ND13 cells were either transduced with empty vector or with MN1, and double-positive cells were selected for further analysis. ND13 + MN1 and ND13 + CTL cells were incubated for 3 days without or with various concentrations of ATRA to determine the concentration of ATRA that inhibits proliferation to 50% (IC50). The IC50 for ND13 + CTL was 0.013 μM ATRA, but 42 μM ATRA for ND13 + MN1 (Figure 2A; P = .007). Thus, expression of MN1 in ND13 cells increased the IC50 by 3230-fold to suprapharmacologic levels of ATRA. This effect was independent of ND13, as the same high resistance was found in cells immortalized by MN1 only (Figure 2A). Proliferation curves without or with 0.1 and 1 μM ATRA show that growth of ND13 + CTL cells is inhibited within 24 hours, and that growth of ND13 + MN1 cells is not affected at these concentrations (Figure S6). Addition of MN1 to ND13 cells changed their immunophenotype toward a more immature myeloid cell with decreased expression of Gr-1 and Mac-1, but increased expression of c-kit (Figure S7). ATRA-induced growth arrest of ND13 cells is accompanied by an increase in myelocytes and mature granulocytes (Figure 2B), and increased Gr-1 and Mac-1 expression and reduced c-kit expression (Figure 2C-E), whereas differentiation of ND13 + MN1 cells is significantly less pronounced or absent (Figure 2B-E). We next evaluated the effect of ATRA on gene expression in these cell lines. Genes known to be up-regulated during ATRA-induced growth arrest (p21,34 p2735 ) or differentiation (PU.136 ) are expressed at lower levels in ND13 + MN1 compared with ND13 + CTL cells (Figure 2F). Upon ATRA treatment, p27 is significantly more up-regulated in ND13 + CTL compared with ND13 + MN1 cells (Figure 2G). These findings suggest that in hematopoietic cells MN1 does not synergize with ATRA, but in fact opposes the biologic effects of ATRA.
MN antagonizes biologic effects of ATRA in vitro. (A) In vitro cytotoxicity assay. ND13 + MN1, ND13 + CTL, or MN1 cells were incubated at a cell density of 4 × 104 cells/mL in the presence of either DMSO (1/1000th of total volume, equal to control) or increasing concentrations of ATRA. After 72 hours, viable cells were counted. Proliferation inhibition is expressed as percentage growth of control (mean ± SD of 3 independent experiments; P = .007 for ND13 + MN1 vs ND13 + CTL). (B) In vitro differentiation assay with ND13 + MN1 or ND13 + CTL cells using either DMSO (1/1000th of total volume, equal ot control) or ATRA (1 μM). Cell cultures were propagated every 3 days, and DMSO or ATRA were re-added. Morphology was analyzed after 10 days (magnification, × 1000; see “Materials and methods” for more image acquisition information). (C-E) In vitro differentiation assay with ND13 + MN1 or ND13 + CTL cells. Percentage change of cells expressing Gr-1 (C), Mac-1 (D), or c-kit (E) in ATRA-treated (0.1, 1, or 2 μM) compared with DMSO-treated (1/1000th of total volume, equal to control) cells after incubation for 48 hours (mean ± SD of 3 independent experiments; *P < .05 for each comparison). (F) Relative gene expression of p21, p27, PU.1, and C/ebpa in ND13 + MN1 cells at baseline compared with relative gene expression of these genes in ND13 + CTL cells at baseline (mean ± SD of 3 independent experiments). (G) Change of relative gene expression of p27 over time during in vitro differentiation of ND13 + MN1 or ND13 + CTL cells comparing ATRA-treated (1 μM) to DMSO-treated cells (mean ± SD of 3 independent experiments; *P < .01 for each time point).
MN antagonizes biologic effects of ATRA in vitro. (A) In vitro cytotoxicity assay. ND13 + MN1, ND13 + CTL, or MN1 cells were incubated at a cell density of 4 × 104 cells/mL in the presence of either DMSO (1/1000th of total volume, equal to control) or increasing concentrations of ATRA. After 72 hours, viable cells were counted. Proliferation inhibition is expressed as percentage growth of control (mean ± SD of 3 independent experiments; P = .007 for ND13 + MN1 vs ND13 + CTL). (B) In vitro differentiation assay with ND13 + MN1 or ND13 + CTL cells using either DMSO (1/1000th of total volume, equal ot control) or ATRA (1 μM). Cell cultures were propagated every 3 days, and DMSO or ATRA were re-added. Morphology was analyzed after 10 days (magnification, × 1000; see “Materials and methods” for more image acquisition information). (C-E) In vitro differentiation assay with ND13 + MN1 or ND13 + CTL cells. Percentage change of cells expressing Gr-1 (C), Mac-1 (D), or c-kit (E) in ATRA-treated (0.1, 1, or 2 μM) compared with DMSO-treated (1/1000th of total volume, equal to control) cells after incubation for 48 hours (mean ± SD of 3 independent experiments; *P < .05 for each comparison). (F) Relative gene expression of p21, p27, PU.1, and C/ebpa in ND13 + MN1 cells at baseline compared with relative gene expression of these genes in ND13 + CTL cells at baseline (mean ± SD of 3 independent experiments). (G) Change of relative gene expression of p27 over time during in vitro differentiation of ND13 + MN1 or ND13 + CTL cells comparing ATRA-treated (1 μM) to DMSO-treated cells (mean ± SD of 3 independent experiments; *P < .01 for each time point).
MN1-induced inhibition of myeloid differentiation can be selectively abolished
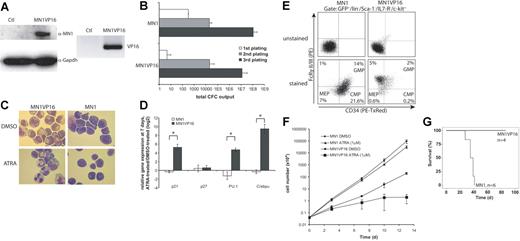
Based on the finding of reduced expression of genes associated with differentiation and cell-cycle arrest, we hypothesized that MN1 blocks myeloid differentiation by repression of gene transcription. We cloned a transcriptional activation domain (VP16) to the C-terminus of MN1 to test whether it abolishes the inhibitory effect of MN1 on differentiation and gene expression (Figures S1, 3A). MN1VP16 immortalized BM cells, which could be serially replated in colony assays (Figure 3B), and grew in culture for more than 80 days. However, their proliferation rate was lower compared with MN1 cells (data not shown). In contrast to MN1 cells, MN1VP16 cells were susceptible to differentiation. In colony assays there were more colonies of terminally differentiated macrophages, more cells expressed Gr-1 and Mac-1, and fewer cells expressed c-kit compared with MN1 cells (data not shown). Strikingly, MN1VP16 cells differentiated to mature granulocytes upon treatment with 1 μM ATRA (Figure 3C). Consistent with this observation, RARα target genes responsible for differentiation (C/ebpa, PU.1) and cell-cycle arrest (p21) were up-regulated upon ATRA treatment in MN1VP16 but not in MN1 cells (Figure 3D). As MN1VP16 cells could be differentiated by ATRA treatment, we evaluated whether the proportion of MN1VP16 cells is reduced at the CMP, GMP, and MEP levels compared with MN1 cells. The proportion of MN1VP16 cells at all 3 progenitor stages was greatly reduced, and the CMP-dominant distribution in MN1 cells is shifted toward a GMP-dominant distribution in MN1VP16 cells (Figure 3E). MN1VP16 cells were also susceptible to ATRA-induced cell-cycle arrest after treatment with 1 μM ATRA for 13 days (Figure 3F). Interestingly, mice that received transplants of MN1VP16-transduced cells remained healthy for more than 3 months after transplantation, suggesting that the leukemogenic impact of MN1 is eliminated or severely blunted when fused to VP16 (Figure 3G). Mice showed an engraftment of GFP+ cells of 56% in PB (mean of 4 mice; range, 42%-88%), decreased RBC counts, normal WBC counts in 3 mice, and signs of myeloproliferation in 1 mouse. In summary, fusion of a strong transcriptional activator to MN1 released (at least in part) the differentiation block induced by MN1. Conversely, these data suggest that wild-type MN1 confers transcriptional repressor activity to differentiation-associated genes, thus inhibiting myeloid differentiation.
MN1-induced inhibition of myeloid differentiation can be selectively abolished. (A) Western blot using an antihuman MN1 and antimouse Gapdh antibody in lysates of control (ctl)- and MN1VP16-transduced BM cells (left panels). RT-PCR using VP16 specific primers in the same cells used in the Western blot (right panel). (B) Total number of colonies in serial replating. Cumulative yield is shown for an initial plating of 4000 transduced BM cells (mean ± SD of 3 independent experiments). (C) In vitro differentiation assay with MN1 or MN1VP16 cells using either DMSO (1/1000th of total volume, equal to control) or ATRA (1 μM). Cell cultures were propagated every 3 days, and DMSO or ATRA were re-added. Morphology was analyzed after 10 days (magnification, × 1000; see “Materials and methods” for more image acquisition information). (D) Relative gene expression of p21, p27, PU.1, and C/ebpa in MN1 or MN1VP16 cells comparing ATRA-treated (1 μM) to DMSO-treated cells after 7 days of treatment (mean ± SD of 3 independent experiments; *P < .02). (E) FACS analysis of CMP-, GMP-, and MEP-like populations in MN1 and MN1VP16 cells (representative blot from 2 independent experiments). (F) In vitro cytotoxicity assay with 1 μM ATRA over time. Cumulative cell numbers were determined at the given time points for MN1 and MN1VP16 cells (mean ± SD of 3 independent experiments). (G) Survival curves for mice given transplants of MN1- or MN1VP16-transduced BM cells (n = 6 and 4, respectively; P = .004).
MN1-induced inhibition of myeloid differentiation can be selectively abolished. (A) Western blot using an antihuman MN1 and antimouse Gapdh antibody in lysates of control (ctl)- and MN1VP16-transduced BM cells (left panels). RT-PCR using VP16 specific primers in the same cells used in the Western blot (right panel). (B) Total number of colonies in serial replating. Cumulative yield is shown for an initial plating of 4000 transduced BM cells (mean ± SD of 3 independent experiments). (C) In vitro differentiation assay with MN1 or MN1VP16 cells using either DMSO (1/1000th of total volume, equal to control) or ATRA (1 μM). Cell cultures were propagated every 3 days, and DMSO or ATRA were re-added. Morphology was analyzed after 10 days (magnification, × 1000; see “Materials and methods” for more image acquisition information). (D) Relative gene expression of p21, p27, PU.1, and C/ebpa in MN1 or MN1VP16 cells comparing ATRA-treated (1 μM) to DMSO-treated cells after 7 days of treatment (mean ± SD of 3 independent experiments; *P < .02). (E) FACS analysis of CMP-, GMP-, and MEP-like populations in MN1 and MN1VP16 cells (representative blot from 2 independent experiments). (F) In vitro cytotoxicity assay with 1 μM ATRA over time. Cumulative cell numbers were determined at the given time points for MN1 and MN1VP16 cells (mean ± SD of 3 independent experiments). (G) Survival curves for mice given transplants of MN1- or MN1VP16-transduced BM cells (n = 6 and 4, respectively; P = .004).
MN1 expression levels predict ATRA resistance and sensitivity in patients with AML
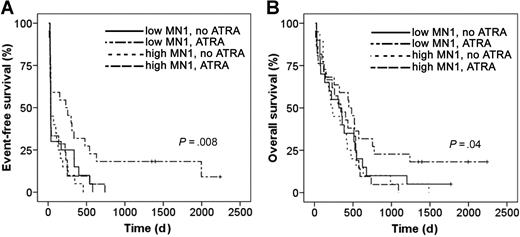
We evaluated whether MN1 expression levels and ATRA-related treatment outcome are associated in patients with AML older than 60 years treated with standard chemotherapy with or without ATRA. All patients were uniformly treated within treatment protocol AML HD98-B23 . All patients from whom RNA was available were included in the analysis. Of a total of 83 patients, 40 were treated with standard chemotherapy (idarubicin, cytarabin, and etoposide), and 43 patients were treated with standard chemotherapy plus ATRA (45 mg/m2 on days 3-5 and 15 mg/m2 on days 6-28). MN1 expression levels at diagnosis were measured by quantitative RT-PCR. Patient characteristics were equally distributed between ATRA versus no-ATRA-treated patients, including EFS and OS, except for a higher proportion of secondary AMLs in the ATRA-treated group (Table 2). Both groups were subdivided according to MN1 expression levels above or below the median MN1 expression (Table S4 for patient characteristics according to MN1 expression levels and treatment group). EFS was equally poor for patients with high MN1 expression whether they were treated with ATRA or not. EFS for patients with low MN1 expression but no ATRA treatment was as poor as for patients with high MN1 expression. However, patients with low levels of MN1 who were treated with ATRA had a significantly improved EFS compared with all other patients (P = .008, log-rank test; Figure 4A). Similarly, OS was significantly longer for patients with low MN1 expression and ATRA treatment compared with all other groups (P = .04, log-rank test; Figure 4B). Thus, high MN1 expression in patients with AML could be associated with resistance to ATRA treatment. Importantly, patients with low MN1 expression constitute a subgroup of AML that seems to benefit from the addition of ATRA to standard chemotherapy compared with standard chemotherapy alone, both in terms of EFS and OS. These findings support our preclinical data showing that MN1 induces resistance against the biologic effects of ATRA.
Patient characteristics
| . | ICE . | A-ICE . | P . |
|---|---|---|---|
| n | 40 | 43 | NA |
| Age, mean y (range) | 67.9 (60.2-84.5) | 67.6 (60.6-76.8) | .78 |
| Sex, no. M/F | 24/16 | 21/22 | .38 |
| Leukocytes, no. patients (mean ×106/mL) | 39 (33.9) | 41 (50.1) | .12 |
| LDH, no. patients (mean U/L) | 39 (620) | 39 (564) | .66 |
| Type of AML, no. patients | .03 | ||
| De novo | 36 | 30 | |
| Other | 4 | 13 | |
| Cytogenetic risk (AMLSG), no. patients | .81 | ||
| Standard | 24 | 28 | |
| High | 13 | 13 | |
| Remission, no. patients | .5 | ||
| CR | 26 | 24 | |
| No CR | 14 | 19 | |
| EFS, median d | 36 | 37.5 | .09 |
| OS, median d | 238 | 400 | .22 |
| . | ICE . | A-ICE . | P . |
|---|---|---|---|
| n | 40 | 43 | NA |
| Age, mean y (range) | 67.9 (60.2-84.5) | 67.6 (60.6-76.8) | .78 |
| Sex, no. M/F | 24/16 | 21/22 | .38 |
| Leukocytes, no. patients (mean ×106/mL) | 39 (33.9) | 41 (50.1) | .12 |
| LDH, no. patients (mean U/L) | 39 (620) | 39 (564) | .66 |
| Type of AML, no. patients | .03 | ||
| De novo | 36 | 30 | |
| Other | 4 | 13 | |
| Cytogenetic risk (AMLSG), no. patients | .81 | ||
| Standard | 24 | 28 | |
| High | 13 | 13 | |
| Remission, no. patients | .5 | ||
| CR | 26 | 24 | |
| No CR | 14 | 19 | |
| EFS, median d | 36 | 37.5 | .09 |
| OS, median d | 238 | 400 | .22 |
NA indicates not applicable.
MN1 expression levels predict ATRA resistance and sensitivity in patients with AML. (A) EFS and (B) OS of patients with AML older than 60 years treated with standard chemotherapy with or without ATRA and stratified for MN1 expression above or below the median expression (Kaplan-Meier survival analysis; log-rank test).
MN1 expression levels predict ATRA resistance and sensitivity in patients with AML. (A) EFS and (B) OS of patients with AML older than 60 years treated with standard chemotherapy with or without ATRA and stratified for MN1 expression above or below the median expression (Kaplan-Meier survival analysis; log-rank test).
Discussion
We found that MN1 overexpression rapidly induces lethal AML in mice. MN1 was first identified as the fusion partner of TEL in AML with t(12;22).4 Previous studies have therefore focused on the oncogenic potential of the fusion protein. Whereas the fusion protein transformed fibroblasts, MN1 alone did not.37 When expressed in hematopoietic cells under the control of the AML1 regulatory sequences, MN1-TEL caused T-lymphoid tumors with a long latency,6 or, in cooperation with HOXA9, AML.5 If MN1/TEL was retrovirally overexpressed, AML developed within 3 months, even if the DNA binding domain of TEL was mutated.38 Our data suggest that MN1 by itself is sufficient to cause leukemia. It is one of the most potent hematopoietic oncogenes described today. Our data suggest that MN1 has at least 2 functions: (1) to promote self-renewal and proliferation; and (2) to block differentiation. We found that the block in differentiation is associated with repression of genes associated with differentiation and cell-cycle arrest. This block could at least in part be released by masking the repressive function of MN1 with a strong transcriptional activator. Interestingly, the block in differentiation is selectively abolished, as MN1 is still able to immortalize BM cells when fused to the transcriptional activator, and these cells engraft in lethally irradiated mice. Fusion of the transcriptional repression domain of the polycomb group gene M33 to the C-terminus of MN1 aggravated the leukemic phenotype compared with MN1, thus excluding the possibility that simply the fusion of VP16 rather than its transcriptional activation function is responsible for the described effects (data not shown). This suggests that the self renewal-promoting function of MN1 is due to its ability to activate transcription, a function of MN1 described previously.7 So far, we have not determined whether these functions are encoded by different domains of MN1, or by its ability to bind different proteins, or both. MN1-transduced cells represent a powerful model to study leukemogenesis, as 2 separate functions are combined in 1 molecule and can be manipulated separately. Although MN1 appears to induce leukemia by a single hit, it does not contradict the 2-hit model of leukemogenesis due to its duplicate function of promoting self-renewal and blocking differentiation.
We found that MN1 highly increases resistance to ATRA-induced differentiation and cell-cycle arrest. Van Wely et al first described that MN1 acts as a transcriptional cofactor locating to RAREs.7 They found 1 such element in the MSV-LTR and used it as a reporter system. MN1 activated transcription synergistically with ATRA by recruiting p300 and RAC3.7 Transcriptional activation was highly dependent on context, as MN1 did not activate transcription from its binding site when it was cloned in front of the basal promoter of the herpes simplex virus TK gene. Our findings suggest that MN1 can activate transcription thereby promoting self-renewal of hematopoietic cells, although this was not studied in detail. However, we found that in MN1-overexpressing hematopoietic cells, several genes regulated by RARα (p21, p27, PU.1) are repressed and cannot be up-regulated by ATRA treatment. This finding resembles the oncogenic activity of RARα fusion proteins responsible for APL, rendering RARα less sensitive (PML-RARα) or insensitive (PLZF-RARα) to ATRA-induced differentiation.39 An appealing hypothesis is that MN1 represses RARα target genes either by directly binding to their regulatory sequences or by binding to RARα. However, the hypothesis that dysregulation of RARα target genes is driving leukemogenesis in APL has been challengend recently,40 and the exact mechanism how MN1 constitutes the block in differentiation has to be resolved in future studies. We explain the difference between our findings and the findings of van Wely et al by the context in which MN1 was investigated and the different end points used (biologic vs reporter assay). Our findings rather extend than contradict findings made by van Wely et al. Exposure to ATRA during development causes craniofacial defects like a cleft palate and hypoplastic bones.41,42 Interestingly, MN1 knockout mice develop the same craniofacial defects.43 In light of our findings, it seems likely that MN1 inhibits RARα signaling during osteogenesis to prevent detrimental effects of ATRA.
In patients with AML, high MN1 expression was associated with resistance to ATRA treatment, supporting our preclinical findings on the function of MN1. However, ATRA treatment significantly improved the outcome of patients with low MN1 expression compared with standard chemotherapy alone. Addition of ATRA to standard chemotherapy has been investigated in 4 randomized trials including adult patients with non-APL AML. Three of these trials did not detect a benefit with ATRA.18,21,22 All trials included only high-risk patients (relapsed, refractory, or poor-risk cytogenetics). As high MN1 expression is associated with treatment resistance and relapse,1 it is likely that most of these patients expressed high levels of MN1, thus preventing an effect of ATRA. The fourth study found a significant benefit of ATRA in the primary treatment of elderly patients of all risk groups.23 We analyzed a subset of these patients in which a benefit of ATRA was not yet evident (Table 1). Only after stratification for MN1 expression levels did the survival benefit in the ATRA-treated group with low MN1 expression become clear. Thus, MN1 expression levels allow selection of a target population likely to benefit from ATRA treatment. On the other hand, this approach spares patients from treatment with potentially life-threatening adverse effects who will not benefit from this treatment. Predictive markers are increasingly used in different oncology settings to increase the effectiveness of targeted therapies (eg, breast44 and lung45 cancers). Previously, markers have been identified in AML that describe the prognosis of a patient under certain treatment regimes. Unlike a prognostic marker, we describe here the use of MN1 expression as a predictive marker able to predict sensitivity to a specific drug and to guide the choice of treatment. Importantly, our findings need to be confirmed in an independent patient cohort.
In summary, we show that MN1 is a very potent oncogene in murine hematopoiesis that both promotes proliferation/self-renewal, and blocks differentiation by repressing transcription of differentiation-associated genes. Furthermore, we demonstrate that high expression of MN1 is associated with ATRA resistance in vitro and in human AML, and that patients with low MN1 expression may benefit from treatment with ATRA.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank Dr Andrew Weng for the review of histologic slides, and Patty Rosten and Adrian Wan for technical assistance.
This work was supported by grants from the National Cancer Institute of Canada with funds from the Terry Fox Foundation, Canada, the German Competence Network “Acute and Chronic Leukemia,” and the Gottfried-Arndt Stiftung Hannover, Germany. M.H. is supported by the Deutsche Forschungsgemeinschaft Germany (grant no. He 5240/1-1). B.A. is a recipient of a Leukemia Research Fund of Canada fellowship. F.K. is supported by the Deutsche Forschungsgemeinschaft Germany (grant no. Ku 2288/1-1). E.Y. is a recipient of postdoctoral fellowships from the Michael Smith Foundation for Health Research and the Canadian Institute of Health Research.
Authorship
Contribution: M.H., B.A., C.B., H.D., A.G., and R.K.H. designed the research; M.H., B.A., F.K., E.Y., J.P., S.F., K.D., C.R., T.H., and A.S. performed the research; M.H., R.F.S., C.R., B.S., and R.K.H. analyzed the data; and M.H., B.A., E.Y, and R.K.H wrote the paper. All authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: R. Keith Humphries, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, V5Z 1L3 Canada; e-mail: khumphri@bccrc.ca.

![Figure 1. MN1 immortalizes murine BM cells and induces AML in mice. (A) Western blot using an antihuman MN1 and antimouse Gapdh antibody in lysates of control (ctl)- and MN1-transduced BM cells. (B) Total number of GMPs in serial replating. Cumulative yield is shown for an initial plating of 4000 transduced BM cells (mean ± SD of 3 [MN1] or 2 [control] independent experiments). (C) Survival curves for mice that received transplants of MN1-transduced (n = 18, 5 independent experiments) or GFP control-transduced (n = 7, 3 independent experiments) BM cells. (D) Representative Wright-Giemsa–stained cytospin preparation of BM cells from a leukemic mouse (MN1). (E,F) Representative hematoxylin-eosin–stained tissue sections from spleen (E) and liver (F) of leukemic mice (MN1). See “Materials and methods” for image acquisition information. (G) Immunophenotype of leukemic BM cells at time of death gated on GFP+ cells (mean ± SD of 9 analyzed mice). (H) Southern blot analysis using 10 μg genomic DNA of bulk BM from leukemic MN1 mice (4 mice each of 2 independent BM transductions; panels B,C). The cDNA for eGFP was used for probing.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/5/10.1182_blood-2007-03-080523/6/m_zh80160706010001.jpeg?Expires=1769096887&Signature=evPEUj0L5PYYFHx-Hn3dSaEpp7WzwuUA~GdmJ9SU8F0eIHuP3LJAcV1oMzlSBpjdYcFeXSAAhIFLRGpj0KSTzuArwHmXpjNgxcvVOUL6GsXUV-V8Eoz5Vza0d3IsqyuzM9VLR9z4dZ-X7PMkAM36S5h-FK5I~vK64NzEcpLoxvXUxTFbkJLZbitNSt~BqmxLOX1niBu5apWfRYq0ck-7Gg0mcErr1dpfAtDbaZQxIYRK2n5CXe3WTjuGeXN4tL6xQo32Y6x44sR9UYnf4Mv-ngC-zzkSneBTxyUgiN-~uTGQn1ke43q0qCFxcN~kw-DX4WergE8SE8FnXKBcuaAumA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal