Abstract
To identify novel predictors of outcome in childhood acute lymphoblastic leukemia (ALL), we analyzed gene expression in the leukemic cells of 187 children with newly diagnosed ALL and compared the findings with minimal residual disease (MRD) results obtained on day 19 of remission induction treatment. Genes that showed a significant relationship to MRD were then tested for their capacity to predict leukemic relapse in an independent cohort of 99 patients. We identified 674 probe sets that were associated with MRD on day 19 (P < .006); 40 of the identified genes predicted relapse (P < .03). Among these, 14 showed independent prognostic significance after adjustment for age, leukocyte count at diagnosis, and genetic subtype. More than half of the 40 genes and nearly all of the 14 genes were functionally related, as indicated by their roles in the regulation of cell proliferation. Underexpression of genes promoting cell proliferation was associated with resistance to chemotherapy. The biologic processes regulated by the genes we identified appear to be key determinants of the early cytoreductive response to remission induction therapy and subsequent clinical outcome in childhood ALL. Incorporation of the expression levels of these genes into existing strategies of risk classification could improve clinical management.
Introduction
Precise assessment of the relapse hazard for individual patients with childhood acute lymphoblastic leukemia (ALL) followed by adjustment of treatment intensity is central to the successful management of this pediatric cancer.1 Risk classification in ALL tends to be based on the presenting clinical characteristics of the patient, such as age and leukocyte count, and the biologic features of the leukemic cells, such as immunophenotype, karyotype, and molecular genetics.1 Another useful prognostic indicator is the extent to which the leukemic cells are cleared by induction chemotherapy,2–5 a factor whose clinical importance has been confirmed many times, especially with methods capable of detecting minimal residual disease (MRD).6,7 Indeed, patients with the same risk classification based on presenting clinical and biologic features may have markedly different MRD findings after induction therapy,8–11 underscoring the need for greater refinement of risk stratification schemes.
Genome-wide gene-expression profiling offers a powerful new approach to the study of leukemia cell biology and promises to contribute significantly to the molecular classification of leukemic cells. The gene-expression profiles of leukemic lymphoblasts differ from those of other cell lineages12 and are strongly associated with the distinct immunophenotypic and genetic subtypes.13–17 Recent comparisons between gene-expression profiling and response to therapy in vitro provided clues to the genes that participate in drug resistance and influence treatment outcome.18,19 We reasoned that in vivo drug response as determined by MRD levels should be useful to identify single genes or sets of genes with a significant impact on prognosis. Here we show that a small group of genes involved in the regulation of cell proliferation and associated with MRD during early remission induction therapy independently predict outcome in childhood ALL.
Patients, materials, and methods
Patients and treatment
Bone marrow samples were collected at diagnosis from 286 children with ALL who were enrolled in St Jude Total Therapy Studies XIII, XIV, or XV20–22 and on day 19 of remission induction chemotherapy from 187 patients for MRD studies. The immunophenotypic and karyotypic features of the diagnostic samples were determined by standard techniques. BCR-ABL, E2A-PBX1, TEL-AML1 fusion, or MLL gene rearrangement were detected by reverse transcriptase-polymerase chain reaction (RT-PCR). Among the 286 ALL cases studied (187 in an initial cohort to identify genes associated with MRD and 99 in an independent cohort to test the clinical significance of the identified genes), 46 were classified to have T-lineage ALL and 240 were classified to have B-lineage ALL. The latter included 16 cases with BCR-ABL, 22 with E2A-PBX1, 17 with MLL rearrangements, 57 with TEL-AML1, 52 with hyperdiploidy (> 50 chromosomes), and 76 with other genetic abnormalities.
Patients received methotrexate alone followed 4 days later by remission induction therapy with prednisone, vincristine, daunorubicin, asparaginase, and then etoposide plus cytarabine or cyclophosphamide plus cytarabine plus mercaptopurine.20–22 After achieving complete remission, all patients received consolidation therapy with high-dose methotrexate and mercaptopurine, followed by risk-directed continuation therapy. The studies were approved by the St Jude Institutional Review Board, with informed consent obtained in accordance with the Declaration of Helsinki from the parents or guardians and assent from the patients as appropriate.
Gene-expression profiling and MRD studies
Gene-expression profiling studies were performed as previously described.13,15 Intensity values for a total of 22 283 probe sets on the U133A microarray were obtained. The same gene-expression data set was used in a previous study.23
MRD was measured by flow cytometry after bone marrow mononuclear cells were labeled with combinations of monoclonal antibodies conjugated to fluorescein isothiocyanate, phycoerythrin, peridinin chlorophyll protein, and allophycocyanin, as previously described.8,10,24 Cell staining was analyzed with a dual-laser FACSCalibur flow cytometer equipped with Cell Quest software (Becton Dickinson, San Jose, CA), according to a protocol described elsewhere.10,24 These detection methods allow the identification of 1 leukemic cell among 10 000 or more normal bone marrow cells.
Statistical analysis
The expression of individual genes was first adjusted for lineage and genetic subtypes with an analysis-of-variance (ANOVA) model; then, the t test was applied to the residual expression levels from the ANOVA model to identify genes associated with MRD without adjustment for other factors. Statistical significance in this part of the analysis was determined with the profile information criterion, an error index developed for microarray data that determines the P-value threshold that best balances false-positive and false-negative errors.25
Correlation among expression of different genes was analyzed with the Spearman correlation and simple linear regression. Correlations among gene expression, clinicobiologic features of ALL, and MRD status were performed with the Kruskal-Wallis test for multiple samples or Wilcoxon Mann-Whitney tests for 2 samples. Event-free survival and the cumulative incidence of relapse (where death in remission and second malignancy were treated as competing risks) were analyzed by a proportional-hazards regression model and by the log-rank and Gray test, respectively. The cumulative incidence of relapse, adjusting for recognized competing prognostic factors, was analyzed with a Fine and Gray model.26 All analyses were performed with SAS Version 9.1 (SAS Institute, Cary, NC) and S-plus Version 7.0.6 (Insightful, Seattle, WA) programs. Heat maps and principal component scatter plots were generated using Spotfire Decision Site 9.0 software (Spotfire, Somerville, MA).
Results
Probe sets associated with MRD on day 19 of remission induction therapy
We first compared gene-expression data for the diagnostic bone marrow samples of 187 patients enrolled on Total Therapy Study XIV or XV with the results of MRD measurements made on day 19 of remission induction therapy. In 109 patients (58.3%), at least 0.01% bone marrow mononuclear cells expressed the leukemia-associated immunophenotype identified at diagnosis. To facilitate the identification of genes associated with MRD on day 19, we first eliminated all genes whose expression correlated strongly with genetic subtypes of ALL known to influence treatment response (BCR-ABL, MLL gene rearrangements, TEL-AML1, and hyperdiploidy > 50 chromosomes) or whose maximum expression signal was below 500. We then applied a P-value threshold of .006 by t test analysis.25 By this procedure, we identified 674 probe sets whose expression was associated with MRD: 348 probe sets were overexpressed in diagnostic samples from patients with MRD on day 19, and 326 were underexpressed. We compared these results with those reported in 2 previous publications that identified genes associated with in vitro drug resistance in childhood ALL.18,19 Fourteen of the previously reported genes were also present in our series. Those with higher expression in patients with MRD on day 19 included HNPRF, HMGB2, H2AFZ, KPNA2, SNRPG, KIAA0922, U2AF2, and CCDC109B, found to be overexpressed in cases with in vitro drug resistance. Genes with lower expression in patients with MRD on day 19 included SLC2A3, MAFF, CD69, EGR1, CKS1B, and OTUB1, all underexpressed in ALL cells with in vitro drug resistance.
Probe sets that predict leukemic relapse
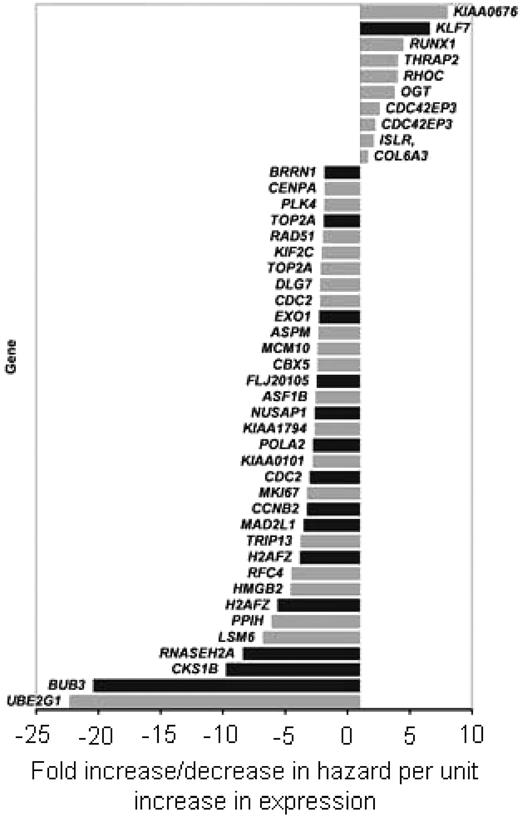
We previously found that leukemia relapse occurs in approximately one third of patients with detectable MRD on day 19.24 To identify predictors of relapse among the 674 probe sets associated with MRD on day 19, we tested each set both as a continuous variable and as a categoric variable (ie, by dividing patients into 2 groups based on gene expression above or below the median value) in a separate group of 99 patients enrolled in Total Therapy Study XIII (median follow-up, 9.8 years; range, 4.5 to 14.3 years).20 Forty-four of the 674 probe sets (corresponding to 40 genes) were significantly associated with ALL relapse (P < .03), regardless of whether they were analyzed as a continuous or categoric variable (Figure 1; Table S1, available on the Blood website; see the Supplemental Materials link atthe top of the online article). Importantly, genes expressed at lower levels in patients with MRD on day 19 were expressed at lower levels in patients with a higher risk of relapse.
Genes significantly associated with minimal residual disease (MRD) on day 19 and with leukemic relapse. Among the 674 probe sets that were significantly associated with MRD on day 19 in 187 patients (P < .006), 44 were significantly associated with leukemic relapse (P < .03) in an independent cohort of 99 patients, regardless of whether they were analyzed as a continuous or categoric variable. Bars indicate the fold increase/decrease in hazard per unit increase in expression of each gene, calculated with the proportional hazards regression model.26 Black bars correspond to genes that were independent predictors of outcome after adjustment for known prognostic features of childhood acute lymphoblastic leukemia (ALL).
Genes significantly associated with minimal residual disease (MRD) on day 19 and with leukemic relapse. Among the 674 probe sets that were significantly associated with MRD on day 19 in 187 patients (P < .006), 44 were significantly associated with leukemic relapse (P < .03) in an independent cohort of 99 patients, regardless of whether they were analyzed as a continuous or categoric variable. Bars indicate the fold increase/decrease in hazard per unit increase in expression of each gene, calculated with the proportional hazards regression model.26 Black bars correspond to genes that were independent predictors of outcome after adjustment for known prognostic features of childhood acute lymphoblastic leukemia (ALL).
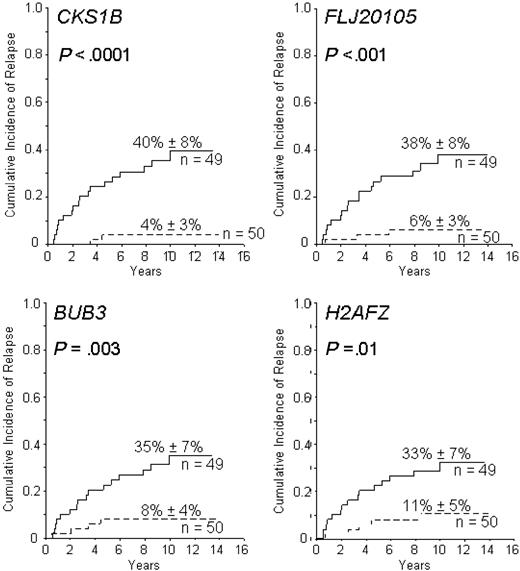
Using the same cohort of 99 patients, we next searched for probe sets whose expression remained prognostically significant after adjustment for known risk factors. In addition to the 44 probe sets, univariate analysis identified 3 conventional features (BCR-ABL, age, and leukocyte count) that predicted relapse (data not shown). When the cumulative incidence of relapse was examined with a multivariate model that treated the 44 probe sets (40 genes) as continuous variables with adjustment for BCR-ABL expression, age (< 1, 1 to 10, and ≥ 10 years), and leukocyte count (< or ≥ 50 × 109/L), 15 probe sets (corresponding to 14 genes) retained statistical significance (Table 1; Figure 1). When used as a categoric variable (ie, dividing patients into 2 groups using the median expression value), the probe sets proved to be highly informative. As shown in Figure 2 for 4 of the 14 genes, the majority of relapses occurred in the groups identified by levels of gene expression associated with MRD in the separate cohort. These genes (as a continuous variable) were also significantly associated with outcome in analyses restricted to the 90 B-lineage ALL patients (P < .005), but their predictive power among individual genetic subsets could not be determined with certainty because of the low number of patients in each subset.
Probe sets identifying independent predictors of relapse in childhood ALL
| Probe set Id . | Gene name . | Gene symbol . | Biologic process . | P* . |
|---|---|---|---|---|
| 201291_s_at | Topoisomerase (DNA) II alpha 170kDa | TOP2A | DNA replication, DNA topologic change, DNA repair | .014 |
| 203022_at | Ribonuclease H2, large subunit | RNASEH2A | DNA replication, RNA catabolism, RNA metabolism | .032 |
| 204441_s_at | Polymerase (DNA directed), alpha 2 (70-kDa subunit) | POLA2 | DNA replication | .016 |
| 201897_s_at | CDC28 protein kinase regulatory subunit 1B | CKS1B | Cell cycle | < .001 |
| 202705_at | Cyclin B2 | CCNB2 | Cell cycle | .010 |
| 203214_x_at | Cell division cycle 2, G1 to S and G2 to M | CDC2 | Cell cycle | .016 |
| 203362_s_at | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 | Cell cycle | .010 |
| 212949_at | Barren homolog 1 (Drosophila) | BRRN1 | Cell cycle | .021 |
| 204334_at | Kruppel-like factor 7 (ubiquitous) | KLF7 | Cell cycle, transcription | .015 |
| 201457_x_at | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | BUB3 | Mitosis | < .001 |
| 218039_at | Nucleolar and spindle associated protein 1 | NUSAP1 | Mitosis | .022 |
| 200853_at | H2A histone family, member Z | H2AFZ | Nucleosome assembly, chromosome organization | < .001 |
| 213911_s_at | H2A histone family, member Z | H2AFZ | .004 | |
| 204603_at | Exonuclease 1 | EXO1 | DNA repair | .004 |
| 219650_at | FLJ20105 protein | FLJ20105 | Unknown | < .001 |
| Probe set Id . | Gene name . | Gene symbol . | Biologic process . | P* . |
|---|---|---|---|---|
| 201291_s_at | Topoisomerase (DNA) II alpha 170kDa | TOP2A | DNA replication, DNA topologic change, DNA repair | .014 |
| 203022_at | Ribonuclease H2, large subunit | RNASEH2A | DNA replication, RNA catabolism, RNA metabolism | .032 |
| 204441_s_at | Polymerase (DNA directed), alpha 2 (70-kDa subunit) | POLA2 | DNA replication | .016 |
| 201897_s_at | CDC28 protein kinase regulatory subunit 1B | CKS1B | Cell cycle | < .001 |
| 202705_at | Cyclin B2 | CCNB2 | Cell cycle | .010 |
| 203214_x_at | Cell division cycle 2, G1 to S and G2 to M | CDC2 | Cell cycle | .016 |
| 203362_s_at | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 | Cell cycle | .010 |
| 212949_at | Barren homolog 1 (Drosophila) | BRRN1 | Cell cycle | .021 |
| 204334_at | Kruppel-like factor 7 (ubiquitous) | KLF7 | Cell cycle, transcription | .015 |
| 201457_x_at | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | BUB3 | Mitosis | < .001 |
| 218039_at | Nucleolar and spindle associated protein 1 | NUSAP1 | Mitosis | .022 |
| 200853_at | H2A histone family, member Z | H2AFZ | Nucleosome assembly, chromosome organization | < .001 |
| 213911_s_at | H2A histone family, member Z | H2AFZ | .004 | |
| 204603_at | Exonuclease 1 | EXO1 | DNA repair | .004 |
| 219650_at | FLJ20105 protein | FLJ20105 | Unknown | < .001 |
Association with cumulative incidence of relapse by Fine and Gray method. The expression of each gene was treated as a continuous variable and adjusted for age, leukocyte count at diagnosis, and genetic subtype.
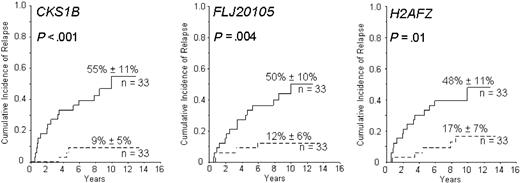
Cumulative incidence of leukemic relapse according to the expression of CKS1B, FLJ20105, BUB3, and H2AFZ, as detected by probe sets 219650_at, 201897_s_at, 201457_x_at, and 200853_at on the Affymetrix 133A gene chip, respectively. Solid lines represent patients with low gene expression and dashed lines those with expression at or above the median value; 10-year cumulative incidence of relapse (± SD) is indicated. In a multivariate analysis, each of these genes was a stronger predictor of leukemic relapse than any competing covariate included in the model.
Cumulative incidence of leukemic relapse according to the expression of CKS1B, FLJ20105, BUB3, and H2AFZ, as detected by probe sets 219650_at, 201897_s_at, 201457_x_at, and 200853_at on the Affymetrix 133A gene chip, respectively. Solid lines represent patients with low gene expression and dashed lines those with expression at or above the median value; 10-year cumulative incidence of relapse (± SD) is indicated. In a multivariate analysis, each of these genes was a stronger predictor of leukemic relapse than any competing covariate included in the model.
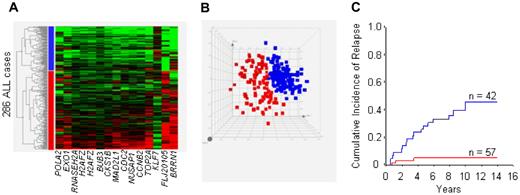
An unsupervised analysis based on the collective signals of all 15 probe sets could identify 2 distinct clusters among the 286 patients included in this study (Figure 3A,B). In an analysis performed with the cohort of 99 patients enrolled in Total XIII, the risk of leukemia relapse was markedly different among patients grouped by this gene-expression profile (P < .001; Figure 3C).
Clustering by gene-expression profiling and principal component analysis of 286 diagnostic samples of childhood ALL in relation to risk of relapse. Signals represent the 15 probe sets (14 genes) that were associated with MRD on day 19, and independently predicted outcome. (A,B) Heat map and principal component analysis. Clusters are indicated in blue and red. (C) Cumulative incidence of relapse among the 99 patients enrolled on Total XIII belonging to each cluster (P < .001).
Clustering by gene-expression profiling and principal component analysis of 286 diagnostic samples of childhood ALL in relation to risk of relapse. Signals represent the 15 probe sets (14 genes) that were associated with MRD on day 19, and independently predicted outcome. (A,B) Heat map and principal component analysis. Clusters are indicated in blue and red. (C) Cumulative incidence of relapse among the 99 patients enrolled on Total XIII belonging to each cluster (P < .001).
Relation of the 15 probe sets to CASP8AP2 expression
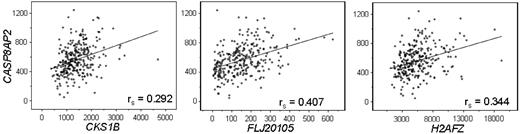
In a previous study, we found that low expression of CASP8AP2, a gene encoding a positive regulator of apoptosis, was strongly associated (P < .001) with the presence of MRD on day 46, the end of induction therapy; it was also associated with MRD on day 19, although less strongly (P = .019; a P value beyond the .006 threshold used in this study).23 The signal intensity for all 15 probe sets was directly related with that of CASP8AP2 expression (P < .01 by Spearman correlation analysis), but the relationship was far from absolute, as illustrated by the scatter plots for H2AFZ, BUB3, CKS1B, and FLJ201053 (Figure 4). We therefore asked whether the 15 probe sets identified in this study would be significantly associated with MRD after adjustment for CASP8AP2 expression in a cohort of patients with available MRD data on day 19 (n = 205) and/or on day 46 (n = 221). All 15 sets showed a significant relationship to MRD on day 19, whereas only 1 (specific for the DNA repair gene EXO1) retained this association on day 46. Conversely, CASP8AP2 expression was not significantly associated with MRD on day 19 after adjustment for any of the 15 probe sets but retained significance (P < .05) on day 46 after adjustment for expression of all but 6 of the 15 sets (data not shown).
Correlation between expression of CASP8AP2 and that of 3 of the 14 independent predictors of leukemic relapse. Plots show signal intensities as measured by the Affymetrix 133A gene chip. Spearman correlation coefficients are shown.
Correlation between expression of CASP8AP2 and that of 3 of the 14 independent predictors of leukemic relapse. Plots show signal intensities as measured by the Affymetrix 133A gene chip. Spearman correlation coefficients are shown.
High CASP8AP2 expression is strongly associated with a low risk of relapse, and intermediate and low expression are associated with a higher risk.23 If, as our findings suggest, there is only minimal overlap between the 15 probe sets and CASP8AP2 expression in MRD predictive capacity, it should be possible to demonstrate their prognostic significance in patients with defined levels of CASP8AP2 expression. Figure 5 shows the cumulative incidence of relapse, analyzed separately for each of the 4 probe sets, in 66 patients from Study XIII with low or intermediate CASP8AP2 expression. In each comparison, low expression of the candidate gene identified patients at a high risk of relapse. Thus, measurements of these genes and of CASP8AP2 provide complementary information.
Cumulative incidence of leukemic relapse according to the expression of CKS1B, FLJ20105, and H2AFZ among 66 patients with low or intermediate levels of CASP8AP2 expression. Solid lines represent patients with low gene expression and dashed lines those with expression above the median value; 10-year cumulative incidence of relapse (± SD) is indicated.
Cumulative incidence of leukemic relapse according to the expression of CKS1B, FLJ20105, and H2AFZ among 66 patients with low or intermediate levels of CASP8AP2 expression. Solid lines represent patients with low gene expression and dashed lines those with expression above the median value; 10-year cumulative incidence of relapse (± SD) is indicated.
Genes associated with MRD and treatment outcome have related functions
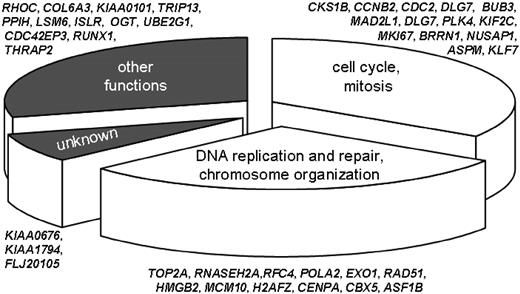
The 44 probes sets found to be associated with MRD and capable of predicting treatment outcome in the univariate analysis represented 40 discrete genes, 4 of which (CDC2, TOP2A, H2AFZ, and CDC42EP3) were duplicated (Table S1). Ten of the probe sets yielded higher signals in the cells of patients who had MRD on day 19, whereas the remaining 34 probe sets yielded lower signals in such cells. Remarkably, more than half of the 40 genes encoded proteins with closely related functions. Thus, 13 (33%) of the 40 gene products regulated the cell cycle or mitosis, and another 12 (30%) regulated DNA replication, DNA repair, or chromosome assembly (Figure 6). Overall, the genes whose expression is associated with cell proliferation were underexpressed in cases with MRD on day 19 and with a higher risk of relapse (Figure 1). Among the remaining 15 genes, 12 encode molecules with a variety of known functions, including signal transduction, transcription, and ubiquitination; only 3 encode hypothetical proteins.
Biologic processes influenced by the 40 gene products associated with MRD on day 19 and with leukemic relapse
Biologic processes influenced by the 40 gene products associated with MRD on day 19 and with leukemic relapse
The 15 probe sets that were independent predictors of outcome in the multivariate analysis corresponded to 13 named genes: 2 probe sets hybridized to the same gene (H2AFZ) and 1 hybridized to the gene encoding the FLJ20105 hypothetical protein. These 13 genes encoded proteins with known regulatory functions in cell-cycle progression, such as cyclin B2 (CCNB2), cell division cycle 2 (CDC2) kinase, CDC28 protein kinase regulatory subunit 1B (CKS1B), and Kruppel-like factor 7 (KLF7); chromatin condensation and chromosome organization, such as barren homolog 1 (BRRN1), topoisomerase II alpha (TOP2A), and H2A histone family member Z (H2AFZ); mitotic spindle formation, such as mitotic arrest deficient-like 1 (MAD2L1), budding uninhibited by benzimidazoles 3 homolog (BUB3), nucleolar and spindle-associated protein 1 (NUSAP1); and DNA replication, recombination, and repair, such as polymerase alpha 2 70kD subunit (POLA2), exonuclease 1 (EXO1), and ribonuclease H2 large subunit (RNASAH2) (Table 1). All of these genes, with the exception of KLF7, were down-regulated in cases at higher risk for suboptimal responses to remission induction therapy and subsequent leukemic relapse.
To determine whether the genes associated with treatment outcome in our correlative studies were aberrantly expressed in leukemic cells, we compared their expression in 286 cases of B-lineage ALL with that in normal CD19+CD10+ cells purified from the bone marrow mononuclear cells of 4 healthy individuals using antibodies to CD19 and CD10 and high-speed cell sorting. Such cells are normal B-cell precursors and have immunophenotypic and genotypic features closely resembling those of leukemic B-lineage lymphoblasts.27 Many leukemic samples appeared to aberrantly express these genes (Table 2). For example, expression of H2FAZ, TOP2A, CCNB2, MADL2, and NUSAP1 was lower than the normal range in more than 200 of the 286 ALL samples studied.
Probe set signal in 286 ALL samples and in normal CD19+CD10+cells
| Probe set . | Gene . | Signal expression range in normal CD19+ CD10+ cells, n=4 . | No. of ALL samples (of 286)* . | |
|---|---|---|---|---|
| Lower expression . | Higher expression . | |||
| 200853_at | H2AFZ | 7862-11838 | 245 | 8 |
| 201291_s_at | TOP2A | 1215-4378 | 262 | 0 |
| 201457_x_at | BUB3 | 3965-4452 | 168 | 85 |
| 201897_s_at | CKS1B | 1256-2012 | 127 | 32 |
| 202705_at | CCNB2 | 1661-2492 | 256 | 9 |
| 203022_at | RNASEH2A | 491-808 | 24 | 146 |
| 203214_x_at | CDC2 | 598-827 | 65 | 162 |
| 203362_s_at | MADL2 | 849-1115 | 211 | 30 |
| 204334_at | KLF7 | 887-1263 | 197 | 26 |
| 204441_s_at | POLA2 | 247-422 | 85 | 97 |
| 204603_at | EXO1 | 141-495 | 30 | 36 |
| 212949_at | BRRN1 | 184-375 | 117 | 57 |
| 213911_s_at | H2AFZ | 9096-10558 | 229 | 16 |
| 218039_at | NUSAP1 | 3722-4736 | 261 | 9 |
| 219650_at | FLJ20105 | 21-123 | 15 | 131 |
| Probe set . | Gene . | Signal expression range in normal CD19+ CD10+ cells, n=4 . | No. of ALL samples (of 286)* . | |
|---|---|---|---|---|
| Lower expression . | Higher expression . | |||
| 200853_at | H2AFZ | 7862-11838 | 245 | 8 |
| 201291_s_at | TOP2A | 1215-4378 | 262 | 0 |
| 201457_x_at | BUB3 | 3965-4452 | 168 | 85 |
| 201897_s_at | CKS1B | 1256-2012 | 127 | 32 |
| 202705_at | CCNB2 | 1661-2492 | 256 | 9 |
| 203022_at | RNASEH2A | 491-808 | 24 | 146 |
| 203214_x_at | CDC2 | 598-827 | 65 | 162 |
| 203362_s_at | MADL2 | 849-1115 | 211 | 30 |
| 204334_at | KLF7 | 887-1263 | 197 | 26 |
| 204441_s_at | POLA2 | 247-422 | 85 | 97 |
| 204603_at | EXO1 | 141-495 | 30 | 36 |
| 212949_at | BRRN1 | 184-375 | 117 | 57 |
| 213911_s_at | H2AFZ | 9096-10558 | 229 | 16 |
| 218039_at | NUSAP1 | 3722-4736 | 261 | 9 |
| 219650_at | FLJ20105 | 21-123 | 15 | 131 |
Lower refers to values below the lowest value measured in normal CD19+ CD10+ cells; higher refers to values above the highest value in normal cells.
Discussion
We used a 2-step strategy to identify new predictors of outcome in childhood ALL. In the first step, we found a group of 674 probe sets (approximately 3% of the total number in the microarray) whose expression was significantly related to the presence or absence of MRD on day 19 of remission induction therapy in 187 patients. Because the early response to therapy as measured by MRD assays is a strong prognostic indicator in ALL,24,28 we reasoned that some of the genes in this restricted list might exert considerable prognostic strength by themselves. In tests performed with a separate cohort of 99 patients, we identified 40 genes that predicted clinical outcome; 14 were independent risk factors for leukemic relapse. Strikingly, the proteins encoded by these 14 genes had highly interrelated functions in cell proliferation and could readily discriminate between 2 distinct clusters of ALL cases in an analysis extended to 286 patients. Comparison of the expression profiles of the same 14 genes in leukemic versus nonleukemic lymphoid progenitors revealed their widespread aberrant expression in the leukemic cells.
The treatment that the patients received before the MRD measurement on day 19 consisted of 5 drugs (methotrexate, prednisone, vincristine, daunorubicin and asparaginase) whose mechanisms of action provide a functional basis for several of the genes we identified. For example, our result that lower expression of TOP2A, the target of daunorubicin activity, is associated with positive MRD and a higher risk of leukemic relapse agrees well with the report that a decrease in TOP2A expression in a leukemia cell line caused a reduction in sensitivity to daunorubicin.29 In addition, low TOP2A expression was associated with an inferior outcome in breast cancer patients receiving anthracyclins30,31 and with high-risk features in childhood ALL,32 although no significant relation was found between TOP2A expression and in vitro resistance to daunorubicin and teniposide in diagnostic and relapsed childhood ALL samples.33 BUB3, MAD2L1, and NUSAP1, which participate in mitotic spindle assembly, were all expressed at lower levels in patients with MRD and an inferior outcome in our study, consistent with the spindle poison activity of vincristine. Indeed, overexpression of MAD2 has been shown to increase the cellular sensitivity to vincristine,34 and inhibition of BUBR1, another component of the mitotic spindle complex, enhances cellular resistance to this drug.35 Hence, defective mitotic spindle assembly due to underexpression of these genes in leukemic cells would likely contribute to vincristine resistance and decreased leukemia cytoreduction during remission induction therapy. Of note, lower expression of MAD2L1 was also associated with high levels of MRD at week 12 in an analysis performed in children with ALL enrolled in the ALL-Berlin-Frankfurt-Münster (BFM) 2000 study,36 in spite of the fact that this study differed from ours in the therapy administered, the timing of MRD measurement, and microarray platform used.
A substantial proportion of the genes that were underexpressed in cases with a higher likelihood of MRD and leukemic relapse in our study, such as CCNB2, CDC2, and CKS1B, are essential for cell-cycle progression, a function with direct bearing on the drug sensitivity of cancer cells. For example, in patients with follicular lymphoma who received combination chemotherapy that included vincristine, prednisone, and an anthracycline, lower expression of CDC2 and CKS1B was associated with an inferior outcome,37 in agreement with our results. Overexpression of the transcription factor KLF7, which leads to G1 growth arrest in fibroblasts and neuroblastoma cells,38 was also associated with MRD and relapse in our study. Thus, abnormal gene expression that hinders the replication of transformed lymphoid cells appears to impart resistance to chemotherapy, especially to drugs such as methotrexate, vincristine, daunorubicin, thioguanines, and cytarabine, which are less effective against nonproliferating cells.39 Possibly, the impact of these genes on chemosensitivity can only be detected by measuring the effects of multiagent chemotherapy in vivo, as genes regulating cell proliferation did not appear to have a strong association with drug sensitivity in vitro in previous studies.18,19 Nevertheless, Holleman et al18 found CSK1B to be underexpressed in cells with poorer in vitro responses to prednisolone, and Lugthart et al19 reported that low expression of H2AFZ was associated with chemotherapy cross-resistance in vitro.
In a previous study, we found that decreased expression of CASP8AP2, a positive regulator of apoptosis, was strongly related to MRD at the end of remission induction therapy (day 46) and with leukemic relapse.23 Although expression levels of the 14 genes that independently predicted outcome in this study were statistically related to CASP8AP2 expression, this link was rather tenuous in most cases. After adjustment for CASP8AP2 expression, each of these genes remained strongly associated with MRD on day 19, but only 1 showed this correlation on day 46. In the converse situation, CASP8AP2 lost its significant association with MRD on day 19 while retaining it on day 46. These results suggest that response to the first phase of remission induction therapy (when decrease in leukemic burden is greatest)40 depends on the proliferative propensity of the leukemic cells, and propensity to apoptosis assumes prominence during the subsequent weeks. Of note, Bhojwani et al41 recently reported that leukemic lymphoblasts at relapse expressed higher levels not only of antiapoptotic genes but also of genes associated with cell proliferation, including TOP2A. One possible explanation is that leukemia relapse in the cases studied by Bhojwani et al41 originated from a small cell subpopulation with a pattern of gene expression distinct from that of the presenting leukemia and undetectable at diagnosis. Alternatively, leukemic cells resistant to initial therapy may change their pattern of gene expression during the course of the disease. Since we focused our analysis on genes strongly associated with early treatment response and MRD on day 19, it is possible that other important molecular determinants of treatment outcome could have been missed in this analysis. For example, the strength of the association of CASP8AP2 expression with MRD on day 19 (P = .019) was below the threshold that we used for selecting the initial 674 probe sets for further analysis.
How could the genes showing independent prognostic significance in this study be effectively incorporated into existing systems of risk classification? One attractive strategy would be to include them with other informative genes in a leukemia chip array. If the expression of the encoded proteins correlates well with that of the corresponding transcripts, one could also measure their expression using specific antibodies and flow cytometry. The information gained should augment the predictive power of recognized clinical and biologic risk factors in childhood ALL and, when used in conjunction with MRD testing during early treatment, should increase the accuracy of risk classification and hence the selection of appropriate therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Chris Clark, Peixin Liu, and Mo Mehrpooya for technical assistance; Meyling Cheok for providing data on gene expression and in vitro drug resistance; and John Gilbert for editorial suggestions.
This work was supported by grants CA60419 and CA21765 from the National Cancer Institute and by the American Lebanese Syrian Associated Charities (ALSAC).
National Institutes of Health
Authorship
Contribution: C.F. analyzed microarray data; E.C.-S. performed MRD analysis; D.P. performed statistical analysis; C.C. performed statistical analysis; G.S. contributed to microarray analysis; C.-H.P. directed the clinical protocols supporting the study; J.R.D. directed the acquisition and analysis of microarray data; and D.C. initiated the study, coordinated the analysis, and prepared the manuscript with the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: D. Campana, Department of Oncology, St Jude Children's Research Hospital, 332 North Lauderdale, Memphis, TN 38105; e-mail: dario.campana@stjude.org.