Abstract
The inability of myeloid chronic myelogenous leukemia blast crisis (CML-BC) progenitors to undergo neutrophil differentiation depends on suppression of C/EBPα expression through the translation inhibitory activity of the RNA-binding protein hnRNP-E2. Here we show that “oncogene dosage” is a determinant factor for suppression of differentiation in CML-BC. In fact, high levels of p210-BCR/ABL are required for enhanced hnRNP-E2 expression, which depends on phosphorylation of hnRNP-E2 serines 173, 189, and 272 and threonine 213 by the BCR/ABL-activated MAPKERK1/2. Serine/threonine to alanine substitution abolishes hnRNP-E2 phosphorylation and markedly decreases its stability in BCR/ABL-expressing myeloid precursors. Similarly, pharmacologic inhibition of MAPKERK1/2 activity decreases hnRNP-E2 binding to the 5′UTR of C/EBPα mRNA by impairing hnRNP-E2 phosphorylation and stability. This, in turn, restores in vitro and/or in vivo C/EBPα expression and G-CSF–driven neutrophilic maturation of differentiation-arrested BCR/ABL+ cell lines, primary CML-BCCD34+ patient cells and lineage-negative mouse bone marrow cells expressing high levels of p210-BCR/ABL. Thus, increased BCR/ABL oncogenic tyrosine kinase activity is essential for suppression of myeloid differentiation of CML-BC progenitors as it is required for sustained activation of the MAPKERK1/2-hnRNP-E2-C/EBPα differentiation-inhibitory pathway. Furthermore, these findings suggest the inclusion of clinically relevant MAPK inhibitors in the therapy of CML-BC.
Introduction
Impaired differentiation is a common feature of many hematologic malignancies including chronic myelogenous leukemia blast crisis (CML-BC). CML is a myeloproliferative disorder caused by the constitutive tyrosine kinase activity of p210-BCR/ABL oncoprotein, the product of the t(9;22)(q34;q11) translocation.1 In the initial chronic phase (CML-CP), BCR/ABL provides a survival advantage but does not affect the differentiation of myeloid progenitors.1 However, progression from chronic phase to the acute and fatal blast crisis (CML-BC) stage impairs the ability of myeloid progenitors to terminally differentiate into mature neutrophils.1 Myeloid differentiation is controlled by a coordinate network of several transcription factors that regulate the expression of important differentiation-related genes, including those encoding growth factors and their receptors.2,3 The transcriptional factor CCAAT/enhancer-binding protein α (C/EBPα) is crucial for the differentiation of multipotent myeloid progenitors into granulocytic precursors,4-6 a process that depends, in part, on the C/EBPα-mediated transcriptional regulation of genes essential for granulocytic differentiation (eg, G-CSF receptor, myeloperoxidase, and neutrophil elastase).7-10
We recently reported that down-regulation of C/EBPα expression occurs in Ph1(+) myeloid progenitors from CML-BC patients.11 The importance of loss of C/EBPα activity as a central mechanism leading to differentiation arrest of CML-BC myeloid blasts is supported by 2 lines of evidence: (a) ectopic C/EBPα expression induces maturation of differentiation-arrested BCR/ABL+ myeloid precursors11-14 ; and (b) a CML-BC–like process emerges in mice that receive a transplant of BCR/ABL-transduced Cebpa-null, but not heterozygous or wild-type fetal liver cells.15
Genetic or functional inactivation of C/EBPα is also a common event occurring in differentiation-arrested acute myeloid leukemia blasts.16,17 Dominant-negative mutations that abrogate transcriptional activation of C/EBPα have been found in 10% of acute myeloid leukemia (AML) with normal cytogenetics.18,19 By contrast, no mutation of the CEBPA gene was detected in a screening of 95 CML-BC patients.20 In fact, loss of C/EBPα in CML-BC depends on the BCR/ABL-regulated activity of the RNA-binding protein hnRNP-E2 (PCBP2; poly(rC)-binding protein 2) that, upon interaction with the 5′ untranslated region of CEBPA mRNA, inhibits CEBPA translation.11 C/EBPα protein but not mRNA expression is downmodulated in primary bone marrow cells from CML-BC patients and inversely correlates with BCR/ABL levels.11 Accordingly, hnRNP-E2 expression inversely correlates with that of C/EBPα11 , as hnRNP-E2 levels are abundant in CML-BC but undetectable in CML-CP mononuclear marrow cells. Furthermore, in myeloid precursors expressing high levels of p210-BCR/ABL, hnRNP-E2 levels are downmodulated by imatinib treatment,11 suggesting that BCR/ABL-generated signals suppress differentiation by affecting hnRNP-E2 expression/function.
Herein, we show that hnRNP-E2 expression is induced by BCR/ABL in a dose- and kinase-dependent manner through constitutive activation of MAPKERK1/2. This, in turn, posttranslationally increases hnRNP-E2 protein stability. Furthermore, in vitro and in vivo suppression of hnRNP-E2 phosphorylation/expression by inhibition of MAPKERK1/2 activity restores C/EBPα expression and rescues G-CSF–driven differentiation of 32D-BCR/ABL cells, patient-derived CML-BCCD34+ progenitors, and primary lineage-negative (Lin−) bone marrow cells ectopically expressing high levels of p210-BCR/ABL oncoprotein.
Materials and methods
Cell cultures and primary cells
Philadelphia1 -positive K562 and EM-3 cell lines were maintained in culture in IMDM supplemented with 10% FBS and 2 mM l-glutamine. IL-3–dependent 32Dcl3 myeloid precursor and its derivative cell lines were maintained in culture in IMDM supplemented with 10% FBS, 2 mM l-glutamine, and 10% WEHI-conditioned medium as a source of mIL-3. For assays requiring cell starvation, cells were washed 4 times in PBS and incubated for 8 hours in IMDM supplemented with 10% FBS or 0.1% FBS and 2 mM l-glutamine. 32Dcl3- and 32D-BCR/ABL–derived cell lines were generated by retroviral infections followed by antibiotics-mediated selection or fluorescence-activated cell sorting (FACS)–mediated sorting of GFP+ cells as described.11 Newly established 32D-BCR/ABLhigh cells were grown in the presence of IL-3 during selection and the cultures of 17 to 25 days after retroviral transduction were used in differentiation assays. Normal murine hematopoietic marrow cells were obtained from the femurs of untreated or 5-FU–treated C57BL/6 mice after hypotonic lysis. Mononuclear cells (from 5-FU–treated mice) or lineage-negative cells (Lin−) (Miltenyi Biotech isolation protocol, Auburn, CA) were kept for 2 days in IMDM supplemented with 10% FBS, 2 mM l-glutamine, and murine recombinant cytokines (2 ng/mL IL-3, 2 ng/mL IL-6, 10 ng/mL SCF, 5 ng/mL GM-CSF, and 5 ng/mL Flt3) (R&D systems, Minneapolis, MN) before infection with MigRI-GFP or MigRI-GFP-BCR/ABL (W. Pear, University of Pennsylvania, Philadelphia, PA). Frozen samples of CD34+ bone marrow cells (NBMs) from different healthy donors were purchased from Cincinnati Children's Hospital, Division of Experimental Hematology (Cincinnati, OH). All studies performed with human specimens obtained from the Ohio State University (OSU) Leukemia Tissue Bank (Columbus, OH), and the Division of Hematology, Maisonneuve-Rosemont Hospital (Montreal, QC) were done with approval from the OSU institutional review board. Mononuclear hematopoietic cells of CML patients were ficoll-separated and cultured in IMDM supplemented with 30% FBS, 2 mM l-glutamine, and a recombinant human cytokine cocktail (IL-3 [20 ng/mL], IL-6 [20 ng/mL], Flt3-ligand [100 ng/mL], SCF [100 ng/mL]) (Stem Cell Technologies, Vancouver, BC). The CD34+ fraction was enriched by using a magnetic-activated cell sorting (MACS) CD34 kit (Miltenyi Biotec). Where indicated, cells were treated for the indicated time with the following enzyme inhibitors: 1 to 2 μM imatinib mesylate (STI571; kindly provided by Novartis Oncology, East Hanover, NJ), 25 μM U0126 (Promega, Madison, WI), 3 to 10 μM CI-1040 (kindly provided by Pfizer, New York, NY), 2 μM U73122 (Biomol, Plymouth Meeting, PA), 50 μM PD98059, 20 nM rapamycin, 50 μM LY294002, 25 μM ALLN, and 25 μM ALLM (Calbiochem, San Diego, CA).
Plasmids
pMAL-PCBP2/hnRNP-E2 was a kind gift of R. Andino (University of California, San Francisco). pMAL-hnRNP-E2S173A, pMAL-hnRNP-E2S189A, pMAL-hnRNP-E2T213A, pMAL-hnRNP-E2S272A, and pMAL-hnRNP-E2S173A,S189A,T213A,S272A, which carry single or quadruple serine/threonine to alanine mutations in the hnRNP-E2 consensus ERK phosphorylation sites [(S/T)P] were generated by site-directed mutagenesis of pMAL-hnRNP-E2 with Quikchange Mutagenesis system (Stratagene, La Jolla, CA). To construct plasmids MigRI-HA-hnRNP-E2 and MigRI-HA-hnRNP-E2S173A,S189A,T213A,S272A, wild-type hnRNP-E2 and hnRNP-E2S173A,S189A,T213A,S272A cDNAs were PCR-amplified using an upstream primer containing a BamHI site followed by the hemagglutinin (HA) epitope sequence at the 5′ end of hnRNP-E2, and a downstream primer containing a XhoI site. The PCR products were digested with BamHI and XhoI and subcloned into the MigRI vector. MigRI-HA-hnRNP-E2S173D,S189D,T213E,S272D was generated by site-directed mutagenesis of MigRI-HA-hnRNP-E2. All regions mutagenized or amplified by PCR were verified by DNA sequencing. MigRI-ERK1, MigRI-ERK1K71R, MigRI-ERK2, and MigRI-ERK2K52R were previously described.21
Differentiation assays
For induction of neutrophilic differentiation, cells were washed 3 times in PBS and seeded in IMDM supplemented with 10% FBS, 2 mM l-glutamine, and 10% U87MG-conditioned medium as a source of crude G-CSF or 25 ng/mL human recombinant G-CSF (Abazyme, Needham, MA). For differentiation assay of Lin− BMCs, cells were seeded in IMDM supplemented with 10% FBS, 2 mM l-glutamine, 0.2 ng/mL IL-3, 5 ng/mL GM-CSF, 5 ng/mL Flt3-ligand, and 25 ng/mL G-CSF. Morphologic differentiation was assayed by May-Grumwald/Giemsa staining or flow cytometric detection of the differentiation marker Gr-1 with a specific phycoerythrin (PE)–conjugated mouse monoclonal antibody (Pharmingen, San Diego, CA). The microscopy was performed using a Zeiss Axioskop 2 Plus microscope (Thornwood, NY) equipped with a Plan-Neo 40 ×/0.75 NA objective and a Canon Powershot A70 digital camera (Lake Success, NY). Images were acquired using Canon Remote Capture software and prepared for figures in Adobe Photoshop CS (San Jose, CA).
Western blot and immunoprecipitation
Cells (1 × 107) were harvested, washed with ice-cold PBS, and lysed in 100 μL 50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride supplemented with a protease inhibitor cocktail (Roche, Indianapolis, IN). For C/EBPα detection, cells (1 × 106) were washed with ice-cold PBS, lysed directly in 50 μL Laemmli buffer, and denatured (5 minutes, 100°C) prior to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (4%-15%). Antibodies used were as follows: polyclonal anti–hnRNP-E2 (R. Andino, University of California, San Francisco, CA), rabbit polyclonal anti-C/EBPα, anti-G-CSF, monoclonal anti-Abl(24–11) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phosphotyrosine, clone 4G10 (Upstate, Lake Placid, NY), anti-HSP90, anti-GRB2 (BD transduction laboratories, Lexington, KY), monoclonal anti-MBP (New England Biolabs, Beverly, MA), anti-HA (Convance, Princeton, NJ), anti-ERK1/2, and anti-pERKThr202/Tyr204 (Cell Signaling, Danvers, MA). Immunoprecipitations were performed in 20 mM HEPES, pH 7.0, 150 mM NaCl, 0.2% NP-40 supplemented with protease and phosphatase inhibitors. Lysates were precleared with protein G–agarose beads and immunoprecipitated with antibody-coated beads for 3 hours at 4°C. After washings, immunoprecipitates were subjected to Western blot analysis.
Recombinant MBP-fusion protein purification and in vitro kinase assay
Overnight BL21 cultures were diluted and grown in LB + AMP at 30°C to an OD600 of 0.5. Protein expression was IPTG induced for 3 hours at 30°C. Cells were harvested, washed, resuspended in MBP buffer (20 mM Tris-Cl, 200 mM NaCl, 1 mM EDTA, 1 mM DTT) supplemented with 0.1 M PMSF, 5 mM DTT, and a protease inhibitor cocktail, and lysed by sonication. The lysates were incubated with amylose beads (Sigma, St Louis, MO) for 4 hours at 4°C. After washings, MBP-fusion proteins were eluted with 100 mM maltose in MBP buffer. To detect phosphorylation of MBP-hnRNP-E2 by ERK1/2 in vitro, immunoprecipitated ERK1/2 or recombinant ERK1/2 (Upstate) was used in kinase assay. Kinase reactions were initiated by addition of 50 μM ATP, 20 μCi (0.74 MBq) γ-32P ATP (Perkin-Elmer, Shelton, CT), and 1 μg affinity-purified MBP fusion proteins in a final volume of 30 μL. After incubation (30 minutes; 30°C), reactions were terminated by adding 4 × Laemmli buffer.
Pulse-chase experiments
32D-BCR/ABL cells (2 × 107 cells) expressing HA-hnRNP-E2, HA-hnRNP-E2S173A,S189A,T213A,S272A, or HA-hnRNP-E2S173D,S189D,T213E,S272D were washed (4 ×) and incubated for 60 minutes in methionine-free RPMI 1640 containing 10% dialyzed FBS. Cells (5 × 106 cells/mL) were labeled in medium containing [35S]-methionine (250 μCi [9.25 MBq]/mL; 1.5 hours). After labeling, cells were washed (2 ×) with methionine-containing RPMI and cultured (105 cells/mL) for 24 hours in RPMI containing l-methionine (15 μg/mL). At different times, cells (5 × 106 cells) were harvested and lysed in isotonic buffer and immunoprecipitations were performed as described in “Western blot and immunoprecipitation” under “Materials and methods.” The immunoprecipitated proteins were visualized by autoradiography or Western blot analysis and analyzed by densitometry. The values of radioactivity were normalized.
In vivo 32P labeling
32D-BCR/ABL cells (1.5 × 107 cells) expressing HA-hnRNP-E2 or HA-hnRNP-E2S173A,S189A,T213A,S272A were washed (3 ×) with 0.9% NaCl, pH 7.4, resuspended in phosphate-free DMEM containing 0.1% bovine serum albumin and 10 mM HEPES, pH 7.4, and labeled with [32P]orthophosphate (18.5 MBq/mL; 3 hours). Following labeling, cells were washed (3 ×) with 0.9% NaCl, pH 7.4, and lysed in isotonic buffer. HA-hnRNP-E2 immunoprecipitations were carried out as described. Immunoprecipitated proteins were visualized by autoradiography or Western blot analysis.
RNA electrophoretic mobility shift assay (REMSA)
Cytoplasmic extracts from parental and BCR/ABL-expressing 32Dcl3 cells were prepared and used in REMSA as described previously.11,22 Cells (107) were harvested and lysed in 100 μL binding buffer (10 mM HEPES-KOH, pH 7.5, 14 mM KCl, 3 mM MgCl2, 5% glycerol, 0.2% NP-40, 1 mM DTT, 1 mM PMSF, 5 mM benzamidine, 1 mM Na3VO4, 50 mM NaF, 10 mM β-glycerophosphate) supplemented with protease inhibitors, and the cytoplasmic fraction was purified by centrifugation (2 minutes, at 8500g, 4°C). For REMSA, 20 μg cytoplasmic proteins was incubated with 50 000 cpm 32P-labeled WT-uORF of CEBPA ribo-oligonucleotide for 30 minutes at room temperature (RT). After an additional 20-minute incubation with heparin, the reactions were resolved on 5% PAGE.
Reverse transcription–polymerase chain reaction (RT-PCR)
Nuclear RNA (1 μg) obtained from sucrose gradient-purified nuclei by acid-phenol/guanidinium–mediated extraction was treated with DNAse I and subjected to reverse transcription using 200 U Moloney murine leukemia virus (M-MLV) reverse transcriptase (Gibco-BRL, Grand Island, NY), 200 μM dNTPs, 0.25 U/mL random hexamers (Pharmacia, Piscataway, NJ), and sets of primers specific for hnRNP-E2 or hnRNP-E1 or that recognize both hnRNP-E2 and E1 mRNA. As an internal control, all cDNA samples were adjusted to yield relatively equal amplification of β-actin.
In vivo effect of CI-1040 on neutrophilic maturation of differentiation-arrested BCR/ABL+ cells
6.15-MigR1 and 6.15-WT-uORF cells were injected (5 × 105 cells/mouse) intravenously into 10 week-old severe combined immunodeficient (SCID) mice (n = 6 per group). Mice were monitored for engraftment a week after cell injection by nested RT-PCR-mediated detection of BCR/ABL transcripts.23 CI-1040 was dissolved in 10% Cremophor EL (Sigma), 10% ethanol, and 80% water as previously reported.24,25 Mice were divided into 2 groups; one received for 2 weeks intraperitoneal injections of G-CSF (filgrastim; Amgen, Thousand Oaks, CA) (100 μg/kg/day)26,27 and CI-1040 (100 mg/kg per mouse twice a day)24 using dosage and schedules previously described by other groups, whereas the other received G-CSF and vehicle only for the same time period. After the end of treatment, mice were killed and tissue sections from spleens were fixed in formalin and embedded in paraffin blocks. Slides were stained with hematoxylin/eosin (H&E) or with chloroacetate esterase (Leder staining) to assess presence of differentiated myeloid cells. The microscopy was performed using a Zeiss Axioskop 2 Plus microscope equipped with a Plan-Neo 40 ×/0.75 NA objective and a Canon Powershot A70 digital camera. Images were acquired using Canon Remote Capture software and prepared for figures in Adobe Photoshop CS.
Results
BCR/ABL posttranslationally enhances hnRNP-E2 expression
HnRNP-E2 expression is induced upon retroviral transduction of the 32Dcl3 myeloid cells with p210-BCR/ABL and directly correlates with BCR/ABL levels and kinase activity in primary CML progenitors.11 Similarly, doxycycline treatment (1 μg/mL, 24 hours) enhances hnRNP-E2 expression in the TonB210.1 lymphoid precursor cells expressing a tetracycline-inducible p210-BCR/ABL (not shown).
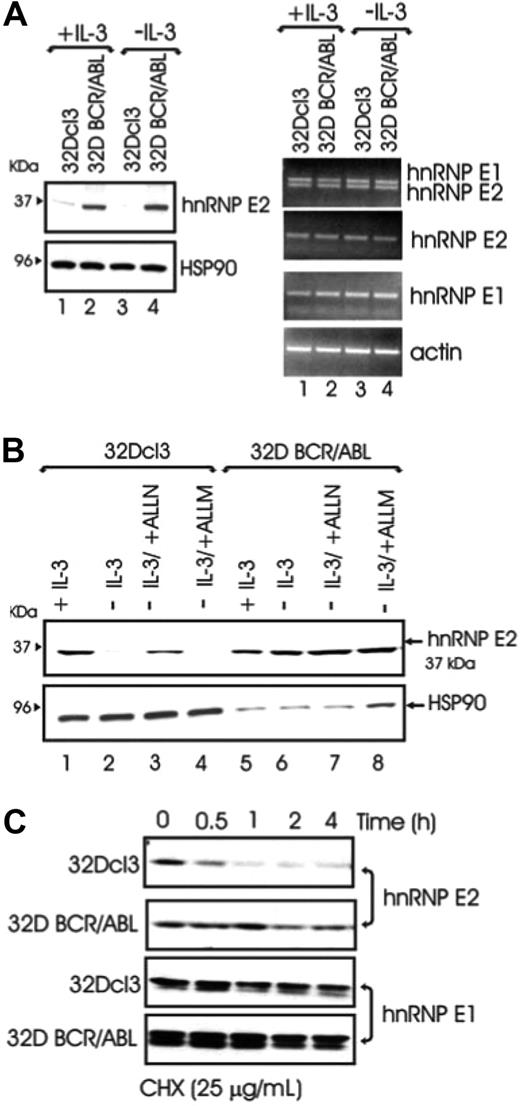
To determine the molecular mechanism(s) whereby BCR/ABL increases hnRNP-E2 levels, hnRNP-E2 mRNA and protein expression was assessed in parental and BCR/ABL-expressing 32Dcl3 cells cultured in the presence of IL-3 or IL-3 deprived for 8 hours. Regardless of the presence or absence of IL-3, hnRNP-E2 protein but not mRNA levels were enhanced in 32D-BCR/ABL cells when compared with parental cells (Figure 1A). Notably, 32Dcl3 exhibited high levels of hnRNP-E2 mRNA but low levels of protein that further decreased upon IL-3 deprivation (Figure 1A). These data suggest that the primary mechanism by which myeloid cells regulate hnRNP-E2 expression is posttranscriptional. Interestingly, treatment of IL-3-deprived 32Dcl3 cells with the proteosome/calpain inhibitor ALLN restored, albeit not totally, hnRNP-E2 expression (Figure 1B lanes 1-3). Because the calpain inhibitor ALLM was unable to rescue hnRNP-E2 expression in parental cells (Figure 1B lane 4), and neither ALLN nor ALLM affected hnRNP-E2 expression in 32D-BCR/ABL (Figure 1B lanes 5-8), increased protein stability may account for hnRNP-E2 overexpression in BCR/ABL cells. Indeed, a more rapid decline of hnRNP-E2 levels was observed in parental 32Dcl3 than 32D-BCR/ABL cells treated with the protein synthesis inhibitor cycloheximide (CHX, 25 μg/mL) (Figure 1C). As expected,11 mRNA and protein expression of the closely related hnRNP-E1 protein28 was not affected by BCR/ABL expression (Figure 1A,C). Thus, p210-BCR/ABL enhances hnRNP-E2 expression by inducing posttranslational modifications that increase hnRNP-E2 stability most likely by delaying its proteasome-dependent turnover.
BCR/ABL posttranslationally enhances hnRNP-E2 expression by increasing its protein stability. (A) Western blot (left panel) and RT-PCR (right panel) analyses of hnRNP-E2 expression in parental and BCR/ABL-transduced 32Dcl3 cells cultured in the presence of IL-3 or IL-3 deprived for 8 hours. (B) Effect of ALLN (25 μM) and ALLM (25 μM) on hnRNP-E2 levels. Note that 100 μg and 25 μg lysates from 32Dcl3 and 32D-BCR/ABL cells were loaded for each lane, respectively. (C) Effect of the protein synthesis inhibitor cycloheximide (CHX) on hnRNP-E2 and hnRNP-E1 levels in parental and BCR/ABL-transduced 32Dcl3 cells.
BCR/ABL posttranslationally enhances hnRNP-E2 expression by increasing its protein stability. (A) Western blot (left panel) and RT-PCR (right panel) analyses of hnRNP-E2 expression in parental and BCR/ABL-transduced 32Dcl3 cells cultured in the presence of IL-3 or IL-3 deprived for 8 hours. (B) Effect of ALLN (25 μM) and ALLM (25 μM) on hnRNP-E2 levels. Note that 100 μg and 25 μg lysates from 32Dcl3 and 32D-BCR/ABL cells were loaded for each lane, respectively. (C) Effect of the protein synthesis inhibitor cycloheximide (CHX) on hnRNP-E2 and hnRNP-E1 levels in parental and BCR/ABL-transduced 32Dcl3 cells.
Increased hnRNP-E2 expression requires phosphorylation by the BCR/ABL-activated MAPKERK1/2
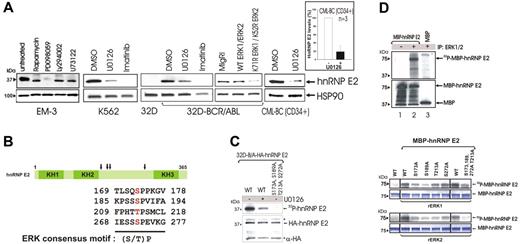
To determine the mechanism(s) whereby BCR/ABL induces hnRNP-E2 up-regulation, the CML-BC–derived EM-3 myeloid cell line was exposed to various inhibitors of BCR/ABL-activated pathways.1,29-33 HnRNP-E2 levels were markedly reduced by treatment with the MEK inhibitor PD098059 (50 μM) but not with inhibitors of PI-3K–dependent (LY294002, 50 μM), PLCγ-dependent (U73122, 2 μM), or mTOR/S6K-dependent (rapamycin, 20 nM) signals (Figure 2A), suggesting that enhanced hnRNP-E2 expression may depend on BCR/ABL-induced activation of MAPK (ie, ERK1/2). Indeed, hnRNP-E2 levels were also inhibited by treatment with the MEK1/2 inhibitor U0126 (25 μM) in K562 and 32D-BCR/ABL cells and by ectopic expression of dominant-negative ERK1K71R and ERK2K52R mutants34 (Figure 2A). Furthermore, decreased hnRNP-E2 levels (∼ 80% reduction) were observed upon treatment of primary CML-BCCD34+ progenitors (n = 3) with U0126 (Figure 2A). As expected, inhibition of BCR/ABL activity by imatinib also led to suppression of hnRNP-E2 expression (Figure 2A).
The BCR/ABL-activated MAPKERK1/2 is responsible for enhanced hnRNP-E2 expression. (A) Effect of different chemical kinase inhibitors on hnRNP-E2 protein levels in Ph1(+) EM-3, K562, 32D-BCR/ABL, and primary CML-BCCD34+ progenitors. Graph shows mean plus and minus standard deviation (SD) of hnRNP-E2 levels after normalization with HSP90 in untreated and U0126-treated primary CML-BCCD34+ progenitors (n = 3). Effect of wild-type and dominant-negative K71R ERK1 and K52R ERK2 expression on hnRNP-E2 levels in 32D-BCR/ABL cells. (B) Schematic diagram shows ERK phosphorylation sites (S/T-P) in hnRNP-E2. (C) In vivo MAPK-dependent phosphorylation of hnRNP-E2. Autoradiography (top) shows phosphorylated HA-hnRNP-E2 in 32P-labeled 32D-BCR/ABL cells ectopically expressing HA-hnRNP-E2 (untreated and U0126-treated) and HA-hnRNP-E2 S173A, S189A, T213A, S272A. Anti-HA Western blot (bottom) shows expression of HA-hnRNP-E2 and HA-hnRNP-E2 S173A, S189A, T213A, S272A. Asterisks indicate IgG chains. (D) In vitro MAPK-dependent phosphorylation of hnRNP-E2. (top panel). Bacterially expressed and purified wild-type MBP-hnRNP-E2 was subjected to an in vitro kinase assay and Western blot analysis. Autoradiography (top) shows phosphorylated MBP-hnRNP-E2 by the anti-ERK1/2 immunoprecipitates. Western blot (bottom) shows expression of MBP-hnRNP-E2 and MBP. Effect of serine/threonine to alanine mutations on hnRNP-E2 phosphorylation (bottom panel). Wild-type MBP-hnRNP-E2 and its serine/threonine to alanine mutants were subjected to a kinase assay with recombinant ERK1 (rERK1) and ERK2 (rERK2), respectively. Autoradiography show phosphorylated MBP-hnRNP-E2 by ERK1 and ERK2, respectively. Coomassie blue–stained gels were used as controls for equal loading.
The BCR/ABL-activated MAPKERK1/2 is responsible for enhanced hnRNP-E2 expression. (A) Effect of different chemical kinase inhibitors on hnRNP-E2 protein levels in Ph1(+) EM-3, K562, 32D-BCR/ABL, and primary CML-BCCD34+ progenitors. Graph shows mean plus and minus standard deviation (SD) of hnRNP-E2 levels after normalization with HSP90 in untreated and U0126-treated primary CML-BCCD34+ progenitors (n = 3). Effect of wild-type and dominant-negative K71R ERK1 and K52R ERK2 expression on hnRNP-E2 levels in 32D-BCR/ABL cells. (B) Schematic diagram shows ERK phosphorylation sites (S/T-P) in hnRNP-E2. (C) In vivo MAPK-dependent phosphorylation of hnRNP-E2. Autoradiography (top) shows phosphorylated HA-hnRNP-E2 in 32P-labeled 32D-BCR/ABL cells ectopically expressing HA-hnRNP-E2 (untreated and U0126-treated) and HA-hnRNP-E2 S173A, S189A, T213A, S272A. Anti-HA Western blot (bottom) shows expression of HA-hnRNP-E2 and HA-hnRNP-E2 S173A, S189A, T213A, S272A. Asterisks indicate IgG chains. (D) In vitro MAPK-dependent phosphorylation of hnRNP-E2. (top panel). Bacterially expressed and purified wild-type MBP-hnRNP-E2 was subjected to an in vitro kinase assay and Western blot analysis. Autoradiography (top) shows phosphorylated MBP-hnRNP-E2 by the anti-ERK1/2 immunoprecipitates. Western blot (bottom) shows expression of MBP-hnRNP-E2 and MBP. Effect of serine/threonine to alanine mutations on hnRNP-E2 phosphorylation (bottom panel). Wild-type MBP-hnRNP-E2 and its serine/threonine to alanine mutants were subjected to a kinase assay with recombinant ERK1 (rERK1) and ERK2 (rERK2), respectively. Autoradiography show phosphorylated MBP-hnRNP-E2 by ERK1 and ERK2, respectively. Coomassie blue–stained gels were used as controls for equal loading.
Consistent with the existence of a BCR/ABL-MAPK pathway that posttranslationally regulates hnRNP-E2 expression, sequence analysis of hnRNP-E2 revealed the presence of 4 consensus ERK phosphorylation sites (S/T-P)35,36 at amino acid residues 173, 189, 213, and 272 (Figure 2B). Thus, wild-type and serine/threonine to alanine mutant HA-tagged and MBP-fusion hnRNP-E2 proteins were generated and used to assess in vivo and in vitro MAPK-dependent phosphorylation. In [32P]-labeled 32D-BCR/ABL cells, wild-type HA-tagged hnRNP-E2 was abundantly phosphorylated (Figure 2C lane 1). By contrast, alanine substitution of residues 173, 189, 213, and 272 completely abolished hnRNP-E2 in vivo phosphorylation (Figure 2C lane 3), which was also markedly reduced by inhibition of MAPK activity with U0126 (Figure 2C lane 2). Furthermore, recombinant MBP-hnRNP-E2 fusion protein was efficiently phosphorylated in vitro by the BCR/ABL-activated ERK1/2 immunoprecipitated from lysates of 32D-BCR/ABL cells (Figure 2D top, upper panel, lane 2). Likewise, in vitro kinase assays showed that ERK1/2-dependent phosphorylation of rMBP-hnRNP-E2 was significantly reduced by single serine/threonine to alanine substitutions and almost totally abolished in the quadruple hnRNP-E2 mutant (Figure 2D bottom panels). Note that the effect of ERK2 on hnRNP-E2 phosphorylation was more pronounced than that of ERK1 (Figure 2D bottom panel). Because ERK2 activity is higher than that of ERK1 in CML-BCCD34+ progenitors,21 these data demonstrate that serines/threonine at residues 173, 189, 213, and 272 are crucial for ERK-dependent phosphorylation of hnRNP-E2 and suggest that MAPKERK2 is the kinase responsible for hnRNP-E2 phosphorylation in BCR/ABL+ cells.
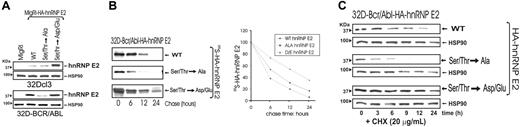
To determine whether ERK phosphorylation accounts for increased hnRNP-E2 protein stability, expression of HA-tagged wild-type hnRNP-E2, nonphosphatable hnRNP-E2S173A,S189A,T213A,S272A (Ser/Thr→Ala), and phospho-mimetic hnRNP-E2S173D,S189D,T213E,S272D (Ser/Thr→Asp/Glu) was assessed in parental and BCR/ABL-transduced 32Dcl3 cells that were IL-3 deprived for 8 hours. Expression of wild-type and nonphosphatable hnRNP-E2 was low or undetectable in 32Dcl3 cells (Figure 3A top panel). Likewise, hnRNP-E2S173A,S189A,T213A,S272A expression was also less abundant than wild-type in BCR/ABL-expressing cells (Figure 3A bottom panel). Conversely, levels of the phosphomimetic hnRNP-E2S173D,S189D,T213E,S272D were abundant in 32Dcl3 cells and comparable with those of wild type in 32D-BCR/ABL cells (Figure 3A), further suggesting that increased hnRNP-E2 stability depends on ERK-mediated phosphorylation at these indicated amino acids. Indeed, pulse chase experiments revealed that the half-life of newly synthesized 35S-hnRNP-E2S173A,S189A,T213A,S272A (t1/2 ≅ 5.5 hours) was shorter than that of wild-type (t1/2 ≅ 7 hours) and phosphomimetic (t1/2 ≅ 14 hours) hnRNP-E2 proteins in metabolically labeled 32D-BCR/ABL cells (Figure 3B). Accordingly, expression of the hnRNP-E2S173A,S189A,T213A,S272A mutant was more rapidly downmodulated than that of wild-type and phosphomimetic hnRNP-E2 in CHX-treated 32D-BCR/ABL–expressing cells (Figure 3C). All together, these data indicate that ERK-dependent phosphorylation of hnRNP-E2 at serines 173, 189, and 272, and threonine 213 is responsible for increased hnRNP-E2 protein stability in BCR/ABL-transformed cells.
ERK-dependent phosphorylation controls hnRNP-E2 protein stability. (A) hnRNP-E2 expression in parental and BCR/ABL-transduced 32Dcl3 cells expressing an empty vector, HA-hnRNP-E2, HA-hnRNP-E2S173A,S189A,T213A,S272A (Ser/Thr→Ala), and HA-hnRNP-E2S173D,S189D,T213E,S272D (Ser/Thr→Asp/Glu). Cells were IL-3 deprived for 8 hours. (B) Stability of newly synthesized HA-tagged hnRNP-E2, hnRNP-E2S173A,S189A,T213A,S272A, and hnRNP-E2S173D,S189D,T213E,S272D in 35S-labeled 32D-BCR/ABL cells. The half-life of hnRNP-E2 was assessed by pulse-chase assay and quantified by densitometry. Each point on the graph represents the relative normalized amounts of hnRNP-E2 during the chase period. (C) Stability of HA-tagged hnRNP-E2, hnRNP-E2S173A,S189A,T213A,S272A, and hnRNP-E2S173D,S189D,T213E,S272D in 32D-BCR/ABL cells treated with cycloheximide (CHX). HSP90 levels were used as control for equal loading.
ERK-dependent phosphorylation controls hnRNP-E2 protein stability. (A) hnRNP-E2 expression in parental and BCR/ABL-transduced 32Dcl3 cells expressing an empty vector, HA-hnRNP-E2, HA-hnRNP-E2S173A,S189A,T213A,S272A (Ser/Thr→Ala), and HA-hnRNP-E2S173D,S189D,T213E,S272D (Ser/Thr→Asp/Glu). Cells were IL-3 deprived for 8 hours. (B) Stability of newly synthesized HA-tagged hnRNP-E2, hnRNP-E2S173A,S189A,T213A,S272A, and hnRNP-E2S173D,S189D,T213E,S272D in 35S-labeled 32D-BCR/ABL cells. The half-life of hnRNP-E2 was assessed by pulse-chase assay and quantified by densitometry. Each point on the graph represents the relative normalized amounts of hnRNP-E2 during the chase period. (C) Stability of HA-tagged hnRNP-E2, hnRNP-E2S173A,S189A,T213A,S272A, and hnRNP-E2S173D,S189D,T213E,S272D in 32D-BCR/ABL cells treated with cycloheximide (CHX). HSP90 levels were used as control for equal loading.
In vitro and/or in vivo suppression of ERK-dependent hnRNP-E2 phosphorylation restores C/EBPα expression and rescues G-CSF–driven neutrophilic maturation of BCR/ABL+ cell lines, primary Lin− bone marrow cells, and CML-BCCD34+ progenitors
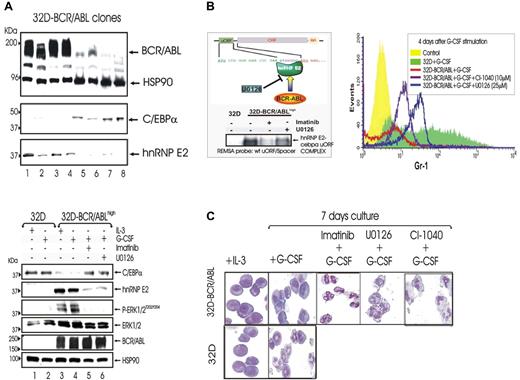
To further assess the importance of BCR/ABL expression in the hnRNP-E2–mediated suppression of myeloid differentiation, 32D-BCR/ABL single-cell clones expressing low or high levels of p210-BCR/ABL were isolated upon transduction of 32Dcl3 cells with the MigR1-p210-BCR/ABL expression plasmid. 32D-BCR/ABL cells expressing high BCR/ABL levels showed a marked increase of hnRNP-E2 levels and suppression of C/EBPα expression (Figure 4A top panel, lanes 1-4). This correlated with the development of growth factor independence and differentiation arrest (not shown). By contrast, low BCR/ABL-expressing cells exhibited high C/EBPα expression and barely detectable hnRNP-E2 levels (Figure 4A top panel, lanes 5-8), were IL-3 dependent, and underwent differentiation upon treatment with G-CSF (not shown).
Effect of ERK-dependent hnRNP-E2 down-regulation on C/EBPα expression and neutrophilic differentiation of 32D-BCR/ABLhigh cells.(A) Protein levels of C/EBPα and hnRNP-E2 in newly established 32Dcl3 cell clones expressing different levels of p210-BCR/ABL (top panel). Levels of C/EBPα, hnRNP-E2, phospho-ERK1/2, total ERK1/2, and BCR/ABL in 32Dcl3 cells and untreated, imatinib-treated, and U0126-treated 32D-BCR/ABL cells expressing high levels of the p210-BCR/ABL oncoprotein (32D-BCR/ABLhigh) (bottom panel). Cells were cultured in the presence of IL-3 or G-CSF. (B) REMSA with 32P-labeled WT-uORF/spacer CEBPA oligoribonucleotide probe and cytosolic lysates from 32Dcl3 cells and untreated, imatinib-treated, and U0126-treated 32D-BCR/ABLhigh cells (left panel). Expression of Gr-1 on 32Dcl3 and untreated, U0126-treated, and CI-1040–treated 32D-BCR/ABLhigh cells upon G-CSF stimulation for 4 days (right panel). (C) May-Grunwald/Giemsa staining of 32Dcl3 and 32D-BCR/ABLhigh cells cultured with IL-3 or G-CSF for 7 days in the presence or absence of imatinib, U0126, or CI-1040.
Effect of ERK-dependent hnRNP-E2 down-regulation on C/EBPα expression and neutrophilic differentiation of 32D-BCR/ABLhigh cells.(A) Protein levels of C/EBPα and hnRNP-E2 in newly established 32Dcl3 cell clones expressing different levels of p210-BCR/ABL (top panel). Levels of C/EBPα, hnRNP-E2, phospho-ERK1/2, total ERK1/2, and BCR/ABL in 32Dcl3 cells and untreated, imatinib-treated, and U0126-treated 32D-BCR/ABL cells expressing high levels of the p210-BCR/ABL oncoprotein (32D-BCR/ABLhigh) (bottom panel). Cells were cultured in the presence of IL-3 or G-CSF. (B) REMSA with 32P-labeled WT-uORF/spacer CEBPA oligoribonucleotide probe and cytosolic lysates from 32Dcl3 cells and untreated, imatinib-treated, and U0126-treated 32D-BCR/ABLhigh cells (left panel). Expression of Gr-1 on 32Dcl3 and untreated, U0126-treated, and CI-1040–treated 32D-BCR/ABLhigh cells upon G-CSF stimulation for 4 days (right panel). (C) May-Grunwald/Giemsa staining of 32Dcl3 and 32D-BCR/ABLhigh cells cultured with IL-3 or G-CSF for 7 days in the presence or absence of imatinib, U0126, or CI-1040.
Since increased hnRNP-E2 expression depends on its phosphorylation by the BCR/ABL-activated MAPKERK1/2 kinases, we investigated the in vitro and in vivo effects of MEK1/2 inhibition by U0126 or by the clinically relevant CI-104037-40 (PD184352; Pfizer) on the BCR/ABL-MAPKERK1/2-hnRNP-E2-C/EBPα axis and granulocytic differentiation of myeloid progenitors (primary and cell lines) expressing high levels of p210-BCR/ABL.
In newly established 32D-BCR/ABLhigh cells cultured in the presence of G-CSF, inhibition of MAPKERK1/2 activity by U0126 (32 hours) (Figure 4A bottom panel) and by CI-1040 (48 hours) (not shown) abolished the expression of hnRNP-E2 and rescued that of C/EBPα. Accordingly, REMSA assays with the 32P-labeled C-rich uORF/spacer region of CEBPA mRNA and cytoplasmic lysates of U0126-treated 32D-BCR/ABLhigh cells revealed that inhibition of MAPK activity decreases binding of hnRNP-E2 to the translation inhibitory element contained in the 5′UTR of CEBPA mRNA (Figure 4B left panel). Furthermore, 32D-BCR/ABLhigh cells treated with U0126 (25 μM) or CI-1040 (10 μM) in the presence of G-CSF underwent terminal granulocytic differentiation as indicated by increased expression of the myeloid differentiation marker Gr-1 after 4-day culture in G-CSF and by the appearance of polymorphonuclear cells after 7-day culture in G-CSF (Figures 4B right panel and 5C). Likewise, suppression of BCR/ABL activity by imatinib caused molecular and morphologic effects similar to those induced by MEK1/2 inhibition (Figure 4B-C). As expected, parental 32Dcl3 cells expressed C/EBPα but not hnRNP-E2 and underwent G-CSF–driven granulocytic differentiation (Figure 4).
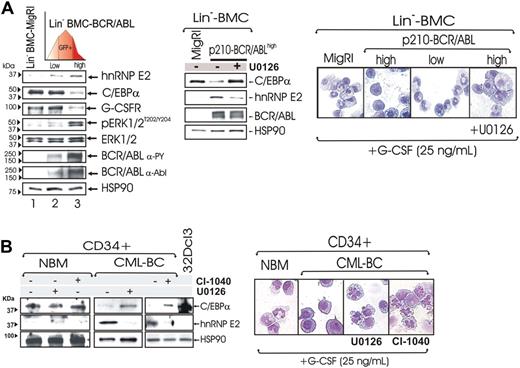
Effect of ERK-dependent suppression of hnRNP-E2 on C/EBPα expression and neutrophilic differentiation of Lin− BMC-BCR/ABLhigh cells and patient-derived CML-BCCD34+ progenitors. (A) Levels of hnRNP-E2, C/EBPα, G-CSFR, phospho-ERK1/2, and total ERK1/2 in lineage-negative (Lin−) mouse bone marrow cells (BMCs) transduced with the empty vector (Lin−BMC-MigRI) or with a p210-BCR/ABL retrovirus and sorted for low and high GFP expression (Lin−BMC-BCR/ABLlow and -BCR/ABLhigh) (left panel). Effect of MAPK inhibition by U0126 on hnRNP-E2 and C/EBPα expression and/or G-CSF–driven differentiation in Lin− BMC-BCR/ABLhigh cells (middle and right panels). May-Grumwald/Giemsa–stained representative microphotographs of Lin− BMC-MigRI, BMC-BCR/ABLlow, untreated, and U0126-treated BMC-BCR/ABLhigh cells (right panel). (B) Effect of MAPK inhibition by U0126 and CI-1040 on hnRNP-E2 and C/EBPα expression in primary CD34+ human normal bone marrow cells (NBMCD34+) and CML-BCCD34+ cells (left panel). May-Grumwald/Giemsa staining of NBMCD34+ and untreated, U0126-treated, and CI-1040-treated CML-BCCD34+ cells (right panel).
Effect of ERK-dependent suppression of hnRNP-E2 on C/EBPα expression and neutrophilic differentiation of Lin− BMC-BCR/ABLhigh cells and patient-derived CML-BCCD34+ progenitors. (A) Levels of hnRNP-E2, C/EBPα, G-CSFR, phospho-ERK1/2, and total ERK1/2 in lineage-negative (Lin−) mouse bone marrow cells (BMCs) transduced with the empty vector (Lin−BMC-MigRI) or with a p210-BCR/ABL retrovirus and sorted for low and high GFP expression (Lin−BMC-BCR/ABLlow and -BCR/ABLhigh) (left panel). Effect of MAPK inhibition by U0126 on hnRNP-E2 and C/EBPα expression and/or G-CSF–driven differentiation in Lin− BMC-BCR/ABLhigh cells (middle and right panels). May-Grumwald/Giemsa–stained representative microphotographs of Lin− BMC-MigRI, BMC-BCR/ABLlow, untreated, and U0126-treated BMC-BCR/ABLhigh cells (right panel). (B) Effect of MAPK inhibition by U0126 and CI-1040 on hnRNP-E2 and C/EBPα expression in primary CD34+ human normal bone marrow cells (NBMCD34+) and CML-BCCD34+ cells (left panel). May-Grumwald/Giemsa staining of NBMCD34+ and untreated, U0126-treated, and CI-1040-treated CML-BCCD34+ cells (right panel).
Rescue of C/EBPα expression and restoration of neutrophilic maturation by pharmacologic inhibition of ERK activity were observed not only in differentiation-arrested 32D-BCR/ABLhigh cells but also in lineage-negative (Lin−) mouse bone marrow cells (BMCs) transduced with the MigR1-GFP-BCR/ABL retrovirus11 and sorted for low and high GFP expression. As expected, GFP levels correlated with BCR/ABL expression and activity (Figure 5A left panel). Consistent with the knowledge that differentiation arrest is a characteristic of CML-BC myeloid progenitors expressing high BCR/ABL and hnRNP-E2 levels,11 graded p210-BCR/ABL expression resulted in a dose-dependent induction of MAPKERK1/2 activity and hnRNP-E2 expression that inversely correlated with levels of C/EBPα and of the C/EBPα-regulated G-CSF receptor (G-CSFR) (Figure 5A left panel). Note that total MAPKERK1/2 levels were not affected by BCR/ABL (Figure 5A left panel). Furthermore, Lin− BMC-BCR/ABLlow but not Lin− BMC-BCR/ABLhigh cells underwent granulocytic differentiation within 6 days of culture in the presence of G-CSF (25 ng/mL) (Figure 5A right panel). Notably, down-regulation of hnRNP-E2 levels and restoration of C/EBPα expression were also observed in G-CSF–cultured mouse Lin− BMC-BCR/ABLhigh progenitors (Figure 5A middle panel) and patient-derived CML-BCCD34+ cells (n = 3) (Figure 5B left panel) exposed for 48 hours to the MEK1/2 inhibitor CI-1040 and/or U0126. Moreover, U0126- or CI-1040–treated differentiation-arrested Lin− BMC-BCR/ABLhigh and CML-BCCD34+ developed into mature neutrophils upon exposure to G-CSF (25 ng/mL) (Figure 5A-B right panels).
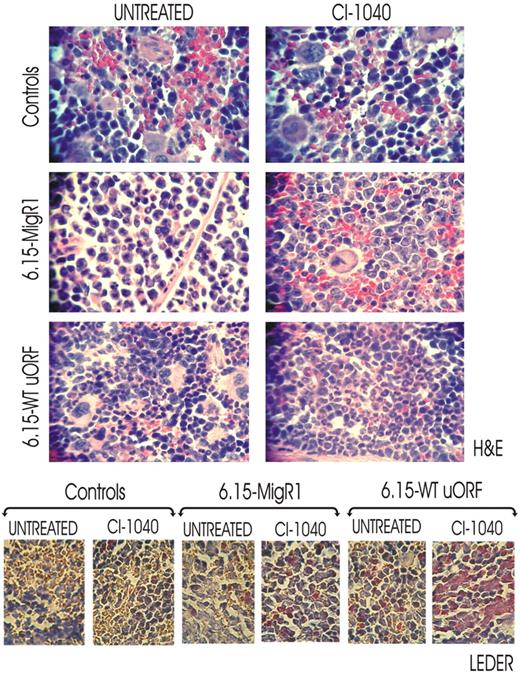
To assess whether inhibition of MAPKERK1/2 activity rescues in vivo neutrophilic maturation of differentiation-arrested BCR/ABL+ myeloid blasts, we used a 32D-BCR/ABL long-term cultured cell clone (6.15 cells) that exhibits abnormally high levels of BCR/ABL and hnRNP-E2 expression but is unable to undergo G-CSF–driven differentiation because of transcriptional suppression of CEBPα expression.11 Thus, 6.15 cells were retrovirally transduced with either a MigRI-GFP-wt-uORF/spacer-C/EBPα (6.15-WT-uORF) plasmid containing the hnRNP-E2–negative translational regulatory element11 or with an empty MigRI-GFP vector (6.15-MigRI). SCID mice (n = 6 per group) were thereafter intravenously injected with 6.15-wt-uORF and 6.15-MigRI cells (5 × 105 GFP-sorted cells/mouse), and engraftment was assessed one week after cell injection by nested RT-PCR–mediated detection of p210-BCR/ABL transcripts in peripheral blood cells (not shown). Thereafter, mice were cotreated with either G-CSF (100 μg/kg per day)26,27 and CI-1040 (100 mg/kg twice a day)24 or G-CSF and vehicle for 14 days. At the end of treatment, spleen tissue sections from untreated, CI-1040–treated, and control (age-matched) mice were subjected to histopathologic examination. Consistent with the almost complete lack of translational control of C/EBPα expression in 6.15 cells,11 hematoxylin/eosin-stained sections of spleens from untreated 6.15-MigRI–injected mice showed massive infiltration of myeloid blasts with a low degree of myeloid differentiation (≤ 1% neutrophils), which was further confirmed by the scarce chloroacetate esterase (Leder staining) positivity (Figure 6 bottom panels). Terminally differentiated myeloid cells were occasionally observed in spleens from CI-1040–treated 6.15-MigRI–injected mice (2%-5% neutrophils) (Figure 6 bottom panels), suggesting the involvement of MAPK in additional mechanisms leading to suppression of myeloid differentiation. By contrast, spleens from CI-1040–treated 6.15-wt-uORF–injected mice showed marked infiltration of mature neutrophils (40%-70% neutrophils) and myeloid precursors at the postmitotic stages of differentiation (Figure 6 top panels). Accordingly, a high degree of chloroacetate esterase positivity was observed in spleen section of CI-1040–treated 6.15-wt-uORF–injected mice (Figure 6 bottom panels). However, 5% to 10% of terminally differentiated myeloid cells were also observed in spleens of vehicle (DMSO)–treated 6.15-wt-uORF–injected mice (Figure 6 top panels) as a consequence of the leaky nature of the MigR1-wt-uORF-C/EBPα plasmid that allows low levels of C/EBPα expression. Note that histopathologic analysis of H&E- and Leder-stained spleen sections from untreated and CI-1040–treated age-matched mice did not show signs of myeloid cell infiltration (Figure 6). All together, these results suggest that in vivo rescue of granulocytic maturation of differentiation-arrested BCR/ABL+ cells by chemical ERK1/2 inactivation is likely due to decreased hnRNP-E2 expression that, in turn, reverses translation inhibition of C/EBPα mRNA.
In vivo effect of pharmacologic inhibition of ERK-dependent hnRNP-E2 expression on neutrophilic maturation of differentiation-arrested myeloid 32D-BCR/ABL cells. H&E (top panels) and Leder (bottom panels) staining of the spleen sections of untreated and CI-1040–treated control and 6.15-MigRI and 6.15-WT-uORF cell–injected mice. In addition, all mice were cotreated with G-CSF. Microphotographs are representative of data obtained by analyzing multiples sections of spleens from 3 mice per group.
In vivo effect of pharmacologic inhibition of ERK-dependent hnRNP-E2 expression on neutrophilic maturation of differentiation-arrested myeloid 32D-BCR/ABL cells. H&E (top panels) and Leder (bottom panels) staining of the spleen sections of untreated and CI-1040–treated control and 6.15-MigRI and 6.15-WT-uORF cell–injected mice. In addition, all mice were cotreated with G-CSF. Microphotographs are representative of data obtained by analyzing multiples sections of spleens from 3 mice per group.
p53 status does not affect BCR/ABL-dependent regulation of C/EBPα levels
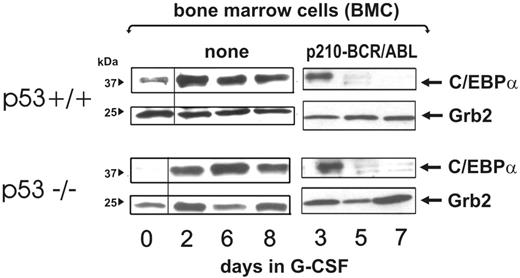
Genetic (mutations) or functional (MDM2-dependent degradation) inactivation of p53 is a common feature of CML-BC.41,42 Because it has been reported that C/EBPα is positively regulated by wild-type p53,43 and loss of p53 activity is associated with and may promote blastic transformation,44,45 we asked whether BCR/ABL-dependent down-regulation of C/EBPα levels may be affected by the p53 status. Thus, bone marrow mononuclear cells were isolated from 5-FU–treated (4 days) wild-type or p53−/− C57BL/6 mice and left untreated or transduced with the MigRI-p210-BCR/ABL retrovirus. After sorting, GFP-positive cells were expanded for 2 days in the presence of IL-3, KL, IL-6, and FLT3 ligands, washed, and cultured in the presence of G-CSF for 3, 5, and 7 days. Levels of C/EBPα were readily detectable after 3 days of culture in the presence of G-CSF but rapidly declined in both cell cultures with similar kinetics (Figure 7). By contrast, in both p53−/− and p53+/+ mononuclear cells maintained in culture for the same length of time, expression of C/EBPα was barely detectable at time 0 but increased with similar kinetics upon G-CSF treatment for 2 to 8 days (Figure 7). All together, these findings suggest that the p53 status does not directly affect C/EBPα expression and does not cooperate with BCR/ABL in promoting its down-regulation.
Effect of the p53 genetic background on BCR/ABL-dependent regulation of C/EBPα expression. C/EBPα levels at a different time course in nontransduced (none) and BCR/ABL-transduced wild-type and p53−/− mouse bone marrow cells (BMCs) cultured in the presence of G-CSF for the indicated time.
Effect of the p53 genetic background on BCR/ABL-dependent regulation of C/EBPα expression. C/EBPα levels at a different time course in nontransduced (none) and BCR/ABL-transduced wild-type and p53−/− mouse bone marrow cells (BMCs) cultured in the presence of G-CSF for the indicated time.
Discussion
The mechanisms responsible for transition of CML chronic phase into blast crisis remain poorly understood, although it appears that the unrestrained activity of BCR/ABL in hematopoietic stem/progenitor cells is the primary determinant of disease progression.1,46 Indeed, BCR/ABL expression specifically increases during blastic transformation in hematopoietic stem cells and committed myeloid progenitor47-53 clones with increasingly malignant characteristics. In fact, augmented BCR/ABL activity alters the expression and/or function of several factors important for proliferation, survival, and maturation of myeloid progenitors.1,46
Despite the molecular heterogeneity of CML-BC, a distinctive feature of blast crisis CML is the inability of myeloid progenitors to differentiate into mature granulocytes.1 We recently reported that impaired differentiation of myeloid CML-BC progenitors is caused by the BCR/ABL-dependent suppression of C/EBPα expression through the translation inhibitory activity of the RNA-binding protein hnRNP-E2,11 and that hnRNP-E2 protein levels specifically increase in CML-BC progenitors in a BCR/ABL kinase–dependent manner.11 By progressively increasing BCR/ABL expression in Lin− mouse marrow progenitors and hematopoietic cell lines, we provided evidence that induction of hnRNP-E2 expression requires high levels of p210-BCR/ABL. These findings are consistent with the lack of C/EBPα expression in CML-BC and the low hnRNP-E2 and BCR/ABL expression in CML-CP progenitors.11 In BCR/ABL-expressing myeloid precursors and primary CML-BC progenitors, the molecular mechanism whereby BCR/ABL up-regulates hnRNP-E2 expression is posttranslational and depends on increased hnRNP-E2 protein stability. This is consistent with the observation that enhanced expression of various RNA-binding proteins is among the many changes in gene expression found in bone marrow–derived myeloid progenitors from CML-BC patients and BCR/ABL-transformed murine myeloid progenitors.11,54 This enhanced expression of RNA-binding proteins correlates with the levels of BCR/ABL and is sensitive to imatinib treatment.11,46,54 Molecular mechanisms responsible for such an increase may involve the activation of a cascade of phosphorylation events leading to either enhanced gene transcription (eg, hnRNP K) or increased protein stability (eg, TLS/FUS, hnRNP A1, and La/SSB).11,21,42,55-58 Increased expression of these RNA-binding proteins correlates with enhanced posttranscriptional and/or translational regulatory activity, which is tightly controlled by different BCR/ABL-activated pathways.21,54,55,58 Indeed, we showed that hnRNP-E2 undergoes phosphorylation in BCR/ABL-expressing cells. Furthermore, by treating BCR/ABL+ cell lines, primary BCR/ABL-expressing Lin− mouse bone marrow progenitors, and patient-derived CML-BCCD34+ cells with chemical inhibitors of MEK1/2, including the clinically relevant CI-1040,59,60 and by expressing ERK1 and ERK2 constructs with dominant-negative activity,34 we demonstrated that increased hnRNP-E2 protein stability requires the constitutive activation of MAPKERK1/2. Furthermore, we also identified 4 hnRNP-E2 MAPKERK1/2 phosphorylation sites and demonstrated that hnRNP-E2 is a bona fide MAPKERK1/2 substrate and that MAPKERK1/2-dependent phosphorylation of hnRNP-E2 at these amino acid residues is essential for increased hnRNP-E2 expression in BCR/ABL-expressing cells. The involvement of MAPKERK1/2 in the regulation of hnRNP-E2 expression in CML-BC is not surprising, as constitutive MAPK activation is readily detectable in CML-BCCD34+ progenitors,21 while CML-CP progenitors are capable of only transient MAPK activation in response to mitogenic and survival signals induced by extracellular growth factors.61 In fact, MAPKERK1/2 activation appears to depend on increased levels of BCR/ABL activity, as graded BCR/ABL expression correlates with a progressive increase in MAPKERK1/2 activity.21 Interestingly, in BCR/ABL+ myeloid progenitor cells, MAPKERK1/2 is also responsible for the increased expression and translational-regulatory function of the shuttling RNA-binding protein hnRNP K21 . Like hnRNP-E2, hnRNP K belongs to the subfamily of the K homology (KH) domain containing poly(rC)-binding proteins.62 However, the molecular mechanism whereby MAPKERK1/2 enhances hnRNP K translational-regulatory function appears different from that of hnRNP-E2. In fact, BCR/ABL promotes the cytoplasmic accumulation of hnRNP K by MAPKERK1/2-dependent phosphorylation at serines 284 and 353.21 Conversely, the subcellular localization of hnRNP-E2 does not appear to be regulated by BCR/ABL or MAPKERK1/2 activity, as neither imatinib nor U0126 treatment induced hnRNP-E2 nuclear relocation in BCR/ABL+ myeloid cells (not shown). Thus, enhancement of hnRNP-E2 translational-regulatory activity results largely from the effect of BCR/ABL-activated MAPKERK1/2 on hnRNP-E2 expression. Nonetheless, hnRNP-E2 and hnRNP K cooperate in the positive regulation of MYC translation via binding to the IRES element located in the 5′UTR of MYC mRNA.21,63 Because hnRNP K translationally enhances MYC expression in a BCR/ABL- and MAPKERK1/2-dependent manner,21 and differentiation of myeloid progenitors requires the C/EBPα-induced MYC down-regulation,64,65 it is conceivable that BCR/ABL uses MAPKERK1/2 to activate 2 similar RNA-binding proteins (ie, hnRNP-E2 and hnRNP K) that, in turn, simultaneously inhibit C/EBPα and enhance MYC expression, 2 steps essential for the phenotype of leukemic myeloid progenitors.14,64,66-68 Indeed, C/EBPα expression decreases proportionally with a progressive increase of BCR/ABL levels. Accordingly, suppression of MAPKERK1/2 activity impairs hnRNP-E2 binding to CEBPA mRNA, rescues C/EBPα and G-CSFR expression, and allows in vitro and in vivo G-CSF–driven neutrophil maturation of differentiation-arrested marrow progenitors expressing high levels of p210-BCR/ABL. Thus, constitutive MAPKERK1/2 activation is not only essential for transduction of mitogenic and survival signals but it also promotes the activation of antidifferentiation signals (ie, hnRNP-E2 phosphorylation) leading to loss of C/EBPα expression in CML-BC progenitors. In support of the relevance of MAPKERK1/2 activation in CML-BC rather than CML-CP, it was reported that levels of activated MAPKERK1/2 in the absence of exogenous cytokines were similar in normal and CML-CP cells, and imatinib neither reduced nor altered MAPKERK1/2 activity in both normal and CML-CPCD34+ progenitors.61 Moreover, MAPKERK1/2 activity appears to be required for posttranslational inactivation of C/EBPα.69,70 MAPKERK1/2-mediated phosphorylation of C/EBPα on serine 21 inhibits its transcriptional-activating function and leads to a block of granulocytic differentiation in acute myeloid leukemia cells.69,70 This observation is consistent with the differentiation-promoting effects of MEK1 inhibitors in myeloid progenitors expressing high levels of p210-BCR/ABL oncoprotein; in fact, rescued C/EBPα is not phosphorylated on serine 21 in BCR/ABL+ cells treated with U0126 and G-CSF (not shown). Hence, constitutive activation of MAPKERK1/2 is a feature of CML-BC but not CML-CP in which myeloid progenitors express C/EBPα11 and retain the ability to terminally differentiate into apparently normal neutrophils.
In conclusion, our data formally demonstrate that impaired differentiation of CML-BC myeloid progenitors requires increased expression/activity of p210-BCR/ABL oncoprotein and depends on the constitutive activation of the inhibitory BCR/ABL-MAPKERK1/2-hnRNP-E2-C/EBPα pathway. Moreover, the demonstration that pharmacologic inhibition of MAPKERK1/2 activity rescues neutrophilic maturation of differentiation-arrested BCR/ABL+ myeloid blasts portends the addition of clinically relevant MEK inhibitors to the current imatinib or dasatinib monotherapy for patients with myeloid CML-BC.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Cancer Institute CA095512 (D.P.), CA9555111 (B.C.), CA078890 (B.C.) CA16058, and GRT8230100 (OSU-CCC); and the U.S. Army, CML Research Program DAMD17-03-1-0184 (D.P.). J.S.C. was in part supported by the Up-on-the-Roof Human Cancer Genetics Fellowship, The Ohio State University (Columbus, OH). P.N. is a fellow of the American Italian Cancer Foundation (New York, NY). D.P. is a Scholar of the Leukemia and Lymphoma Society. D.C.R. is supported by the Canadian Institutes of Health Research and Fonds de la Recherche en Sante du Quebec.
We thank Novartis Oncology and Pfizer for providing imatinib mesylate and CI-1040, respectively.
National Institutes of Health
Authorship
Contribution: J.S.C. performed research and wrote the paper; R.S. performed research; R.T., P.N., A.M.E., E.B., and M.R. performed research; D.C.R. contributed vital material; B.C. supervised M.R.'s work and analyzed data; M.A.C. supervised R.T.'s work and contributed vital material; D.P. designed research, analyzed data, wrote the paper, and supervised work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Danilo Perrotti, The Ohio State University Comprehensive Cancer Center, 2001 Polaris Pkwy, Rm 205, Columbus, OH 43240; e-mail: danilo.perrotti@osumc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal