The histo-blood group i and I antigens have been characterized as straight and branched repeats of N-acetyllactosamine, respectively, and the conversion of the straight-chain i to the branched-chain I structure on red cells is regulated to occur after birth. It has been demonstrated that the human I locus expresses 3 IGnT transcripts, IGnTA, IGnTB, and IGnTC, and that the last of these is responsible for the I branching formation on red cells. In the present investigation, the K-562 cell line was used as a model to show that the i-to-I transition in erythroid differentiation is determined by the transcription factor CCAAT/enhancer binding protein α (C/EBPα), which enhances transcription of the IGnTC gene, consequently leading to formation of the I antigen. Further investigation suggested that C/EBPα IGnTC-activation activity is modulated at a posttranslational level, and that the phosphorylation status of C/EBPα may have a crucial effect. Results from studies using adult and cord erythropoietic cells agreed with those derived using the K-562 cell model, with lentiviral expression of C/EBPα in CD34+ hemopoietic cells demonstrating the determining role of C/EBPα in the induction of the IGnTC gene as well as in I antigen expression.
Introduction
The blood group i and I antigens have been characterized as straight and branched repeats of the N-acetyllactosamine (LacNAc) structures, Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAc-R and Galβ1-4GlcNAcβ1-3(Galβ1-4GlcNAcβ1-6)Galβ1-4GlcNAc-R, respectively, carried on glycolipids and glycoproteins.1,,,–5 First identified on human red cells,6,,,–10 their presence was later also observed on the surface of most human cells and on soluble glycoproteins in various body fluids.5,11,–13 The LacNAc repeats are synthesized by the sequential action of β-1,3-N-acetylglucosaminyltransferase and β-1,4-galactosyltransferase. Conversion of the straight-chain i into a branched I-active structure requires the activity of the I β-1,6-N-acetylglucosaminyltransferase (I β6GlcNAcT).2,14,15
It has been demonstrated that the relationship between the expressions of the i and I antigen is reciprocal, and that the expression of I antigen on red cells is developmentally regulated. Adult human red cells fully express I antigens and contain only a few i antigens, while fetal and neonatal red cells predominately express the latter. After birth, the quantity of I antigen gradually increases as the level of i antigen falls, until the normal Ii status of adult red cells is reached after about 18 months of life.10,16 On the other hand, it has been shown that the failure of conversion of i to I is associated with several hematologic disorders, including thalassaemia, hypoplastic anemia, and acute leukemia.17 Thus, the transition between straight and branched poly-LacNAc chains may play a significant role in erythrocyte differentiation. One of the recognized findings with respect to the function of the straight poly-LacNAc structure on fetal and neonatal red cells is its protection against severe hemolytic disorder resulting from ABO-incompatible pregnancy.18,19
Formation of the I branching structure on cell surfaces is determined by an uncommon molecular genetic architecture. The cDNA encoding the I β6GlcNAcT was first identified by Bierhuizen et al in 1993.20 The gene is designated IGnT (approved as GCNT2 by the Human Genome Organisation [HUGO] Gene Nomenclature Committee). In 2003, Yu et al and Inaba et al further elaborated the complexity of the human I locus, demonstrating that it expresses 3 different transcript forms.21,22 These 3 transcripts, designated IGnTA, IGnTB, and IGnTC,21 have different exon 1, but identical exon 2 and exon 3, regions. The 3 IGnT cDNAs do not have a common 5′ region, indicating that their transcriptions may be determined by different DNA regulatory regions. Indeed, the 3 transcripts display differential expression patterns in different human tissues.21,22 This observation suggests that the I antigens in the various cells are synthesized by different IGnT forms whose transcriptions are apparently regulated by specific regulatory mechanisms.
In a previous investigation of samples from i adults, whose red cells (like fetal and cord analogs) are rich in the i antigen but contain little I, we demonstrated that IGnTC is the gene responsible for the expression of the blood group I antigen on red cells.21,23 This conclusion is further supported by the fact that IGnTC is the only 1 of the 3 IGnT transcripts that is expressed in reticulocytes, the red cell precursors.21 The aim of the present investigation was to elucidate the molecular mechanism responsible for the expression of I branching structure on red cells after birth. It was demonstrated that the i-to-I transition during erythroid differentiation is determined by the transcription factor CCAAT/enhancer binding protein α (C/EBPα).
Materials and methods
Our program was approved by the Institutional Review Board at Mackay Memorial Hospital, Taipei, Taiwan. Informed consent was obtained in accordance with the Declaration of Helsinki.
Erythroid differentiation of K-562 cells
Human leukemia cell line K-562 (American Type Culture Collection, Manassas, VA) was grown in 90% RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin (50 U/mL), and streptomycin (50 μg/mL). To induce erythroid differentiation, K-562 cells (5 × 105/mL) were cultured in medium added with 1 mM of sodium butyrate (Sigma, St Louis, MO) for 2 days.
Flow cytometry analysis
To detect the cell-surface I antigen, the cells were incubated with human monoclonal anti-I antibody (mAb OSK14; a generous gift from Dr Yoshihiko Tani, Osaka Red Cross Blood Center, Japan) at 4°C for 2 hours after blocking with 5% bovine serum albumin. The bound monoclonal antibodies (mAbs) were detected by incubation with fluorescein isothiocyanate (FITC)–conjugated goat anti–human IgM (Serotec, Raleigh, NC), and the cells were subjected to flow cytometry (Epics XL; Beckman Coulter, Fullerton, CA).
Quantification of IGnT and C/EBPα transcripts
Preparation of cellular total RNAs and synthesis of cDNAs were performed as described previously.24 The forward IAF16, IBF18, and ICF20 primers (primer sequences are found in Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) were used to amplify IGnTA, IGnTB, and IGnTC cDNAs, respectively, by polymerase chain reaction (PCR). The reverse primer used, IRs, was common for the 3 IGnT cDNAs. PCR for the β-actin cDNA was performed using the ACTF and ACTR primers. The cDNA sample and 2 pmole of each forward and reverse primer were added to 20 μL of the SYBR Green Master Mix (Applied Biosystems, Foster City, CA). Real-time PCR detection (GenAmp 7300 Sequence Detection System; Applied Biosystems) was used for quantification of the transcripts. Serial dilutions of the pGEM-T vector (Promega, Madison, WI) containing IGnTA, IGnTB, IGnTC, or β-actin cDNAs were used to generate standard curves for each respective transcript.
The C/EBPα transcript in the cDNA sample was quantified using the TaqMan gene expression assay kit for C/EBPα (Applied Biosystems). Serial dilution of the pGEM-T vector containing C/EBPα cDNA was used to generate a standard curve.
Reporter assay
The DNA fragments encompassing the −3359- to +62–bp regions of the IGnTC gene (translation initiation codon as nucleotides +1 to +3) were PCR-amplified using the primers IC5′Fe and IC60R. Genomic DNA from K-562 cells served as the template. The amplified fragment was inserted into the KpnI and XhoI sites of the pGL3-basic vector (Promega), which contains the firefly luciferase reporter gene, yielding the −3359/+62-bp reporter construct. The StuI, EcoRV, and NdeI restriction sites, locating at the −1524, −538, and −318 positions, respectively, were used to digest the −3359/+62-bp construct and shorten the encompassed IGnTC 5′ region and, thus, built the −1524/+62-, −538/+62-, and −318/+62-bp constructs, respectively. The −253/+62-bp and −77/+62-bp segments were PCR-amplified using the reverse primer IC60R and forward primers of IC5′−253 and ICF2, respectively, and were then inserted into the XhoI site of the pGL3-basic vector, yielding the −253/+62- and −77/+62-bp constructs. The −322 to −251–bp region was PCR-amplified using the primers IC5′F-322 and IC5′R-251 and inserted into the SmaI site of the −77/+62-bp construct, yielding the −322/−251&−77/+62-bp construct, which had the −322 to −251–bp region inserted immediately upstream of the −77 to +62–bp region.
Transfection of K-562 cells was performed as described previously.24 The cells were subsequently cultured in medium with or without sodium butyrate for 2 days before being harvested to analyze luciferase activity using the Bright-Glo Luciferase Assay System (Promega).
Expression of Oct-2, Sp1, and C/EBPα transcription factors
The full coding cDNAs for the Oct-2, Sp1, and C/EBPα factors were obtained by PCR using the primer pairs OCTF13K/OCTR14, SPF5/SPR6, and CEBPAF9/CEBPAR10, respectively, with the cDNA sample from the K-562 cells serving as the template. Each of the amplified cDNAs was cloned into the pcDNA3.1(−) mammalian expression vector (Invitrogen, Carlsbad, CA). At 72 hours after transfection of the K-562 cells, the transfected cells were enriched under the selection of antibiotic G418 (1 μg/mL) for 72 hours before harvesting.
Nuclear protein extraction, immunoprecipitation, and Western blotting
For nuclear and cytoplasmic protein extractions, the described protocols25 were followed with minor modifications. Protein concentrations were determined using the Protein Assay kit (Bio-Rad, Hercules, CA).
A total of 20 μg of cytoplasmic and nuclear proteins were separated using 10% polyacrylamide gel electrophoresis and transferred to Immobilon-P membrane (Millipore, Billerica, MA). After blocking with 5% bovine serum albumin, the blots were detected with anti-C/EBPα antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The bound antibody was detected with horseradish peroxidase–conjugated secondary antibodies (Leinco Technologies, St Louis, MO), and the signals developed using Western Lighting Chemiluminescence Reagent Plus (PerkinElmer, Waltham, MA). After stripping, the membrane was blotted successively with anti–α-tubulin (Abcam, Cambridge, United Kingdom) and anti–histone H1 (Stressgen, Victoria, BC) antibodies.
For analysis of the phosphorylation status of C/EBPα, nuclear protein (20 μg) was immunoprecipitated with anti-C/EBPα antibody (1 μg) in a final volume of 500 μL at 4°C for 16 hours, and the antibody-bound proteins were then precipitated using protein A–sepharose CL-4B gel (Amersham, Piscataway, NJ). The bound proteins were separated using 10% polyacrylamide gel electrophoresis and transferred to Immobilon-P membrane. The blots were detected with anti-phosphoSer/Thr/Tyr, anti-phosphoSer (Abcam), anti-phosphoTyr, and anti-phosphoThr (Santa Cruz Biotechnology) antibodies. The signals were developed as described. Each blot was then stripped and detected with anti-C/EBPα antibody.
ChIP analysis
Chromatin immunoprecipitation (ChIP) analysis was performed using the ChIP assay kit (Upstate, Charlottesville, VA). The described protocols25 were followed with minor modifications. Briefly, the cells were incubated in 1% formaldehyde for 10 minutes at room temperature to cross-link the transcription factors with chromatin DNA. The cross-linking reaction was quenched by addition of glycine (125 mM, final concentration). Cells were then lysed with ChIP lysis buffer.25 Chromatins in the lysates were sheared by sonication, and bound with anti–Oct-2, anti-Sp1, or anti-C/EBPα antibodies (Santa Cruz Biotechnology) at 4°C for 2 hours. The antibody-bound chromatin DNAs were precipitated by protein A–agarose gel. After elution, the chromatin DNAs were relieved from the cross-linked proteins by heating at 65°C overnight, and precipitated in 70% ethanol. One-thirtieth of the immunoprecipitated DNAs or input DNA controls served as templates in the PCR amplification for the IGnTC −322 to +62–bp region using IC5′F-322 and IC60R primers. The products were analyzed using 2.0% agarose gel electrophoresis.
Isolation of CD34+ and CD71+ cells and E culture
Adult CD34+ and CD71+ progenitor cells were isolated from granulocyte-colony stimulating factor (G-CSF)–mobilized peripheral blood stem cells and general peripheral blood cells, respectively. Neonatal CD34+ and CD71+ cells were isolated from umbilical cord blood cells. Mononuclear cells were first isolated from the blood samples by Ficoll density gradient using Ficoll-Plaque Plus (Amersham). CD34+ and CD71+ cells were purified using the Dynal CD34 Progenitor Cell Selection System (Invitrogen) and CD71 MicroBeads kit (Miltenyi Biotech, Bergisch Gladbach, Germany), respectively, following the manufacturers' protocols.
For erythropoietic (E) culture, the CD34+ cells (1.5 × 105/mL) were cultured in StemSPAN SFEM medium (StemCell Technologies, Vancouver, BC) supplemented with stem cell factor (50 ng/mL; StemCell Technologies) and erythropoietin (3 U/mL; Janssen-Cilag, Titusville, NJ) for 7 days.
Lentiviral expression of C/EBPα
The packaging vector pCMVdeltaR8.91 and the vesicular stomatitis virus G protein (VSV-G) envelope glycoprotein–expressing vector pMD.G were provided by the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). The full coding cDNA of C/EBPα, obtained as described, was cloned into the pLenti6/V5-D-TOPO vector (Invitrogen), constructing the gene transfer vector pLenti6-C/EBPα. HEK 293T cells (American Type Culture Collection), cultured in Dulbecco modified Eagle medium (DMEM) in 6-well plates (105 cells/well), were transfected with the pLenti6-C/EBPα or mock pLenti6 vectors (0.5 μg) together with pCMVdeltaR8.91 (0.5 μg) and pMD.G (0.1 μg) using Lipofectamine Transfection Reagent (Invitrogen). The medium was collected 48 hours after transfection, and the viron suspensions were concentrated through ultracentrifugation. The CD34+ cells, cultured in StemSPAN SFEM medium in 24-well plates (3 × 105 cells/well), were transduced with the produced viron in medium with added polybrene (8 μg/mL). The cells were transferred to 6-well plates 16 hours after transduction, and were cultured for 72 hours before harvest.
Results
Induction of I antigen and IGnTC gene expression in K-562 erythroid differentiation
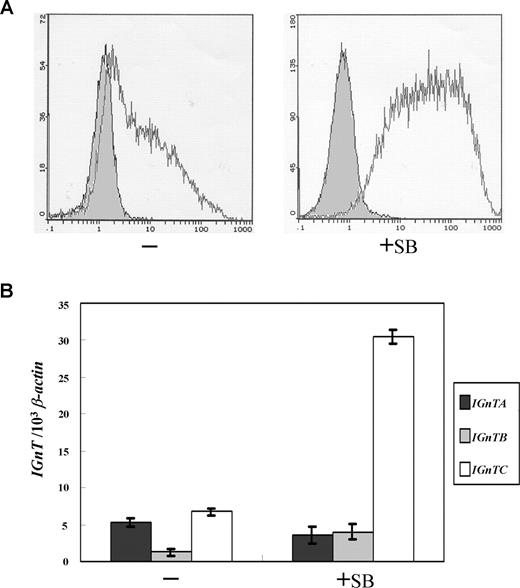
It has been shown that K-562 cells differentiate into erythrocyte lineage when treated with histone deacetylase inhibitors such as sodium butyrate.26,,–29 Further, it has been demonstrated that the treatment of sodium butyrate elicits i-to-I antigen conversion in K-562 cells.30 As shown in Figure 1A, the original K-562 cells weakly express I antigen. This agrees with previous observation that 2% to 5% of K-562 cells are I-antigen positive.30 When K-562 cells were cultured in medium supplemented with sodium butyrate for 2 days, the expression of I antigen is markedly induced. Benzidine staining was used to demonstrate the cells that express hemoglobin.31 The original K-562 cells were virtually devoid of benzidine-positive particles; after sodium butyrate treatment for 2 days, however, around 40% of the cells were positive with benzidine staining (data not shown), indicating induction of hemoglobin expression and erythropoietic differentiation in the K-562 cells.
Expression of I antigen and IGnT genes in erythroid differentiation of K-562 cells. (A) Flow cytometry analysis for I antigen expression. K-562 cells were cultured in medium supplemented with 1 mM of sodium butyrate for 2 days to induce erythroid differentiation. Expressions of the cell-surface I antigens on the K-562 cells with or without sodium butyrate treatment (indicated as +SB and −, respectively) were analyzed using flow cytometry and detected with monoclonal anti-I antibody (mAb OSK14; 1:5 dilution), with the bound mAb on the cell surfaces revealed by FITC-conjugated goat anti-human IgM (1:200 dilution). The open and shaded areas represent results obtained from cells incubated with anti-I mAb and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expression profiles for the IGnT transcripts. Real-time PCR was used to quantify the IGnTA, IGnTB, IGnTC, and β-actin transcripts in the cDNA samples. The quantities of the IGnT transcripts were normalized to that of β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown.
Expression of I antigen and IGnT genes in erythroid differentiation of K-562 cells. (A) Flow cytometry analysis for I antigen expression. K-562 cells were cultured in medium supplemented with 1 mM of sodium butyrate for 2 days to induce erythroid differentiation. Expressions of the cell-surface I antigens on the K-562 cells with or without sodium butyrate treatment (indicated as +SB and −, respectively) were analyzed using flow cytometry and detected with monoclonal anti-I antibody (mAb OSK14; 1:5 dilution), with the bound mAb on the cell surfaces revealed by FITC-conjugated goat anti-human IgM (1:200 dilution). The open and shaded areas represent results obtained from cells incubated with anti-I mAb and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expression profiles for the IGnT transcripts. Real-time PCR was used to quantify the IGnTA, IGnTB, IGnTC, and β-actin transcripts in the cDNA samples. The quantities of the IGnT transcripts were normalized to that of β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown.
To manifest which of the IGnT forms is responsible for the I antigen formation in butyrate-treated K-562 cells, expressions of the IGnTA, IGnTB, and IGnTC transcripts were quantitatively analyzed. The expression of the IGnTC transcript was shown to markedly increase, whereas IGnTA and IGnTB were not altered significantly, in butyrate-treated K-562 cells (Figure 1B), suggesting that the I antigen elevation in the erythroid differentiation of K-562 cells resulted from the activation of IGnTC gene expression. This accords with our previous demonstration that IGnTC is the gene responsible for the I antigen formation in red cells.21
These results strongly suggest the feasibility of using this K-562 erythroid differentiation model for investigation of the molecular mechanism for I antigen formation in red cells.
Identification of the cis DNA element regulating the transcription of the IGnTC gene
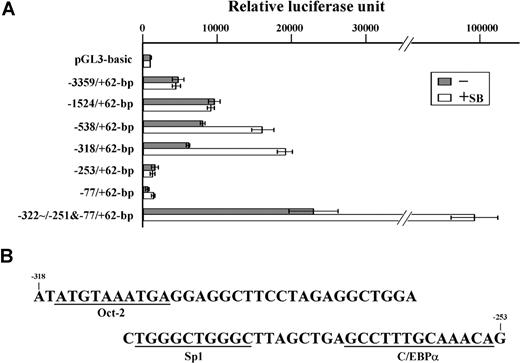
A reporter assay was used to identify the cis-acting DNA element responsible for the regulation of IGnTC gene expression in K-562 erythroid differentiation. When the introduced IGnTC 5′ regions were shortened stepwise from nucleotide −3359 to −318, the transcriptional activities enhanced by sodium butyrate treatment gradually increased (Figure 2A). However, the transcriptional activity of the −253/+62-bp construct was drastically reduced to a level similar to that of the pGL3-basic plasmid. This result suggests that a cis-acting DNA element responsible for the butyrate-induced activation of the IGnTC gene is located in the −318 to −253–bp region. The −322-251−77/+62-bp construct, which had the −322 to −251–bp segment introduced upstream to the −77 to +62–bp region, exhibited strong transcriptional activities, especially when the transfected cells were treated with sodium butyrate, further supporting this suggestion.
Identification of the cis-acting DNA region regulating IGnTC gene transcription. (A) Comparison of transcriptional activities of the IGnTC gene 5′ regions. Different 5′ segments of the IGnTC gene were introduced upstream of the luciferase reporter gene of the pGL3-basic vector. The −322/−251&−77/+62-bp reporter vector had the −322 to −251–bp region inserted immediately upstream of the −77 to +62–bp region. K-562 cells were split into 6-well culture plates at a density of 2 × 105 cells/mL, and transfected with 1.0 μg of the constructed and mock pGL3-basic reporter plasmids. The transcriptional activities of the constructed reporter plasmids in the transfected K-562 cells, with or without sodium butyrate treatment, are schematically represented by □ and ▩, respectively. The results are presented as averages of luciferase activities from 3 repetitions; standard deviations are shown. (B) Nucleotide sequence of the −318 through −253 region of the IGnTC gene. The putative binding motifs for the Oct-2, Sp1, and C/EBPα transcription factors are underlined.
Identification of the cis-acting DNA region regulating IGnTC gene transcription. (A) Comparison of transcriptional activities of the IGnTC gene 5′ regions. Different 5′ segments of the IGnTC gene were introduced upstream of the luciferase reporter gene of the pGL3-basic vector. The −322/−251&−77/+62-bp reporter vector had the −322 to −251–bp region inserted immediately upstream of the −77 to +62–bp region. K-562 cells were split into 6-well culture plates at a density of 2 × 105 cells/mL, and transfected with 1.0 μg of the constructed and mock pGL3-basic reporter plasmids. The transcriptional activities of the constructed reporter plasmids in the transfected K-562 cells, with or without sodium butyrate treatment, are schematically represented by □ and ▩, respectively. The results are presented as averages of luciferase activities from 3 repetitions; standard deviations are shown. (B) Nucleotide sequence of the −318 through −253 region of the IGnTC gene. The putative binding motifs for the Oct-2, Sp1, and C/EBPα transcription factors are underlined.
The −318 to −253–bp nucleotide sequence was analyzed using the Transcription Element Search System utility from the Computational Biology and Informatics Laboratory at the University of Pennsylvania (http://www.cbil.upenn.edu/cgi-bin/tess/tess),32 and the putative binding motifs for the transcription factors of Oct-2, Sp1, and C/EBPα were identified (Figure 2B).
Overexpression of C/EBPα in K-562 cells stimulates I antigen and IGnTC gene expression
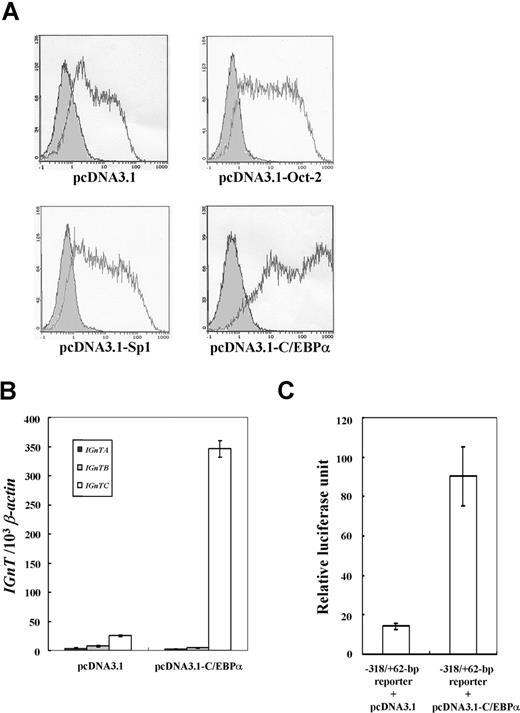
Overexpression experiments in K-562 cells were performed to study whether the transcription factors Oct-2, Sp1, and C/EBPα affected IGnTC gene expression. Flow cytometry analyses showed that the cell surface I antigen on K-562 cells was slightly increased with overexpression of Oct-2 and Sp1, whereas it was markedly enhanced by C/EBPα overexpression (Figure 3A). Coexpression experiments further demonstrated that Oct-2 and Sp1 do not act in synergy with C/EBPα and have little effect in the induction of I antigen expression (Figure S1).
Overexpression of Oct-2, Spl, and C/EBPα in K-562 cells. (A) Expressions of I antigen in the K-562 cells with overexpression of Oct-2, Sp1, and C/EBPα. The pcDNA3.1 expression vector harboring the respective full coding cDNAs of Oct-2, Sp1, or C/EBPα was transfected into the K-562 cells. K-562 cells were split into 6-well culture plates at a density of 2 × 105 cells/mL, and transfected with 1.0 μg of the constructed and mock pcDNA3.1 expression vector. The antibiotic-selected cells were then analyzed for the expression of I antigen using flow cytometry. Open and shaded areas are representative of the cells detected with anti-I mAb (1:5 dilution) and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expression profiles for IGnT transcripts in the K-562 cells with C/EBPα overexpression. The expressions of the IGnTA, IGnTB, and IGnTC transcripts in the K-562 cells transfected with the C/EBPα expression vector or mock pcDNA3.1 plasmid were analyzed using real-time PCR. Quantities of the IGnT transcripts were normalized to that of β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown. (C) Transcriptional activity of C/EBPα on IGnTC gene 5′ promoter. K-562 cells were cotransfected with the −318/+62-bp reporter construct and the C/EBPα expression vector or with the same reporter vector and mock pcDNA3.1 plasmid. After transfection for 48 hours, the cells were harvested to analyze the activity of the expressed luciferase. Results are presented as averages of luciferase activities from 3 repetitions; standard deviations are shown.
Overexpression of Oct-2, Spl, and C/EBPα in K-562 cells. (A) Expressions of I antigen in the K-562 cells with overexpression of Oct-2, Sp1, and C/EBPα. The pcDNA3.1 expression vector harboring the respective full coding cDNAs of Oct-2, Sp1, or C/EBPα was transfected into the K-562 cells. K-562 cells were split into 6-well culture plates at a density of 2 × 105 cells/mL, and transfected with 1.0 μg of the constructed and mock pcDNA3.1 expression vector. The antibiotic-selected cells were then analyzed for the expression of I antigen using flow cytometry. Open and shaded areas are representative of the cells detected with anti-I mAb (1:5 dilution) and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expression profiles for IGnT transcripts in the K-562 cells with C/EBPα overexpression. The expressions of the IGnTA, IGnTB, and IGnTC transcripts in the K-562 cells transfected with the C/EBPα expression vector or mock pcDNA3.1 plasmid were analyzed using real-time PCR. Quantities of the IGnT transcripts were normalized to that of β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown. (C) Transcriptional activity of C/EBPα on IGnTC gene 5′ promoter. K-562 cells were cotransfected with the −318/+62-bp reporter construct and the C/EBPα expression vector or with the same reporter vector and mock pcDNA3.1 plasmid. After transfection for 48 hours, the cells were harvested to analyze the activity of the expressed luciferase. Results are presented as averages of luciferase activities from 3 repetitions; standard deviations are shown.
The expression profiles of the 3 IGnT forms show that the transcription of the IGnTC gene, but not IGnTA and IGnTB, was induced dramatically in K-562 cells with C/EBPα expression (Figure 3B). This profile of IGnTC gene induction is consistent with that observed in the butyrate-induced erythroid differentiation of the same cell, and indicates that the ability of C/EBPα to enhance I antigen formation in K-562 cells is mediated through the capability to stimulate IGnTC gene expression. The luciferase activity in the K-562 cells cotransfected with the −318/+62-bp reporter construct and the C/EBPα expression plasmid was much higher than that in analogs transfected with the same reporter vector and the mock pcDNA3.1 plasmid (Figure 3C). This demonstrates the enhancer activity of the IGnTC −318 to +62–bp region in the presence of C/EBPα transcription factor.
These results are clear evidence of the functional role of C/EBPα in the stimulation of IGnTC gene expression and subsequent I antigen formation in the erythroid differentiation of K-562 cells.
C/EBPα associates with IGnTC gene promoter after butyrate treatment
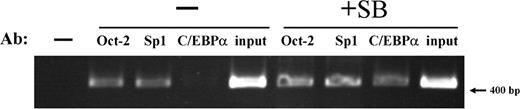
The association of Oct-2, Sp1, and C/EBPα transcription factors with the IGnTC 5′ region (−322 to +62 bp) in K-562 cells was examined using ChIP analysis. The results reveal that Oct-2 and Sp1 associated with the IGnTC 5′ region; however, there was no definite change in association status comparing the butyrate-treated and untreated cells (Figure 4). By contrast, there appeared to be almost no association between C/EBPα and the −322 to +62–bp region in the untreated K-562 cells; however, there was significant association after butyrate treatment.
Association of Oct-2, Sp1, and C/EBPα transcription factors with the IGnTC gene 5′ promoter region. The association of Oct-2, Sp1, and C/EBPα transcription factors with the 5′ promoter region (−322 to +62 bp) of the IGnTC gene in K-562 cells was examined using ChIP analysis. A total of 1 × 107 K-562 cells with or without sodium butyrate treatment (indicated as +SB and −, respectively) were used. The chromatin DNAs immunoprecipitated with 1:500 dilutions of anti–Oct-2, anti-Sp1, or anti-C/EBPα antibodies (Ab) and the input DNA controls were used for PCR amplification. Butyrate-treated cells were used for the no-antibody control in PCR amplification (leftmost lane).
Association of Oct-2, Sp1, and C/EBPα transcription factors with the IGnTC gene 5′ promoter region. The association of Oct-2, Sp1, and C/EBPα transcription factors with the 5′ promoter region (−322 to +62 bp) of the IGnTC gene in K-562 cells was examined using ChIP analysis. A total of 1 × 107 K-562 cells with or without sodium butyrate treatment (indicated as +SB and −, respectively) were used. The chromatin DNAs immunoprecipitated with 1:500 dilutions of anti–Oct-2, anti-Sp1, or anti-C/EBPα antibodies (Ab) and the input DNA controls were used for PCR amplification. Butyrate-treated cells were used for the no-antibody control in PCR amplification (leftmost lane).
Induction of the IGnTC gene is accompanied by C/EBPα dephosphorylation
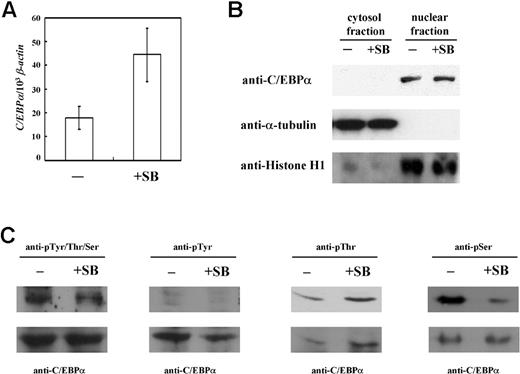
The results of the overexpression experiment make evident the determining role of C/EBPα in the induction of I antigen formation in K-562 cells, while ChIP analysis shows that the association of C/EBPα with the IGnTC 5′ promoter region is induced by sodium butyrate treatment. The expressed mRNA and protein levels of C/EBPα in butyrate-treated and untreated K-562 cells were subsequently examined further. The quantity of C/EBPα transcript in the butyrate-treated cells increased to about 2.5-fold that of the untreated cells (Figure 5A); however, there was no noticeable change in the expressed C/EBPα protein, observed exclusively in the nuclear fractions but not in the cytoplasmic fractions, comparing the butyrate-treated and untreated cells (Figure 5B). Only the 42-kDa products of C/EBPα were observed; the 30-kDa alternative-translated product33 was not detected in our Western blot analysis.
Expression and phosphorylation status of C/EBPα in K-562 cells. (A) Expression of C/EBPα transcript. Expressions of C/EBPα transcript in sodium butyrate-treated and untreated K-562 cells (+SB and −, respectively) were compared using real-time PCR. The quantity of the C/EBPα transcript was normalized to that of the β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown. (B) Western blotting for C/EBPα. C/EBPα protein levels in the cytoplasmic and nuclear fractions of sodium butyrate-treated and untreated K-562 cells were analyzed using Western blotting with anti-C/EBPα antibody (1:500 dilution). After stripping, the membrane was detected successively with the antibodies against α-tubulin (1:5000 dilution) and histone H1 (1:500 dilution), which are exclusively present in the cytosol and nucleus, respectively. (C) Phosphorylation status of C/EBPα. Nuclear proteins, prepared from K-562 cells with or without sodium butyrate treatment were immunoprecipitated using anti-C/EBPα antibody. The bound proteins were then analyzed with Western blotting using the following antibodies: anti-phosphoTyr/Thr/Ser (1:4000 dilution), anti-phosphoTyr (1:2000 dilution), anti-phosphoThr (1:1000 dilution), and anti-phosphoSer (1:2000 dilution) (top row of panels). Each blot was then stripped and detected with anti-C/EBPα antibody (bottom row of panels).
Expression and phosphorylation status of C/EBPα in K-562 cells. (A) Expression of C/EBPα transcript. Expressions of C/EBPα transcript in sodium butyrate-treated and untreated K-562 cells (+SB and −, respectively) were compared using real-time PCR. The quantity of the C/EBPα transcript was normalized to that of the β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown. (B) Western blotting for C/EBPα. C/EBPα protein levels in the cytoplasmic and nuclear fractions of sodium butyrate-treated and untreated K-562 cells were analyzed using Western blotting with anti-C/EBPα antibody (1:500 dilution). After stripping, the membrane was detected successively with the antibodies against α-tubulin (1:5000 dilution) and histone H1 (1:500 dilution), which are exclusively present in the cytosol and nucleus, respectively. (C) Phosphorylation status of C/EBPα. Nuclear proteins, prepared from K-562 cells with or without sodium butyrate treatment were immunoprecipitated using anti-C/EBPα antibody. The bound proteins were then analyzed with Western blotting using the following antibodies: anti-phosphoTyr/Thr/Ser (1:4000 dilution), anti-phosphoTyr (1:2000 dilution), anti-phosphoThr (1:1000 dilution), and anti-phosphoSer (1:2000 dilution) (top row of panels). Each blot was then stripped and detected with anti-C/EBPα antibody (bottom row of panels).
It has been shown that posttranslational modifications of phosphorylation34,,–37 and SUMOylation38 may affect C/EBPα function. Thus, the phosphorylation and SUMOylation status of C/EBPα in K-562 cells was examined. Western blotting with anti–SUMO-1 antibody was used for the detection of the immunoprecipated nuclear C/EBPα proteins, and it appeared that SUMO modification did not take place on C/EBPα in butyrate-treated/untreated K-562 cells (data not shown). In contrast to SUMOylation, phosphorylation was demonstrated on C/EBPα (Figure 5C). Detection with anti-phosphoTyr/Thr/Ser antibody revealed slightly decreased phosphorylation status in the butyrate-treated K-562 cells compared with the untreated cells. The phosphorylated residues were further dissected using anti-phosphoTyr, anti-phosphoThr, and anti-phosphoSer antibodies. The results suggest phosphorylation occurrence at Thr and Ser residues, but not Tyr, and that the phosphoThr content remained unchanged, while that of phosphoSer was significantly decreased, after butyrate treatment in K-562 cells.
Some elevation of C/EBPα transcript levels was exhibited; however, the protein levels remained unchanged in undifferentiated and erythropoietic-differentiated K-562 cells. Thus, it appears that the C/EBPα binding and induction capabilities for the IGnTC promoter and IGnTC transcription, respectively, are not determined at the transcriptional and translational levels of control, but accompany changes in phosphorylation status. These findings suggest that posttranslational regulation, such as phosphorylation (possibly dephosphorylation of certain Ser residues), may play a crucial role in C/EBPα IGnTC-activation function.
Expression profiles of I antigen, IGnT gene, and C/EBPα, and ChIP analysis in adult and cord CD34+, CD71+, and E-cultured cells
To elucidate whether C/EBPα plays a similar role in in vivo erythropoiesis to that demonstrated using the K-562 cell model, the CD34+ and CD71+ blood progenitor cells and E-cultured cells from adults and umbilical cords were subjected to further investigation.
The CD34+ and CD71+ cells were isolated as described in “Isolation of CD34+ and CD71+ cells and E culture.” Flow cytometry analyses showed that more than 95% of the collected cells possessed the respective CD marker (data not shown). Differentiation of the adult and cord CD34+ cells into erythrocyte lineage was induced in E culture for 7 days, with most (> 99%) becoming CD71-marker positive and with only a trace of glycophorin A detected on the cell surfaces (data not shown), suggesting cellular differentiation into early erythroblasts. When the cells were cultured for 18 days, the CD71 marker declined, and glycophorin A was noticeably expressed, indicating that the cells had further differentiated into late erythroblasts. The early erythroblasts, differentiated from CD34+ cells in E culture for 7 days, were used in the following analyses.
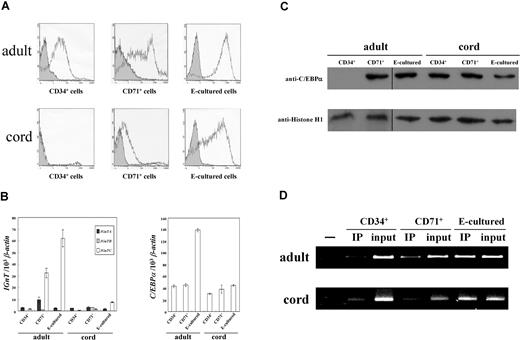
The I antigen was found to weakly express on adult CD34+ cells, and was markedly increased in adult CD71+ cells (Figure 6A). The increase was even more prominent in the E-cultured cells from adults. In the cord cells, I antigen was deficient in the CD34+ cells, with only a trace of I detected on the CD71+ cells. These results agree with previous findings that fetal and cord red cells contained only modest amounts of the I antigen.9,10 When the cord CD34+ cells differentiated into early erythroblasts in E culture, the level of I antigen increased noticeably; however, the amplitude of the increase was less than that for the adult E-cultured cells. Thus, it appears reasonable to suggest that I antigen expression can be induced in cord CD34+ cells using this in vitro E culture system; however, it does not seem to occur in in vivo fetal erythropoiesis, as the cord CD71+ cells do not express sufficient I antigen.
Expression profiles of I antigen, IGnT, and C/EBPα, and ChIP analysis in adult and cord CD34+, CD71+, and E-cultured cells. (A) Flow cytometry analysis for I antigen expression. Expressions of the cell-surface I antigens were analyzed using flow cytometry and detected with anti-I mAb (1:5 dilution). Open and shaded areas represent cells detected with anti-I mAb and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expressions of IGnT and C/EBPα transcripts. Real-time PCR was used to quantify the IGnT and C/EBPα transcripts in the cDNA samples. The quantities of IGnT (left panel) and C/EBPα (right panel) transcripts were normalized to that of the β-actin transcript. Data were obtained from 3 detections; standard deviations are shown. (C) Western blotting for C/EBPα. Nuclear fractions of the adult and cord CD34+, CD71+, and E-cultured cells were analyzed for the expression of C/EBPα protein. After stripping, the membranes were detected with anti–histone H1 antibody. Vertical lines have been inserted to indicate repositioned gel lanes. (D) ChIP analysis. For ChIP analysis, 2 × 106 adult and 1 × 106 cord cells were used (top and bottom panels, respectively). The chromatin DNAs immunoprecipitated with anti-C/EBPα antibody and input DNA controls were used for PCR amplification (indicated as IP and input, respectively). CD71+ cells were used for the no-antibody controls in PCR amplification (leftmost lanes).
Expression profiles of I antigen, IGnT, and C/EBPα, and ChIP analysis in adult and cord CD34+, CD71+, and E-cultured cells. (A) Flow cytometry analysis for I antigen expression. Expressions of the cell-surface I antigens were analyzed using flow cytometry and detected with anti-I mAb (1:5 dilution). Open and shaded areas represent cells detected with anti-I mAb and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expressions of IGnT and C/EBPα transcripts. Real-time PCR was used to quantify the IGnT and C/EBPα transcripts in the cDNA samples. The quantities of IGnT (left panel) and C/EBPα (right panel) transcripts were normalized to that of the β-actin transcript. Data were obtained from 3 detections; standard deviations are shown. (C) Western blotting for C/EBPα. Nuclear fractions of the adult and cord CD34+, CD71+, and E-cultured cells were analyzed for the expression of C/EBPα protein. After stripping, the membranes were detected with anti–histone H1 antibody. Vertical lines have been inserted to indicate repositioned gel lanes. (D) ChIP analysis. For ChIP analysis, 2 × 106 adult and 1 × 106 cord cells were used (top and bottom panels, respectively). The chromatin DNAs immunoprecipitated with anti-C/EBPα antibody and input DNA controls were used for PCR amplification (indicated as IP and input, respectively). CD71+ cells were used for the no-antibody controls in PCR amplification (leftmost lanes).
The expression profiles of IGnT transcripts show that the expression of IGnTC, but not IGnTA and IGnTB, was markedly increased in adult CD71+ cells, and even more so in the adult E-cultured cells, when compared with adult CD34+ cells (Figure 6B left). In the cord cells, expression of the IGnTC gene was slightly enhanced in the E-cultured cells, but not in the CD71+ cells. These profiles demonstrate a good correlation between elevation of IGnTC gene expression and increased I antigen formation in these cells, again illustrating that I antigen formation in erythrocyte-lineage cells is determined by the IGnTC gene.
Little variation was observed in the profiles of the C/EBPα transcript among these cells (Figure 6B right), however, except in the adult E-cultured cells, which exhibited notable elevation of C/EBPα expression. Nevertheless, despite the similar levels of the C/EBPα transcript in adult CD34+ and CD71+ cells, C/EBPα protein was not detected in adult CD34+ cells although it was clearly present in adult CD71+ cells (Figure 6C). Furthermore, increased C/EBPα protein level was not observed in the adult E-cultured cells, their increased transcript levels notwithstanding. It has been shown that translational repression, due to the presence of an upstream open reading frame in the 5′ untranslated region of C/EBPα mRNA, provides 1 avenue for regulating C/EBPα expression.39 This may explain the absence of C/EBPα protein in adult CD34+ cells; nevertheless, further evidence is required to confirm this assumption. In contrast to the adult CD34+ cells, there is marked expression of C/EBPα protein in cord CD34+ cells. The C/EBPα protein levels in all 3 types of cord cells were parallel, and were similar to those levels in adult CD71+ and E-cultured cells, although the IGnTC and I antigen expressions in these cells of different origins (adult and cord) were quite different.
The results of ChIP analysis (Figure 6D) reveals that the profiles of the association between C/EBPα and the IGnTC promoter region paralleled the IGnTC induction in these adult and cord cells, and was similar to those demonstrated using the K-562 cell model; that is, notwithstanding that C/EBPα protein levels were similar, the association between C/EBPα and the IGnTC promoter altered notably and was well correlated with activation of IGnTC expression in the cells. Once again, this suggests that certain posttranslational mechanisms are required for functional activation of the IGnTC gene by C/EBPα.
The effect of sodium butyrate on CD34+ cells was also explored. However, inductions of I antigen and IGnT gene expressions were not detected in the cord and adult CD34+ cells treated with sodium butyrate (for 5 and 10 days; data not shown). It is believed that K-562 and hemopoietic progenitor CD34+ cells are quite different in terms of cellular status. K-562 are leukemia cells with a definite erythroid character. Thus, sodium butyrate treatment alone in CD34+ cells may not be adequate to stimulate similar responses to those observed in the K-562 cells.
Overexpression of C/EBPα in CD34+ cells stimulates I antigen and IGnTC gene expression
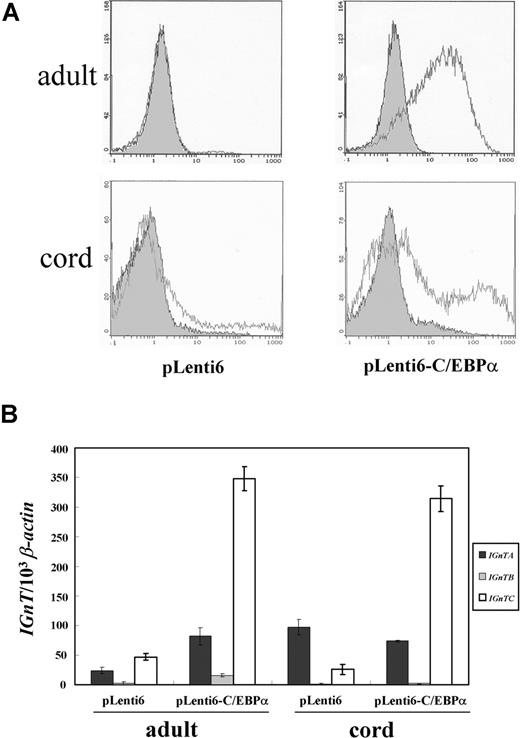
The C/EBPα factor was directly introduced into adult and cord CD34+ cells using the lentiviral expression system. Induction of I antigen expression (Figure 7A), significant in adult cells and to a lesser extent in cord cells, and stimulation of IGnTC gene transcription (Figure 7B) was observed in the cells transduced with virions prepared using the pLenti6-C/EBPα gene transfer vector. This result provides direct evidence of the determining role of C/EBPα in the stimulation of IGnTC gene expression and I antigen formation.
Lentiviral expression of C/EBPα in adult and cord CD34+ cells. (A) Flow cytometry analysis for I antigen expression. Expression of the I antigen on the cells transduced with the virions prepared from the pLenti6-C/EBPα gene transfer vector, which harbored full C/EBPα coding cDNA, and those transduced with the virions prepared from the mock pLenti6 vector were analyzed using flow cytometry and detected with anti-I mAb (1:10 dilution). Open and shaded areas indicate cells detected with anti-I mAb and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expression profiles of IGnT transcripts. Real-time PCR was used to quantify the IGnT and β-actin transcripts. Quantities of the IGnTA, IGnTB, and IGnTC transcripts were normalized to that of β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown.
Lentiviral expression of C/EBPα in adult and cord CD34+ cells. (A) Flow cytometry analysis for I antigen expression. Expression of the I antigen on the cells transduced with the virions prepared from the pLenti6-C/EBPα gene transfer vector, which harbored full C/EBPα coding cDNA, and those transduced with the virions prepared from the mock pLenti6 vector were analyzed using flow cytometry and detected with anti-I mAb (1:10 dilution). Open and shaded areas indicate cells detected with anti-I mAb and FITC-conjugated secondary antibody, and with FITC-conjugated secondary antibody only, respectively. (B) Expression profiles of IGnT transcripts. Real-time PCR was used to quantify the IGnT and β-actin transcripts. Quantities of the IGnTA, IGnTB, and IGnTC transcripts were normalized to that of β-actin transcript in each sample. Data were obtained from 3 detections; standard deviations are shown.
It has been reported that overexpression of C/EBPα in human CD34+ cells leads to granulocytic differentiation.40 In our C/EBPα overexpression experiments in CD34+ and K-562 cells, the possibility of differentiation of the transfected cells into granulocytic lineage, as suggested by the results of previous investigations, cannot be excluded; however, overexpression of C/EBPα did indeed induce IGnTC and I antigen expression in the transfected cells. In combination with other results observed using the K-562 erythroid differentiation model, this evidence demonstrates the determining role of C/EBPα in I antigen formation in erythroid differentiation.
Discussion
Several explanations for the prevention of severe hemolytic disease in the fetus and newborn (HDFN) due to ABO incompatibility have been proposed,4 with the weak expressions of the A and B antigens due to the straight-chain i structure on fetal red cells recognized as 1 of the mechanisms.4,18,19 The I antigen on red cells is controlled to appear after birth, suggesting that the formation of I branching structure should have an elaborate biological significance for postnatal red cells. In the present investigation, it was determined that the phenotypic transition from straight-chain i to branched-chain I on red cells is regulated by the transcription factor C/EBPα, which enhances transcription of the IGnTC gene and consequently leads to the I branching formation.
Despite the presence of IgG anti-A and anti-B antibodies in the serum of group O mothers, severe HDFN caused by these antibodies is rare.41 An IgG antibody molecule is able to attach both combining sites to its binding antigen, a condition called monogamous bivalency,42 with binding of the antibody to 2 or more binding sites favored over analogous single-site attachment by a factor greater than 103 or 104.43,44 The straight and branched poly-LacNAc structures on red cells can be further modified into the blood group A and B antigens. The paucity and spatial configuration of the A and B antigens carried by the straight poly-LacNAc chains restrain the monogamous bivalent binding of IgG anti-A and anti-B antibodies,18 and thus the straight poly-LacNAc structures on the fetal and neonatal red cells lead to very weak reactivity of the cells with anti-A and anti-B antibodies.41 The i-to-I transition in red cells, regulated to take place after birth, occurs at around the same time as the shift from fetal hemoglobin to adult hemoglobin. Furthermore, altered expression patterns for the I and i antigens have often been observed during oncogenetic processes.19,45 Based on these observations, it appears reasonable to infer that the transition between straight and branched poly-LacNAc structures should play a significant, but as yet unknown, cellular role.
The uncommon molecular genetic architecture of the I gene locus, which expresses the 3 IGnT transcripts under the control of specific regulatory mechanisms,21,22 affords the complex regulation of the spatially and temporally specific expression of the I antigen in various cells. For example, the IGnTC gene is induced during the erythroid differentiation processes, as presented in this study, whereas when hemopoietic cells differentiate into monocytic lineage, expression of the IGnTB gene is markedly increased and the cell surface I antigen is elevated (Y.-C.T. and L.-C.Y., unpublished data April 2005). The present study has demonstrated the determining role of C/EBPα in the i-to-I transition in red cells. C/EBPα belongs to a family of leucine-zipper transcription factors.33,46 These proteins share related N-terminal transactivation domains, DNA-binding regions, and C-terminal leucine-zipper protein interaction domains. C/EBPα binds as a homodimer or heterodimer with other C/EBP members or other transcription factors. Further, critical functions have been demonstrated in terms of regulating the balance between cell proliferation and differentiation, particularly in hepatocytes, adipocytes, and hemopoietic cells. In hemopoietic system, it is well known that C/EBPα plays a crucial role in the development of granulocytes and controls granulopoietic differentiation and proliferation in a stage-specific manner.47,48 In addition, it has been demonstrated that C/EBPα regulates the repopulating activity of hemopoietic stem cells.49
With the exception of granulocytic lineage, C/EBPα has not been implicated in the differentiation of other hemopoietic lineages.50 Thus, the finding that I antigen formation in erythroid differentiation is determined by C/EBPα was somewhat unexpected. However, it is well known that C/EBPα is a transcription factor with multiple functions. The variety of C/EBPα functions is manipulated in a number of ways, including translational control by alternative use of initiation codons,51 interaction with various transcription factors, and phosphorylation-mediated changes in DNA-binding activity, transactivation potential, and nuclear localization.33,46,47 Phosphorylation appears to play a critical role in the modulation of C/EBPα function. For example, protein kinase C can phosphorylate C/EBPα at Ser residues, leading to an attenuation of its DNA-binding activity.34 Further, signaling through the phosphatidylinositol 3-kinase (PI3K)–Akt pathway induces protein phosphatase 2A–mediated dephosphorylation of mouse C/EBPα on Ser-193 in liver tumor cells, leading to blockage of C/EBPα-mediated growth inhibition.36 Furthermore, it has been proven that phosphorylation of C/EBPα on Ser-21, mediated by extracellular signal-regulated kinase (ERK) 1/2, inhibits the ability of C/EBPα to induce granulopoiesis.35,37
The IGTC-activation activity of C/EBPα also appears to be modulated at a posttranslational level. Our preliminary results show that dephosphorylation of C/EBPα at certain Ser residues is accompanied by the enhancement of DNA binding of C/EBPα to its binding motif in the IGnTC promoter region and the induction of IGnTC gene expression. Further dissections are required to detailed elaboration of this posttranslational mechanism of C/EBPα IGnTC-activation function. Apart from the phosphorylation modification, it has also been demonstrated that translational repression plays an important role in the modulation of C/EBPα function.39,52 Thus, in spite of the apparent expression of the C/EBPα transcript, the absence of C/EBPα protein in adult CD34+ cells, is both perplexing and intriguing. If C/EBPα deficiency in adult CD34+ cells does result from translational repression, elucidation of the mechanism leading to the relief of this constraint of translational repression after differentiation into CD71+ cells would be of interest.
Overall, the cellular mechanisms in the erythrocyte-lineage cells can modify C/EBPα into the status (phosphorylation status, or some other undetermined modification) possessing the ability to activate the IGnTC gene, leading to I branching formation. Moreover, these cellular mechanisms in the erythrocyte-lineage cells would differ from those in granulocyte-lineage cells, which modify C/EBPα to provide regulation of a different set of genes for granulopoiesis. Further, it appears reasonable to suggest that the cellular mechanisms modifying C/EBPα to provide IGnTC activation may not be present in, or integral to, the fetal erythrocyte-lineage cells, and that these are not regulated to appear until birth. Further detailed elaboration of the C/EBPα modification required for its I antigen-activation activity would be of interest, therefore, together with articulation of the mechanism responsible for this modification in erythroid differentiation. Hence, further investigations elucidating the molecular bases for maintenance of the i phenotype on fetal red cells and leading to the i-to-I postnatal transition could be carried out and should yield answers that will be of particular interest and significance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported in part by grant NSC 96-2320-B-002-074-MY3 from the National Science Council, Taiwan.
Authorship
Contribution: Y.-C.T. designed and performed research, and analyzed data. C.-P.C. contributed vital reagents. C.-Y.H. performed research. C.-H.T. contributed vital reagents. C.-F.S. contributed vital reagents. S.-H.W. performed research. M.-S.C. contributed vital analytical tools. L.-C.Y. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lung-Chih Yu, Institute of Biochemical Sciences, National Taiwan University, PO Box 23-106, Taipei 106, Taiwan; e-mail:yulc@ntu.edu.tw.