Normal prion protein (PrPc), an essential substrate for development of prion disease, is widely distributed in hematopoietic cells. Recent evidence that variant Creutzfeldt-Jakob disease can be transmitted by transfusion of red cell preparations has highlighted the need for a greater understanding of the biology of PrPc in blood and blood-forming tissues. Here, we show that in contrast to another glycosylphosphoinositol-anchored protein CD59, PrPc at the cell surface of cultured human erythroblasts is rapidly internalized through the endosomal pathway, where it colocalizes with the tetraspanin CD63. In the plasma membrane, PrPc colocalizes with the tetraspanin CD81. Cross-linking with anti-PrPc or anti-CD81 causes clustering of PrPc and CD81, suggesting they can share the same microdomain. These data are consistent with a role for tetraspanin-enriched microdomains in trafficking of PrPc. These results, when taken together with recent evidence that exosomes released from cells as a result of endosomal-mediated recycling to the plasma membrane contain prion infectivity, provide a pathway for the propagation of prion diseases.
Introduction
Normal prion protein (PrPc) is a glycosylphosphoinositol (GPI)–linked membrane protein with a very wide tissue distribution. Its function is not clearly understood, although recent evidence suggests a role in self-renewal of hematopoietic stem cells.1 In contrast, abundant evidence supports a pivotal role for PrPc in the propagation of transmissible spongiform encephalopathies (TSEs) through interaction with misfolded prion protein (PrPsc).2 In particular, mice genetically modified to lack PrPc expression in all tissues do not succumb to disease when exposed to PrPsc.3,4 The process whereby PrPsc interacts with host PrPc, converting it to PrPsc and causing disease, is not clear, but a requirement for the GPI membrane anchor of PrPc has been demonstrated in mice engineered to express PrPc without a GPI anchor. When these animals are exposed to PrPsc, there is an extracellular proliferation of the abnormal protein but no disease develops, presumably because of a failure to internalize PrPsc within neural cells.5
Bovine spongiform encephalopathy acquired by oral ingestion of PrPsc present in meat is the most likely cause of variant Creutzfeldt-Jakob disease (vCJD) in humans.6,–8 Recent evidence demonstrates it is highly probable vCJD can be transmitted by transfusion of red cell preparations.9 These data highlight the need for a greater understanding of the distribution and trafficking of PrPc in human hematopoietic cells so that knowledge of the extent to which PrPsc binding and internalization can occur in blood and blood-forming tissues can be used to inform decisions regarding the safety of blood components used for therapy.
In this study, we have examined the expression and trafficking of PrPc in human erythroid cells derived from in vitro culture of CD34+ cells from peripheral blood. The results show that PrPc in erythroblasts behaves differently from other GPI-linked proteins (CD59 and CD55) in that it cycles rapidly from the plasma membrane to internal membrane compartments by a process likely to be clathrin-mediated and involving tetraspanin-enriched microdomains (TEMs).
Materials and methods
Waste buffy-coat material from anonymous blood donors were made available from the National Blood Service (Bristol, United Kingdom). This provision complies with the Nuffield Council on Bioethics Guidance on Human Tissue Ethical and Legal Issues 1954, the Medical Research Council's Operational and Ethical Guidance on Human Tissue and Biological Samples for use in Research 2005, and the Royal College of Pathologists Transitional Guidelines to facilitate changes in procedures for handling “surplus” and archival material from human biological samples.
Antibodies
Monoclonal antibodies to CD63 (H5C6) and CD81 (JS-81) were from BD Biosciences (Oxford, United Kingdom). Anti-CD82 (B-L2) was from Serotec (Oxford, United Kingdom). Monoclonal antibodies to glycophorin A (BRIC163) and CD59 (BRIC229) were from IBGRL (Bristol, United Kingdom). Anti-PrP (ICSM 18) was from D-Gen Ltd (London, United Kingdom). Rabbit polyclonal anti-calnexin was from Stressgen Bioreagents (Ann Arbor, MI), anti-giantin from CRP (San Diego, CA), and anti–LAMP-1 from Affinity Bioreagents (Golden, CO). AffiniPure Fab fragment rabbit anti–mouse IgG and rhodamine (TRITC)–conjugated AffiniPure goat anti–rabbit IgG were from Jackson ImmunoResearch (Newmarket, United Kingdom). FITC-conjugated F(ab′)2 fragment of goat anti–mouse Igs was from DAKO (Ely, United Kingdom). Alexa Fluor 488 goat anti–mouse IgG1, Alexa Fluor 546 goat anti–rabbit IgG, and Alexa Fluor 546 conjugate of anti-transferrin were from Invitrogen (Paisley, United Kingdom).
Erythroid cell culture
Human CD34+ cells were isolated from buffy coats (National Blood Service). Buffy coats were diluted 1:1 with Hanks balanced salt solution (HANKS; Sigma, Poole, United Kingdom) then layered on top of Histopaque 1077 (Sigma). Cells were harvested from the 1077 gradient, washed with HANKS, and then resuspended in red cell lysis buffer (ammonium chloride, 4.15 g/L; EDTA, 0.02 g/L; and potassium bicarbonate, 0.5 g/L). CD34+ cells were isolated by positive selection using the Direct CD34+ progenitor cell isolation kit (Miltenyi Biotech, Bisley, United Kindgom).
Isolated CD34+ cells were cultured under conditions based on the method of Southcott et al10 using serum-free StemSpan Expansion Medium (Stemcell Technologies, London, United Kingdom) supplemented with stem cell factor (SCF; 10 μL/mL; R&D Systems, Abingdon, United Kingdom), erythropoietin (EPO; 3 μL/mL, Roche, Lewes, United Kingdom), low-density lipoprotein (LDL; 8 μL/mL; Calbiochem, Nottingham, United Kingdom), and interleukin-3 (IL-3; 1 μL/mL; R&D Systems). Cells were seeded at 0.5 × 105/mL on day 0 and were maintained at a concentration of 1 × 105/mL thereafter in vented T25 Falcon flasks (Becton Dickinson, Oxford, United Kingdom) in 5% CO2 at 37°C.
Confocal microscopy
Cells were seeded on 0.01% (wt/vol) poly-L-lysine (Sigma) coated coverslips (1 × 106 cells per coverslip) and incubated overnight at 37°C in 5% CO2.
All steps were carried out in 6-well plates, with cells on the top surface of the coverslips for all washes, and with cells facing down on Parafilm M (Lab 3, Bristol, United Kingdom) for all antibody incubations.
Cells were fixed with 3% formaldehyde (TAAB, Aldermaston, United Kingdom) for 20 minutes and permeabilized with 0.05% (wt/vol) digitonin (Sigma) for 5 minutes. Cells were incubated for 15 minutes in 4% bovine serum albumin (BSA; Park Scientific, Northampton, United Kingdom) and then incubated with primary antibodies. Antibodies to glycophorin A or CD59 were used as positive controls, and 4% BSA was used as a negative control. For dual labeling, antibodies were incubated separately with 2 × 5-minute washes in phosphate-buffered saline (PBS) between incubations.
When dual labeling with 2 monoclonal primary antibodies, the weaker antibody was incubated first and then subjected to an extra conversion step by the addition of AffiniPure Fab Fragment Rabbit Anti-Mouse IgG (Jackson ImmunoResearch) at a concentration of 1:10 in 4% BSA. After this conversion step and 2 additional PBS washes, the other monoclonal antibody was added. Goat anti-mouse FITC/Alexa Fluor 488 or goat anti-rabbit TRITC/Alexa Fluor 546 secondary antibodies were diluted in 4% normal goat serum and incubated with the cells for 30 minutes at room temperature in the dark. Coverslips were mounted on Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA) on microscope slides and sealed with nail varnish.
Samples were imaged at 22°C using 63× oil-immersion lenses (magnification = 158.7 μm at zoom 1, 1.32 NA) on a Leica DM RBE upright epifluorescence microscope with phase contrast connected to a Leica TCS-NT confocal laser scanning microscope (Leica, Wetzlar, Germany) at the Medical Research Council (MRC) Imaging Facility, University of Bristol. Images were obtained using Leica software and subsequently processed using Adobe Photoshop (Adobe, San Jose, CA).
For protein internalization experiments, cells were seeded overnight as described and washed once in PBS before incubation with the primary antibody for 20 minutes at 10°C to allow antibody binding. The negative control used for internalization studies was normal mouse IgG (DAKO). Cells were incubated at 37°C for up to 1 hour to allow internalization of the antibody to occur and then fixed and permeabilized as described. After blocking with 4% BSA, either another primary antibody or the secondary antibody was added. All subsequent steps were carried out as described here.
Capping experiments
Erythroid cells were seeded overnight as described, washed once in PBS before incubation with primary antibodies (20 minutes at 10°C), washed twice in PBS, and incubated with secondary antibody (room temperature for 40 minutes) to allow capping to occur. Cells were then washed twice and fixed with 3% formaldehyde for 20 minutes, washed 3 times, and then mounted on slides. For dual labeling using mouse monoclonals, the rabbit conversion antibody was used before capping with secondary antibodies. The second primary antibody was then used after fixation as described.
Quantitation
Internalized PrP was measured by drawing regions of interest (ROIs) both inside the cell under the plasma membrane and around the outside of the cell using Leica quantitation software. The percentage difference in pixels was then calculated between these 2 ROIs.
Immunoprecipitation and Western blotting
Cells from day 6 to day 8 were harvested from culture, washed once in cold PBS, and the cell pellet was put on ice. Lysis buffer (25 mM HEPES buffer, 1 complete protease inhibitor cocktail tablet [Roche, Sussex, United Kingdom], plus 1% detergent: either Triton X-100 [Sigma], Nonidet P40 [NP-40] substitute [USB, Cleveland, OH], Brij97 [Sigma] or CHAPS [Calbiochem] with 0.5% sodium deoxycholate [Sigma] supplementing some 1% detergents) was added to the cell pellet and incubated on ice for 20 minutes. Lysed cells were then centrifuged at maximum speed (11 000g) for 15 minutes at 4°C to pellet debris.
Lysates were precleared with 30 μL protein G–sepharose beads (Amersham Biosciences, Bucks, United Kingdom) for 1 hour at 4°C and subsequently incubated with specific mAbs bound to protein G–sepharose beads overnight at 4°C, rotating. For immunoprecipitations from the cell surface, intact cells were incubated with antibody at 4°C for 1 hour before lysis and then incubated with protein G–sepharose beads overnight. For internalization immunoprecipitations, cells were incubated at 4°C for 30 minutes, then at 37°C for 1 hour to allow endocytosis to occur. After these incubation steps, cells were subject to lysis and then incubated with beads overnight.
Immunoprecipitated proteins were eluted from beads by boiling with sodium dodecyl sulfate (SDS) nonreducing sample buffer, subjected to SDS-PAGE on a 12.5% polyacrylamide gel, and transferred to a nitrocellulose membrane. After blocking in PBS containing 5% (wt/vol) BSA and 0.2% (wt/vol) Tween-20, membranes were probed with either primary mAbs followed by anti–mouse Igs conjugated to horseradish peroxidase (HRP; DAKO) or biotinylated primary mAbs followed by streptavidin peroxidase polymer (Sigma). Primary mAbs were biotinylated using EZ-Link sulfo-NHS-biotin Reagents (Pierce, Rockford, IL).
The presence of secondary protein was detected using the ECL Western Lightning Chemiluminescence Kit (Perkin Elmer, Waltham, MA) and photography film (Kodak BioMax MR film; Kodak, Rochester, NY) or the Kodak 4000R Image station, in accordance with the manufacturer's instructions.
Results
PrPc expression in human erythroid cells cultured in vitro
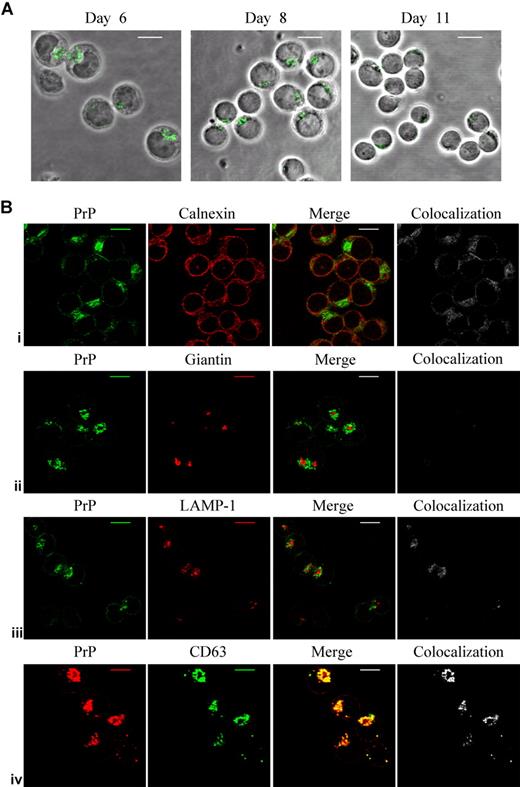
PrPc expression in cultured human erythroblasts was examined using immunofluorescence confocal microscopy. CD34+ cells isolated from peripheral blood were cultured in the presence of SCF, IL-3, and EPO and erythroid cells examined at various stages of maturation. After 6 to 8 days in culture, most cells are at the proerythroblast stage, and there is a large amount of intracellular PrPc with faint cell-surface staining. By day 11, when the cultures contain a heterogeneous population of basophilic erythroblasts and pronormoblasts, PrPc expression is lower and more diffuse with minimal cell-surface staining (Figure 1A).
PrPc expression in human erythroid cells cultured in vitro. (A) Erythroblasts were harvested on days 6, 8, and 11 of culture. After seeding overnight onto poly-L-lysine–coated slips, cells were fixed and permeabilized and then incubated with PrP mAb ICSM18 and subsequently with goat anti-mouse FITC. Scale bars equal 10 μm. (B) Day-6 erythroblasts were fixed and permeabilized. Dual staining of cells with PrP mAb ICSM18 show in green, and ER marker calnexin (i), Golgi marker giantin (ii), and lysosomal marker LAMP-1 (iii) shown in red. Bottom row (iv) shows PrP mAb (red) dual stained with CD63 mAb (green). Colocalization is highlighted in the grayscale image. Scale bars equal 10 μm.
PrPc expression in human erythroid cells cultured in vitro. (A) Erythroblasts were harvested on days 6, 8, and 11 of culture. After seeding overnight onto poly-L-lysine–coated slips, cells were fixed and permeabilized and then incubated with PrP mAb ICSM18 and subsequently with goat anti-mouse FITC. Scale bars equal 10 μm. (B) Day-6 erythroblasts were fixed and permeabilized. Dual staining of cells with PrP mAb ICSM18 show in green, and ER marker calnexin (i), Golgi marker giantin (ii), and lysosomal marker LAMP-1 (iii) shown in red. Bottom row (iv) shows PrP mAb (red) dual stained with CD63 mAb (green). Colocalization is highlighted in the grayscale image. Scale bars equal 10 μm.
The intracellular localization of PrPc in the cultured human erythroblasts was examined by dual staining with markers for organelles using cells from days 6 to 8 of culture (Figure 1B). Only a small proportion of PrPc staining colocalized with the endoplasmic reticulum (ER) marker calnexin, the Golgi marker giantin, and the lysosomal marker LAMP-1, with most PrPc that is present outside these organelles in intracellular membranes most likely to be endosomal (perinuclear recycling compartment) because it shows complete colocalization with the endosomal marker CD63. CD63, a member of the tetraspanin family, is known as a resident of late endosomes and lysosomes, but is also present in secretory vesicles and on the plasma membrane, and it cycles between these compartments.11
PrPc localization in human erythroblasts differs from that of CD59
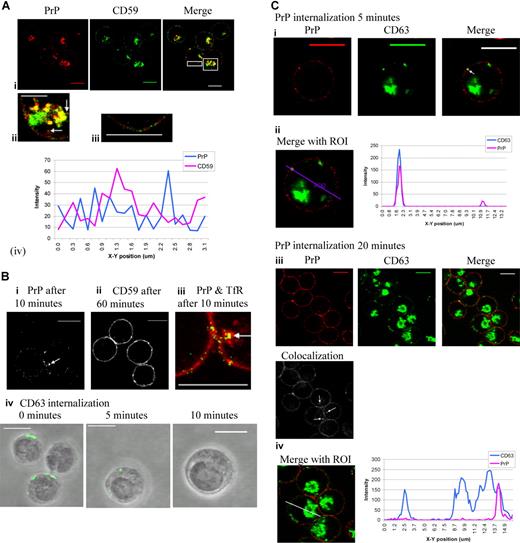
PrPc localization in erythroblasts was compared with that of another GPI-linked protein, CD59. Dual staining of PrPc and CD59 revealed substantial intracellular colocalization in the perinuclear area (Figure 2Ai,ii). However PrPc and CD59 were present in different regions of the plasma membrane (Figure 2Aiii,iv). PrPc was also present in vesicle-like structures just below the plasma membrane, whereas CD59 is not observed in these vesicle-like structures either alone or colocalized with PrPc (Figure 2Aii; arrows highlight PrPc). Another GPI-linked protein, CD55, showed almost complete colocalization with CD59 (data not shown). These results suggest trafficking of PrPc is not typical for GPI-linked proteins.
PrPc and CD59 occupy different microdomains in the plasma membrane, and PrPc cycles rapidly from the plasma membrane to the internal membranes. (Ai) Fixed and permeabilized day-7 erythroblasts were dual stained with PrP mAb (red) after use of conversion antibody 1/10 and CD59 (green) and subsequently stained with FITC and TRITC secondary antibodies. Overlays were prepared and highlighted areas from merge magnified to show (Aii) internal PrP-containing vesicles (arrows) and (Aiii) the plasma membrane. The profile (Aiv) of PrP and CD59 fluorescence colocalization in the surface of the cell membrane was analyzed by quantitation software (Leica). Scale bars equal 10 μm (Ai), 5 μm (Aii,iii). (B) Internalization assays were performed to compare the internalization of PrP and CD59 in erythroid cells. After allowing binding of the relevant antibody to the cell surface, cells were incubated at 37°C for up to 60 minutes to allow endocytosis to occur. (Bi) PrP is internalized within 10 minutes endocytosis (grayscale; arrow). (Bii) CD59 is not internalized after 60 minutes endocytosis (grayscale). (Biii) Dual staining of PrP (green) and TfR (red) after 10 minutes of endocytosis (arrow indicates internal colocalization). (Biv) Internalized CD63 was observed inside the erythroblasts at 5 and 10 minutes. Scale bars equal 10 μm. (C) PrP mAb (red) was subject to the internalization protocol, fixed and permeabilized, incubated with conversion antibody (1/10), and then dual stained with CD63 mAb (green). Cell-surface and internalized PrP only is shown stained in red and total CD63 in green. Areas of colocalization are yellow. (Ci) Internalized PrP after 5 minutes (red) and CD63 (green) colocalize in a vesicle-like structure just under the plasma membrane (arrow). (Cii) A region of interest (ROI) was drawn through the cell to include internalized PrP, and a profile of green and red fluorescence along this ROI was constructed using Leica quantiation software. (Ciii) Internalized PrP after 20 minutes (red) and CD63 (green) colocalize deep within the cell. Grayscale image shows areas of colocalization between PrP and CD63 only (arrows). (Civ) An ROI was drawn through a cell to include internalized PrP, and a profile of green and red fluorescence along this ROI was constructed using Leica quantiation software. Scale bars equal 10 μm.
PrPc and CD59 occupy different microdomains in the plasma membrane, and PrPc cycles rapidly from the plasma membrane to the internal membranes. (Ai) Fixed and permeabilized day-7 erythroblasts were dual stained with PrP mAb (red) after use of conversion antibody 1/10 and CD59 (green) and subsequently stained with FITC and TRITC secondary antibodies. Overlays were prepared and highlighted areas from merge magnified to show (Aii) internal PrP-containing vesicles (arrows) and (Aiii) the plasma membrane. The profile (Aiv) of PrP and CD59 fluorescence colocalization in the surface of the cell membrane was analyzed by quantitation software (Leica). Scale bars equal 10 μm (Ai), 5 μm (Aii,iii). (B) Internalization assays were performed to compare the internalization of PrP and CD59 in erythroid cells. After allowing binding of the relevant antibody to the cell surface, cells were incubated at 37°C for up to 60 minutes to allow endocytosis to occur. (Bi) PrP is internalized within 10 minutes endocytosis (grayscale; arrow). (Bii) CD59 is not internalized after 60 minutes endocytosis (grayscale). (Biii) Dual staining of PrP (green) and TfR (red) after 10 minutes of endocytosis (arrow indicates internal colocalization). (Biv) Internalized CD63 was observed inside the erythroblasts at 5 and 10 minutes. Scale bars equal 10 μm. (C) PrP mAb (red) was subject to the internalization protocol, fixed and permeabilized, incubated with conversion antibody (1/10), and then dual stained with CD63 mAb (green). Cell-surface and internalized PrP only is shown stained in red and total CD63 in green. Areas of colocalization are yellow. (Ci) Internalized PrP after 5 minutes (red) and CD63 (green) colocalize in a vesicle-like structure just under the plasma membrane (arrow). (Cii) A region of interest (ROI) was drawn through the cell to include internalized PrP, and a profile of green and red fluorescence along this ROI was constructed using Leica quantiation software. (Ciii) Internalized PrP after 20 minutes (red) and CD63 (green) colocalize deep within the cell. Grayscale image shows areas of colocalization between PrP and CD63 only (arrows). (Civ) An ROI was drawn through a cell to include internalized PrP, and a profile of green and red fluorescence along this ROI was constructed using Leica quantiation software. Scale bars equal 10 μm.
PrPc cycles rapidly from the plasma membrane to internal membranes and at a rate comparable with that of CD63
In order to compare the trafficking of PrPc and CD59 in erythroid cells, the rates of internalization were compared. After binding of the relevant antibody to the cell surface at 10°C, cells were transferred to 37°C and incubated for up to 60 minutes to allow endocytosis to occur, and the localization of the proteins were determined. The results showed that PrPc is visible inside erythroblasts after 10 minutes of incubation (Figure 2Bi). In contrast, internalized CD59 is not detected after 60 minutes of incubation (Figure 2Bii). These data suggest that PrPc is not internalized by the slower clathrin-independent mechanism used by other GPI-linked proteins.12 PrPc and the transferrin receptor (TfR) were internalized after 10 minutes and colocalized in vesicle-like structures under the plasma membrane (Figure 2Biii arrow). TfR is a prototypical nonraft protein and endocytoses in a clathrin-dependent manner.13 These results indicate that PrPc is subject to clathrin-mediated endocytosis. However, as a GPI-linked protein, PrPc lacks a cytoplasmic domain and so cannot bind directly to clathrin adaptor proteins. PrP colocalizes with the tetraspanin CD63 (Figure 1Biv) which, through its interaction with the adaptor proteins, has a role in protein trafficking.14 Our experiments showed that it internalizes over a similar time-scale to PrP (Figure 2Biv). Very little CD63 is evident at the plasma membrane of fixed and permeabilized erythroblasts. A similar distribution is seen in other cell types, where it is known to cycle to the cell surface.15
When erythroblasts were subjected to the internalization protocol with anti-PrPc, fixed, permeabilized, and stained for CD63, PrPc and CD63 were colocalized in vesicles just under the plasma membrane after 5 minutes (Figure 2Ci arrow). The profile in Figure 2Cii shows that PrPc internalized at 5 minutes is colocalized with CD63. After 20 minutes, PrPc-containing vesicles (corresponding to 22.7% ± 12.5% total PrPc staining; n = 30, results from 4 independent experiments) were colocalized with CD63 deep inside the erythroblasts at the perinuclear recycling area (Figure 2Ciii arrows). The profile in Figure 2Civ shows that all PrPc internalized at 20 minutes is colocalized with CD63.
PrPc colocalizes with tetraspanin CD81 at the plasma membrane
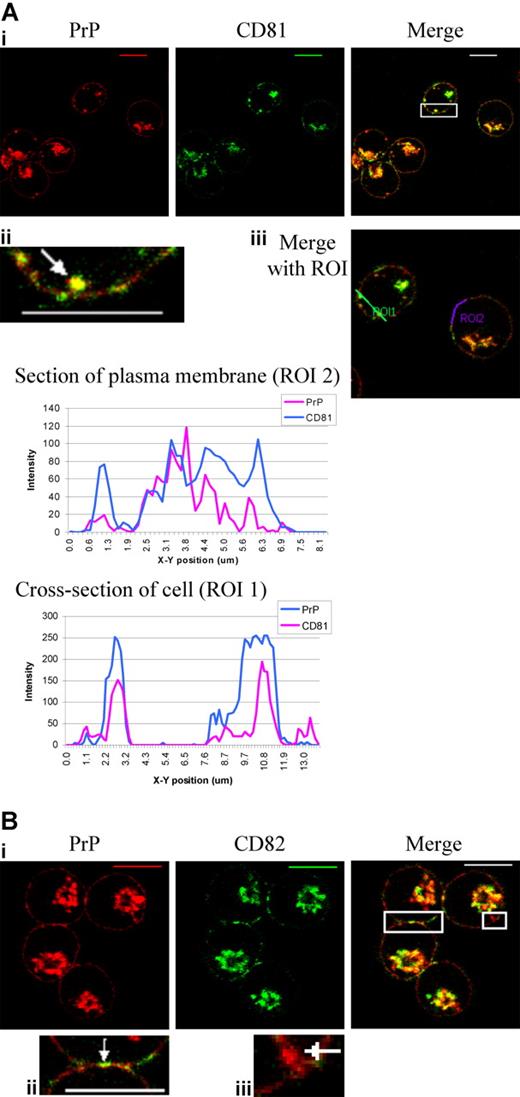
Tetraspanins associate specifically with partner proteins and other members of the tetraspanin family in TEMs. TEMs are distinct from lipid rafts and are thought to organize the plasma membrane and intracellular membranes by selectively concentrating specific membrane proteins.16 Nydegger et al15 demonstrate that in HeLa cells, the small fraction of CD63 found in the plasma membrane is located in microdomains together with other tetraspanins, including CD81 and CD82. In order to address the possible involvement of tetraspanin microdomains in cycling PrPc from the plasma membrane to the endosomal compartment, dual staining of PrPc with tetraspanins CD81 and CD82 was performed, since these are expressed in the plasma membrane of erythroblasts (Figure 3). PrPc and the tetraspanin CD151 were also dual stained but showed only a small amount of intracellular colocalization in the perinuclear area (data not shown).
PrPc colocalizes with tetraspanins CD81 and CD82 at the plasma membrane. (Ai) Fixed and permeabilized day-6 erythroblasts were dual stained with PrPc mAb (red) and CD81 mAb (green). Overlays were prepared and the highlighted area magnified to show (Aii) an internal colocalized vesicle (arrow) and the plasma membrane. (Aiii) ROIs were drawn through a cell (ROI1) and along the cell surface (ROI2), and profiles of green and red fluorescence along these ROIs were constructed using Leica quantiation software. Scale bars equal 10μm. (Bi) Fixed and permeabilized day-6 erythroblasts were dual stained with PrPc mAb (red) and CD82 mAb (green). Overlays were prepared and highlighted areas magnified to show (Bii) plasma membrane at cell-cell contact site and (Biii) internal PrP-containing vesicle (arrow). Scale bars equal 10 μm.
PrPc colocalizes with tetraspanins CD81 and CD82 at the plasma membrane. (Ai) Fixed and permeabilized day-6 erythroblasts were dual stained with PrPc mAb (red) and CD81 mAb (green). Overlays were prepared and the highlighted area magnified to show (Aii) an internal colocalized vesicle (arrow) and the plasma membrane. (Aiii) ROIs were drawn through a cell (ROI1) and along the cell surface (ROI2), and profiles of green and red fluorescence along these ROIs were constructed using Leica quantiation software. Scale bars equal 10μm. (Bi) Fixed and permeabilized day-6 erythroblasts were dual stained with PrPc mAb (red) and CD82 mAb (green). Overlays were prepared and highlighted areas magnified to show (Bii) plasma membrane at cell-cell contact site and (Biii) internal PrP-containing vesicle (arrow). Scale bars equal 10 μm.
At the plasma membrane, PrPc and CD81 are present both separately and colocalized in discrete areas distributed evenly over the cell surface (Figure 3Ai,ii). The profile of red and green fluorescence from the plasma membrane (Figure 3Aiii) shows some colocalized peaks of fluorescence. Colocalization was also observed in vesicles just under the plasma membrane and in the perinuclear area (Figure 3Ai,ii). The profile of red and green fluorescence from the cell cross-section (Figure 3Aiii) shows colocalization of PrPc and CD81 at the cell surface and internally.
Patching experiments suggest CD81 and PrPc can share the same microdomain
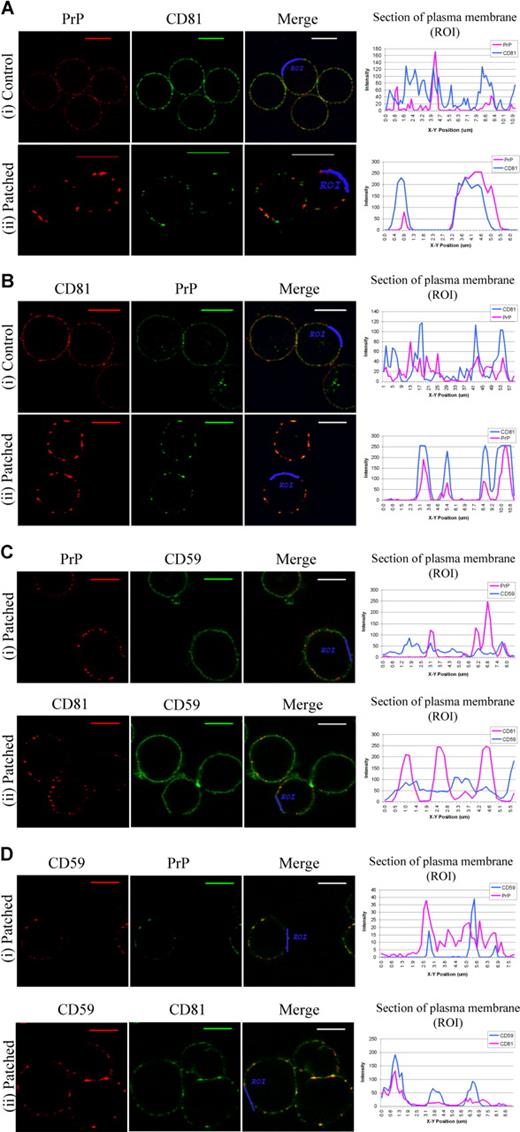
In order to assess whether or not PrPc and CD81 share the same microdomains on the plasma membrane, PrPc was cross-linked with antibody, and the PrPc clusters were examined for the presence of CD81 and vice versa.
Analysis of confocal fluorescence images showed that cross-linking of PrPc led to its redistribution from small, fairly evenly spread dots of fluorescence over the cell surface to larger coalesced patches, and that CD81 redistributed into the PrPc patches. This is demonstrated by the profiles of red and green fluorescence from the plasma membrane of noncapped and capped cells (Figure 4Ai,ii). Similarly, PrPc staining colocalized with CD81-induced patches (Figure 4Bi,ii).
Patching experiments suggest CD81 and PrPc can share the same microdomain. ROIs were drawn along the cell surface of each merge image, and profiles of green and red fluorescence along these ROI were constructed using Leica quantiation software. (Ai) Control cells were fixed then stained for PrP (red) and CD81 (green). (Aii) Cells underwent PrP antibody–induced patching (red) then fixed and dual stained for CD81 (green). Scale bars equal 10 μm. (Bi) Control cells were fixed then stained for CD81 (red) and PrP (green). (Bii) Cells underwent CD81 antibody-induced patching (red), then were fixed and dual stained for PrP (green). (Ci) Cells underwent PrP antibody–induced patching (red), then were fixed and dual stained for CD59 (green). (Cii) Cells underwent CD81 antibody–induced patching (red), then were fixed and then dual stained for CD59 (green). (Di) Cells underwent CD59 antibody–induced patching (red), then were fixed and dual stained for PrP (green). (Dii) Cells underwent CD59 antibody–induced patching (red), then were fixed and dual stained for CD81 (green).
Patching experiments suggest CD81 and PrPc can share the same microdomain. ROIs were drawn along the cell surface of each merge image, and profiles of green and red fluorescence along these ROI were constructed using Leica quantiation software. (Ai) Control cells were fixed then stained for PrP (red) and CD81 (green). (Aii) Cells underwent PrP antibody–induced patching (red) then fixed and dual stained for CD81 (green). Scale bars equal 10 μm. (Bi) Control cells were fixed then stained for CD81 (red) and PrP (green). (Bii) Cells underwent CD81 antibody-induced patching (red), then were fixed and dual stained for PrP (green). (Ci) Cells underwent PrP antibody–induced patching (red), then were fixed and dual stained for CD59 (green). (Cii) Cells underwent CD81 antibody–induced patching (red), then were fixed and then dual stained for CD59 (green). (Di) Cells underwent CD59 antibody–induced patching (red), then were fixed and dual stained for PrP (green). (Dii) Cells underwent CD59 antibody–induced patching (red), then were fixed and dual stained for CD81 (green).
Failure to demonstrate direct binding of PrPc to CD63 or CD81 by immunoprecipitation
Evidence for colocalization of PrPc with CD81 and CD63 raised the possibility that PrPc might bind directly to the tetraspanins. PrPc could be immunoprecipitated from cultured erythroblasts (day 7) after lysis with the zwitterionic detergent CHAPS and nonionic detergents Brij97, NP-40, or TX-100 (all used at 1%). In contrast, internalized PrPc could be immunoprecipitated with NP-40 or Brij97, weakly with CHAPS, but not at all with Brij99, suggesting internalized PrPc is present in a compartment needing more stringent detergent lysis conditions in order to solubilize PrPc (data not shown).
Western blotting of PrPc immunoprecipitates with anti-CD81 or anti-CD63 did not demonstrate coimmunoprecipitation under any of the detergent conditions. We could not demonstrate direct binding of either CD63 or CD81 to PrPc immunoprecipitated from whole cell lysates, the cell surface or internalized PrPc. Neither could we demonstrate the presence of PrPc in immunoprecipitates obtained with anti-CD81 or anti-CD63 (data not shown).
Discussion
Normal prion protein (PrPc) must be present for prion disease to occur.3,4 PrPc is a widely expressed cell-surface GPI-anchored protein that can be converted to a misfolded infectious isoform by interaction with another molecule of misfolded prion protein (PrPsc), thereby initiating an autocatalytic process which ultimately results in prion disease. Compelling evidence that vCJD can be transmitted by transfusion of red cell preparations in man9 has highlighted the need for a greater understanding of the distribution and biology of normal prion protein in human blood and blood-forming tissues so that the risk of disease transmission by individual blood components can be evaluated. In the present study, we examined the distribution and trafficking of PrPc in cultured human erythroblasts. PrPc is found on the cell surface, but most is in the perinuclear region of pronormoblasts, and the amount of PrPc declines as erythroid cells mature. Colocalization experiments using the endosomal marker CD63 suggest most of the PrPc is in the endosomal compartment. The distribution of PrPc in the plasma membrane of pronormoblasts was markedly different from that of other GPI-anchored proteins (CD59, CD55), and the rate of internalization of PrPc was much faster than that of CD59. PrPc colocalized with the transferrin receptor, which is known to be endocytosed in clathrin-coated vesicles. GPI-linked proteins in general occupy “lipid rafts” and are internalized slowly through clathrin-independent mechanisms.12 These data suggest that a significant proportion of PrPc on the plasma membrane of erythroblasts is not located in “lipid rafts.”
These findings are in agreement with observations of PrPc internalization kinetics in primary neurons where PrPc internalizes much faster than Thy-1 and almost as quickly as TfR.13
Sunyach et al13 concluded that PrPc leaves its lipid raft environment while still on the cell surface and moves to a nonraft environment where it enters coated pits and is rapidly endocytosed.
These data raise the question as to the nature of the nonraft environment occupied by PrPc and the mechanism by which it is targeted to coated pits. Our data show that the tetraspanin CD63 colocalizes with PrPc within the cell and is internalized at a similar rate to PrPc. The role of CD63 in protein trafficking has been demonstrated by the interaction of the ion pump H,K-ATPase β-subunit and CD63 at the cell surface.14 Enhanced internalization of the H,K-ATPase β-subunit was shown when it was CD63 associated. This internalization appeared to be mediated in part by μ2 and μ3.14 The μ subunits of the adaptor protein complexes 2 and 3 interact with the tyrosine-based motif in CD63.17 AP-2 participates in clathrin-mediated endocytosis, while AP-3 is involved in lysosomal targeting.17,18 In order to internalize within clathrin-coated pits, PrPc may need a transmembrane protein with which to piggyback since it lacks a cytoplasmic domain. The GPI-linked urokinase receptor piggybacks with the LDL receptor–related protein.19
TEMs form preferentially at and around clathrin-coated pits.15 Therefore, we considered the possible involvement of tetraspanin-enriched domains (TEMs) in the process of PrPc endocytosis and compared the distribution of PrPc with that of the tetraspanins CD81 and CD82, which colocalize with CD63 in HeLa cells.15
Confocal microscopy revealed that a significant proportion of PrPc at the cell surface is associated with CD81. Furthermore patching with anti-CD81 or anti-PrPc increases the degree of association between the proteins, indicating that they can share a similar microenvironment. The role of CD81 as a facilitator of membrane organization has been demonstrated in other systems. It plays a central role as a scaffolding protein for G-protein–coupled receptor transduction,20 and is essential for the raft-stabilizing function of the B-cell coreceptor complex CD19/CD20/CD81.21 However, we were unable to demonstrate a direct interaction between PrPc and CD81 or CD63 by immunoprecipitation.
Endosomal-mediated recycling of proteins to the plasma membrane may also result in the release of exosomes. Exosomes are known to contain PrPc, CD63, and CD81.22 Exosomes bearing PrPsc are infectious.23
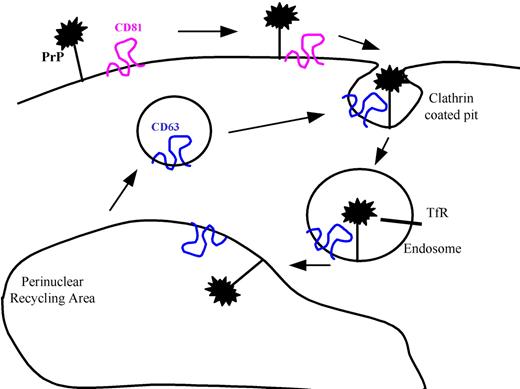
Based on these data, we propose a model (Figure 5) in which PrPc is assembled at the cell surface in tetraspanin microdomains containing CD81 and then internalized in endosomal vesicles containing CD63. This model is analogous to that proposed by Vogt et al24 for MHC class II antigen presentation.
Several lines of evidence suggest that PrPc is resistant to conversion to PrPsc when it is located in lipid rafts (reviewed in Morris et al25 ). It is therefore more likely PrPc that is outside of lipid rafts, perhaps in TEMs, is the focus for PrPsc binding and conversion. Cell-surface TEMs are involved in the pathogenesis of many viral diseases. They are the site at which HIV-1 proteins gather for potential particle exit.15 CD9 and CD81 play an important role in membrane fusion induced by the HIV-1 envelope.26 The HTLV-1 Gag protein is found enriched at TEMs, with the inner loop of CD81 and CD82 mediating the interactions.27 CD81 is a receptor for hepatitis C virus (HCV). The HCV envelope protein E2 binds to the major extracellular loop of CD8128 ; in concert with an unidentified partner, it mediates hepatitis C entry into cells.29
Recent evidence that MoMuLV (Moloney murine leukemia virus) infection strongly enhances the release of scrapie infectivity into the supernatant of coinfected cells led LeBlanc et al30 to propose that retroviruses are cofactors involved in distributing PrPsc to different tissues.
Clearly, further work is necessary to elucidate whether or not TEMs are directly involved in the pathogenesis of prion diseases, but our data suggest this approach represents a promising area for future research. Our observations also show that the mechanism of PrPc cycling in erythroid cells is similar to that in neuronal cells, raising the question as to whether or not erythroid cells can be infected by PrPsc.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Medical Research Council for providing an Infrastructure Award to establish the School of Medical Sciences Cell Imaging Facility, University of Bristol.
This work was supported by the United Kingdom Department of Health.
Authorship
Contribution: R.G. designed and performed research, analyzed data, and wrote the paper. K.H. performed research. D.A. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David J. Anstee, Bristol Institute for Transfusion Sciences, National Blood Service, Southmead Road, Bristol, BS10 5ND United Kingdom; e-mail:david.anstee@nbs.nhs.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal