A longstanding goal for the treatment of hemophilia B is the development of a gene transfer strategy that can maintain sustained production of clotting factor IX (F.IX) in the absence of an immune response. To this end, we have sought to use lentiviral vectors (LVs) as a means for systemic gene transfer. Unfortunately, initial evaluation of LVs expressing F.IX from hepatocyte-specific promoters failed to achieve sustained F.IX expression in hemophilia B mice due to the induction of an anti-F.IX cellular immune response. Further analysis suggested that this may be a result of off-target transgene expression in hematopoietic-lineage cells of the spleen. In order to overcome this problem, we modified our vector to contain a target sequence for the hematopoietic-specific microRNA, miR-142-3p. This eliminated off-target expression in hematopoietic cells, and enabled sustained gene transfer in hemophilia B mice for more than 280 days after injection. Treated mice had more than 10% normal F.IX activity, no detectable anti-F.IX antibodies, and were unresponsive to F.IX immunization. Importantly, the mice survived tail-clip challenge, thus demonstrating phenotypic correction of their bleeding diathesis. This work, which is among the first applications to exploit the microRNA regulatory pathway, provides the basis for a promising new therapy for the treatment of hemophilia B.
Introduction
A long-standing goal for the treatment of hemophilia B has been the development of a strategy that can maintain sustained, endogenous production of coagulation factor IX (F.IX). To this end, there has been a great deal of enthusiasm for using gene therapy.1,2 By introducing the wild-type F.IX cDNA to a patient, it is believed that production of normal F.IX can be established, and blood clotting times can be improved.
A variety of gene delivery vectors and approaches have been evaluated for the treatment of hemophilia B.3,4 In preclinical models, varying degrees of success have been achieved, but no sustained clinical improvement has been realized yet in human trials.5,–7 From the accumulated experience of these studies, the importance of a number of variables has emerged, variables that are crucial for the development of a successful gene transfer strategy.3,7 The vector must have minimal toxicity and low immunogenicity, and, ultimately, should be capable of providing sustained expression of the transgene at physiologically relevant levels.8 The acute toxicity of adenoviral vectors has so far limited their use for hemophilia B gene therapy in humans. AAV vectors have shown great promise in preclinical models of hemophilia B; unfortunately, recent clinical evidence indicates that in humans, stable hepatic delivery of AAV is limited by preexisting immunity to capsid antigens.6 To overcome some of the limitations of current approaches, we developed a strategy for in vivo gene delivery employing lentiviral vectors (LVs).9
LVs have several attractive properties as a gene delivery system.10 Because they are capable of transducing nondividing cells and have a low toxicity profile, LVs can be safely administered systemically to deliver genes to differentiated tissues in adult animals, including the liver.11,,,–15 Their ability to integrate within the genome provides a further advantage in that gene transfer can be maintained even during normal tissue turnover. Moreover, because LVs are derived from HIV, they have a limited exposure within the general population,16 and thus there is a reduced probability of pre-existing immunity to vector components. This improves the prospect of achieving sustained gene transfer with the vector.
Although adaptive immunity to the vector itself might not limit LV-mediated gene transfer, adaptive immunity to transgene-derived antigens may.17 Indeed, studies of intravenously delivered LVs encoding human coagulation factor VIII (hF.VIII) found that mice developed anti-hF.VIII antibodies.18,–20 We and others have reported a similar finding for LV-mediated human F.IX (hF.IX) gene transfer when a ubiquitously expressed promoter was used to drive transgene expression.11,21 To avoid triggering an anti-hF.IX immune response, several groups have employed ex vivo gene transfer of hematopoietic stem cells and bone marrow transplantation.22,23 This approach enabled stable hF.IX expression, even in hemophilia B mice. Myeloablative conditioning was required to achieve sufficient stem cell engraftment, and thus, this procedure carries significant risks as a treatment option for the hemophilias.
An alternative approach to prevent the induction of antitransgene immunity, and enable systemic gene transfer, is to engineer the vector to avoid endogenous transgene expression in antigen presenting cells (APCs) of the immune system.11,24,25 Transduction and endogenous transgene expression in APCs leads to a situation in which transgene-derived peptides are efficiently loaded onto MHC class I and class II molecules, where they are subsequently presented to T cells. Because vector components, including DNA and RNA, can trigger APC maturation,26,27 a context is provided for APC-mediated T-cell activation.28
This approach was recently evaluated in a mouse model of hemophilia B using an LV encoding hF.IX under the control of the hepatocyte-specific apolipoprotein E enhancer/α1-antitrypsin promoter (Apo-HCR/AAT).14 Although gene transfer did not trigger anti-hF.IX antibody formation, the vector was unable to provide therapeutic levels of hF.IX expression (< 1%). Thus, despite its potential, in preclinical evaluation LV-mediated liver gene transfer has been unable to correct the hemophilia B phenotype.
Here, we set out to improve liver-directed LV-mediated gene transfer for the treatment of hemophilia B. Our findings indicate that transcriptional targeting of transgene expression to hepatocytes was insufficient to enable sustained hF.IX expression in hemophilia B mice. Instead, inclusion of a layer of posttranscriptional control, mediated by endogenous microRNA (miRNA) regulation, enabled us to achieve long-term hF.IX gene transfer and rescue the phenotype of adult hemophilia B mice.
Materials and methods
Vector construction and production
For a detailed description of vector construction and production see Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Animal procedures
Hemophilia B mice (ΔF.IX) based on the C57BL/6 mouse strain with disruption of the F.IX gene, were kindly provided by Dr Inder Verma (Salk Institute, La Jolla, CA).29 Balb/c mice were purchased from Charles River Laboratories (Milan, Italy). Vector administration was carried out by tail vein injection on 6- to 8-week-old animals. Whole blood was collected into buffered citrate by phlebotomy of the retro-orbital plexus. All animal procedures were performed according to protocols approved by the Hospital San Raffaele Institutional Animal Care and Use Committee.
Factor IX assays
hF.IX concentration was determined by an enzyme-linked immunosorbent assay (ELISA) specific for hF.IX:Ag (Roche, Milan, Italy). F.IX activity (F.IX:C) was determined in a modified aPTT assay using human F.IX deficient plasma (Diagnostica Stago, Asnieres, France) and a semiautomated coagulometer (Option 2; BioMerieux, Marcy-l'Etoile, France). Calibration curves were constructed with serial dilutions of a pooled normal human plasma collected from 40 healthy volunteers and arbitrarily assigned a value of 100% F.IX activity.
Plasma samples were tested for the presence of hF.IX antibodies by ELISA as previously described.11 Microtiter plates were coated with purified hF.IX from plasma (0.2 μg/well in 0.1 M carbonate buffer, pH 9.6; Sigma, St Louis, MO). A 1:100 dilution of mouse plasma was added, and hF.IX antibodies were detected with peroxidase-conjugated rabbit anti–mouse immunoglobulin (DAKO, Glostrup, Denmark). Plates were reacted with H2O2 and orthophenylenediamine and read at 490 nm.
T-cell proliferation assay
Mice were killed 1 week after vector administration, and spleens were collected and processed into a single cell suspension. Splenocytes (5 × 106) were plated in triplicate in 96-well plates in the presence of hF.IX (10 μg/mL) in complete RPMI. Con A was used (1 μg/mL) as positive control. After 96 hours of culture, 1 μCi (37 000 Bq) of [3H]-thymidine was added, and the plates were harvested after an additional 18 hours. Results are presented as a mean count per minute (CPM).
IFN-γ ELISPOT assay
The frequency of IFN-γ–secreting CD8+ T cells specific for the immunodominant epitope of chicken ovalbumin (OVA257-264) were determined by enzyme-linked immunospot (ELISPOT) assay.30 Briefly, mice were euthanized and CD8+ T cells were magnetically isolated from the spleen (Miltenyi Biotec). Cells (5 × 104) were plated in triplicate in ELISPOT plates (Millipore, Bedford, MA) coated with anti–IFN-γ capture monoclonal antibody (2.5 μg/mL, R46A2; BD Pharmingen, San Diego, CA) in the presence of IL-2 (50 U/mL; BD Pharmingen) and 5 × 104 irradiated (3000 rad) unpulsed EL-4 or OVA257–264 pulsed EL-4 cells. After 42 hours of incubation at 37°C 5% CO2, plates were washed and IFN-γ–producing cells were detected by anti–IFN-γ detection monoclonal antibody (0.5 μg/mL XMG 1.2; BD Pharmingen). Spots were counted by Automated Elisa-Spot Assay Video Analysis System Eli.Expert (A.EL.VIS, Hannover, Germany).
DNA analysis
DNA from cells and tissues was extracted by using the Blood & Cell Culture DNA Midi Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. Vector copies per genome (C/Gs) were quantified by quantitative polymerase chain reaction (qPCR) using the primer and probe set previously described.31 C/Gs were calculated by the formula: (ng LV / ng endogenous DNA) × (no. of LV integrations in the standard curve). The standard curve was generated using a cell line carrying a single vector integrant. Reactions were carried out in triplicate in an ABI Prism 7900 (Applied Biosystems, Weuterstadt, Germany).
Southern blot analysis was carried out using a previously described protocol.9 Briefly, DNA was digested with AflII (or BamHI) and separated on a 1% agarose gel, transferred to a nylon membrane (Hybond-N; Amersham, Arlington Heights, IL), probed with a 32P-labeled Wpre probe, and, following washing, exposed to x-ray film.
microRNA analysis
RNA was extracted from tissues using Tri Reagent (Sigma, St Louis, MO), according to the manufacturer's instructions. For analysis of miRNA expression, the Applied Biosystems Taqman microRNA assay system was used according to the manufacturer's instructions. To determine absolute copy number, a standard curve was generated using a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) purified RNA oligonucleotide corresponding to let-7a (Primm, Milan, Italy). A quantity of 5 ng total yeast RNA was spiked with concentrations of oligonucleotide ranging from 102 to 108 copies, and then analyzed by the Taqman microRNA assay. Similar to Chen et al,32 Taqman cycle threshold (CT) values for each sample reaction were converted to absolute copy number based on this curve. All reactions were carried in duplicate in an ABI Prism 7900.
Fluorescence-activated cell sorting analysis
Single-cell suspensions from the spleen were prepared as previously described,11 and analyzed by flow cytometry on a Beckman Coulter Cytomics FC500 (Miami, FL).
Immunohistochemistry
Immunohistochemical analysis was performed on fixed, frozen sections as previously described.11 Briefly, cryostat sections were permeabilized, blocked in 5% goat serum, and incubated with rabbit anti–green fluorescence protein (GFP; Molecular Probes, Leiden, the Netherlands) and with antimouse CD45 (BD Pharmingen). Nuclei were stained with TOPRO-3 (Molecular Probes). Images were acquired by triple laser confocal microscopy as previously described.26
Results
Transcriptional targeting of transgene expression to hepatocytes is insufficient to mediate stable F.IX gene transfer in hemophilia B mice
Previously, we demonstrated that an LV encoding hF.IX under the control of the albumin promoter (LV.ALB.FIX) could mediate between 1% and 2% of normal hF.IX expression levels following intravenous administration.11 In an effort to improve transgene expression levels, we substituted the ALB promoter with a synthetic, hepatocyte-specific promoter (LV.ET.FIX). This construct was designed to provide enhanced levels of transgene expression and improved specificity over promoters reconstituted from endogenous hepatocyte-specific genes.33
Normal C57BL/6 mice were treated by tail-vein injection with 2 × 108 IU vesicular stomatitis virus glycoprotein (VSV) pseudotyped vectors, and monitored for hF.IX expression by an hF.IX-specific ELISA. While both vectors were capable of mediating hF.IX expression, the LV.ET.FIX vector mediated up to 5-fold-higher levels of hF.IX expression with a comparable vector dose (Figure 1B). These results confirm our previous findings indicating that stable hF.IX expression can be achieved in normal, immunocompetent mice by intravenous administration of LVs. They also indicate that the ET construct can mediate improved levels of transgene expression compared with the endogenous ALB promoter.
Evaluating the effectiveness of hepatocyte-targeted LVs for F.IX gene transfer. (A) Schematic representation of the third-generation self-inactivating LVs used for these studies. The miRNA-regulated LV was generated by incorporating 4 tandem copies of a sequence completely complementary to miR-142-3p. Wpre indicates woodchuck hepatitis post-regulatory element. (B) Measurement of hF.IX:Ag in normal C57BL/6 mice treated with 2 × 108 IU of LV.ALB.FIX (open and filled gray squares) or LV.ET.FIX (open and filled black circles). Results are presented for individual mice. (C) Measurement of hF.IX:Ag in hemophilia B mice treated with 5 × 108 IU of LV.ET.FIX. Results are presented as the mean plus or minus the standard error of the mean (SEM; n = 7). (D) T-cell proliferation response to hF.IX antigen. Splenocytes were isolated from untreated ( ) and LV.ET.FIX-treated (
) and LV.ET.FIX-treated ( ) hemophilia B mice at 7 days after injection, and cultured either in complete RPMI media alone or complete RPMI medium containing hF.IX antigen (10 μg/mL). T-cell proliferation was determined by measurement of thymidine incorporation. Results are presented as the mean plus or minus SEM (n = 3, *P < .05). CPM indicates counts per million.
) hemophilia B mice at 7 days after injection, and cultured either in complete RPMI media alone or complete RPMI medium containing hF.IX antigen (10 μg/mL). T-cell proliferation was determined by measurement of thymidine incorporation. Results are presented as the mean plus or minus SEM (n = 3, *P < .05). CPM indicates counts per million.
Evaluating the effectiveness of hepatocyte-targeted LVs for F.IX gene transfer. (A) Schematic representation of the third-generation self-inactivating LVs used for these studies. The miRNA-regulated LV was generated by incorporating 4 tandem copies of a sequence completely complementary to miR-142-3p. Wpre indicates woodchuck hepatitis post-regulatory element. (B) Measurement of hF.IX:Ag in normal C57BL/6 mice treated with 2 × 108 IU of LV.ALB.FIX (open and filled gray squares) or LV.ET.FIX (open and filled black circles). Results are presented for individual mice. (C) Measurement of hF.IX:Ag in hemophilia B mice treated with 5 × 108 IU of LV.ET.FIX. Results are presented as the mean plus or minus the standard error of the mean (SEM; n = 7). (D) T-cell proliferation response to hF.IX antigen. Splenocytes were isolated from untreated ( ) and LV.ET.FIX-treated (
) and LV.ET.FIX-treated ( ) hemophilia B mice at 7 days after injection, and cultured either in complete RPMI media alone or complete RPMI medium containing hF.IX antigen (10 μg/mL). T-cell proliferation was determined by measurement of thymidine incorporation. Results are presented as the mean plus or minus SEM (n = 3, *P < .05). CPM indicates counts per million.
) hemophilia B mice at 7 days after injection, and cultured either in complete RPMI media alone or complete RPMI medium containing hF.IX antigen (10 μg/mL). T-cell proliferation was determined by measurement of thymidine incorporation. Results are presented as the mean plus or minus SEM (n = 3, *P < .05). CPM indicates counts per million.
Following our studies in normal mice, we set out to evaluate the effectiveness of the LV.ET.FIX vector in treating hemophilia B (ΔF.IX) mice. Mice were injected with 5 × 108 IU LV, and monitored for hF.IX expression (n = 7). One week after vector administration, circulating hF.IX could be detected at up to 1.2% of normal levels. However, expression was not sustained. By 2 weeks after injection, no hF.IX expression could be detected in any of the hemophilia B mice treated with the LV.ET.FIX vector (Figure 1C).
Unlike normal mice, hemophilia B mice do not produce their own F.IX. Therefore, expression of hF.IX by gene transfer introduces a strong neo-antigen, which is expected to be more immunogenic in hemophilic mice than in normal mice. To determine whether an antitransgene immune response was involved in the loss of gene expression, we monitored the animals' immunologic responsiveness to hF.IX. Splenocytes were isolated from untreated and LV.ET.FIX-treated hemophilia B mice, and T-cell proliferation was measured following exposure to hF.IX protein. As shown in Figure 1D, increased proliferation was detected in mice treated with LV.ET.FIX, indicating a prior response to hF.IX antigen. Interestingly, anti-hF.IX antibodies were not detected in treated mice. Thus, the loss of F.IX expression was not due to the development of an anti-hF.IX humoral immune response, but likely due to a cellular immune response, resulting in clearance of cells expressing hF.IX.
Tissue-specific promoters do not always behave tissue-specifically
Since studies by our group and others have indicated that direct expression of an antigen in hematopoietic cells is a major determinant of an immune response,31,34 we decided to ascertain the specificity of the LV.ET vector. Earlier in vitro analysis had shown that the ET promoter was highly specific for hepatocytes35 ; however, until now, in vivo analysis of the promoter has not been reported. To determine the in vivo expression profile of the ET promoter, Balb/c mice (n = 5) were injected intravenously with 5 × 108 transducing units (TUs) of LV.ET.GFP encoding GFP (LV.ET.GFP).
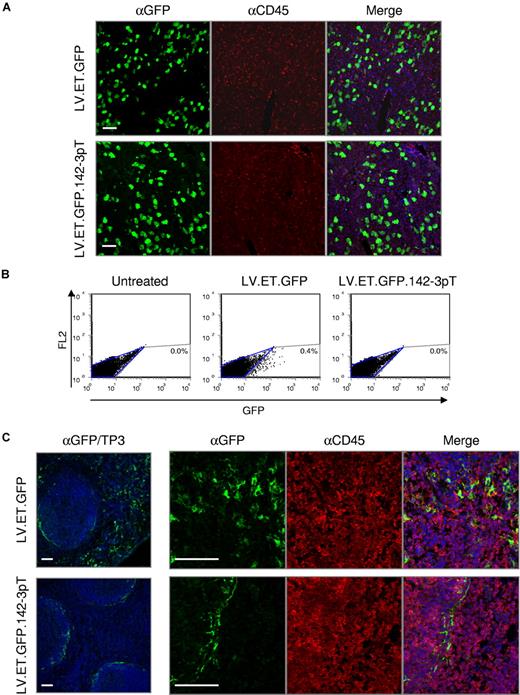
In the liver, we found high levels of GFP expression, which was confined almost exclusively to hepatocytes (Figure 2A). Unexpectedly, however, we also found a significant frequency of GFP-positive cells within the spleen (Figure 2B,C). As indicated by FACS analysis, up to 0.4% of splenocytes were GFP positive. Moreover, confocal fluorescent microscopy revealed that a fraction of the GFP-positive cells were of hematopoietic origin, as indicated by positive costaining for the hematopoietic-specific lineage cell marker CD45. This outcome, which was similarly found for the albumin promoter,31 indicates that even when transgene expression is targeted to hepatocytes by means of transcriptional control, “off-target” transgene expression can still occur.
Endogenous miRNA regulation can be exploited to improve vector targeting. Balb/c mice (n = 5/group) were treated by intravenous administration with 5 × 108 TU of the indicated vector and analyzed at 10 days after injection. (A) Confocal immunofluorescence analysis of the liver. Fixed frozen sections were costained with anti-GFP (green) and anti-CD45 (red) antibodies to monitor the vector expression profile. TO-PRO-3 (blue) was used to stain nuclei. Scale bars = 60μm. (B) FACS analysis of total splenocytes from treated animals were analyzed for GFP expression. (C) Confocal immunofluorescence analysis of the spleen. Fixed frozen sections were costained with anti-GFP (green) and anti-CD45 (red) antibodies to monitor the vector expression profile. TO-PRO-3 (blue) was used to stain nuclei. Scale bars are equal to 60 μm.
Endogenous miRNA regulation can be exploited to improve vector targeting. Balb/c mice (n = 5/group) were treated by intravenous administration with 5 × 108 TU of the indicated vector and analyzed at 10 days after injection. (A) Confocal immunofluorescence analysis of the liver. Fixed frozen sections were costained with anti-GFP (green) and anti-CD45 (red) antibodies to monitor the vector expression profile. TO-PRO-3 (blue) was used to stain nuclei. Scale bars = 60μm. (B) FACS analysis of total splenocytes from treated animals were analyzed for GFP expression. (C) Confocal immunofluorescence analysis of the spleen. Fixed frozen sections were costained with anti-GFP (green) and anti-CD45 (red) antibodies to monitor the vector expression profile. TO-PRO-3 (blue) was used to stain nuclei. Scale bars are equal to 60 μm.
miRNA regulation reduces off-target transgene expression from a hepatocyte-specific promoter
A variety of factors can contribute to aberrant regulation of a vector's promoter. Because these factors operate at the transcriptional level, we speculated that a layer of control at the posttranscriptional level may reduce this nonspecific activity. To do this, we sought to exploit the endogenous miRNA regulatory network. Recently, we demonstrated that inclusion of miRNA target sequences within a ubiquitously expressed vector resulted in detargeted transgene expression from cells that endogenously express the respective miRNA.31 Correspondingly, we modified the LV.ET vector to contain 4 tandem copies of a 23 nucleotide sequence (mirT) with perfect complementarity to the hematopoietic-specific miRNA, miR-142-3p,36 in the 3′-untranslated region (3′UTR) of the transgene expression cassette (LV.ET.GFP.142-3pT).
Balb/c mice were intravenously injected with 5 × 108 TU of LV.ET.GFP.142-3pT (n = 5), and analyzed at 10 days after injection. Similar to LV.ET.GFP-treated mice, high levels of GFP expression were found in a large fraction of hepatocytes within the liver (Figure 2A). In the spleen, however, the pattern of GFP expression was markedly different between the 2 treatment groups. Whereas LV.ET.GFP-treated mice had scattered GFP expression throughout the spleen, including within hematopoietic cells, in LV.ET.GFP.142-3pT–treated mice there was no GFP expression detected in splenocytes (Figure 2B). Interestingly, a small fraction of cells found exclusively within the marginal zone sinus remained GFP-positive, but were CD45-negative, indicating that they were not from the hematopoietic lineage (Figure 2C).
These findings show that inclusion of the miR-142-3p target sequences can prevent “off-target” transgene expression from a tissue-specific promoter specifically within hematopoietic cells. Moreover, the high levels of GFP expression within the liver indicate that the miR-142-3p target sequences do not interfere with transgene expression in hepatocytes.
miRNA-regulated LV does not dysregulate the normal miRNA profile of the liver
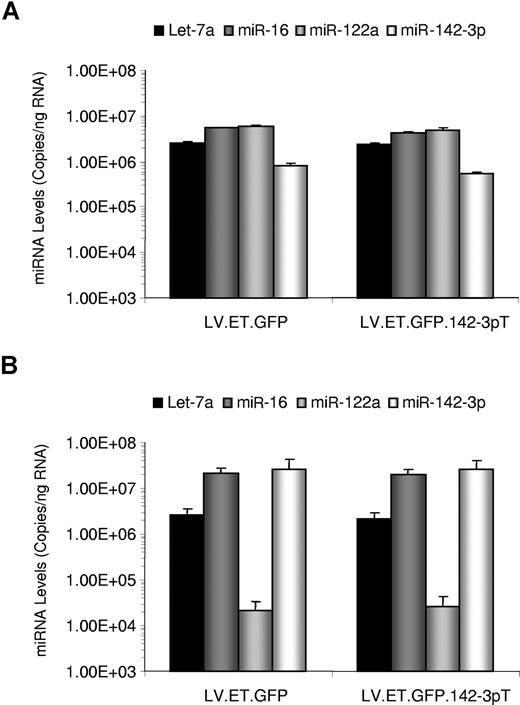
Little is known about the effects of increasing the levels of a miRNA target on miRNA expression. Since miRNAs can function in circuitries that regulate their expression,37 it is possible that perturbation of a miRNA's activities, due to the presence of increased targets, may change the expression level of the given miRNA. To test whether our vector, which contains 4 target sequences for miR-142-3p, alters miRNA expression patterns, we monitored the levels of 2 ubiquitous miRNAs, let-7a and miR-16, a liver-specific miRNA, miR-122a, and miR-142-3p in the liver and spleens of treated mice (n = 5). To quantitate the levels of miRNA expression, we employed a sensitive quantitative reverse-transcription PCR (RT-qPCR) assay, which is capable of detecting the mature form of the miRNAs (Figure 3).
miRNA expression profile analysis of LV-treated mice. RNA was isolated from the liver (A) and spleen (B) of Balb/c mice (n = 5/group) treated by intravenous administration with 5 × 108 TU of the indicated vector. miRNA levels were quantitated by RT-qPCR using primers and probe specific for the indicated miRNAs. Results are presented as the mean plus or minus SEM.
miRNA expression profile analysis of LV-treated mice. RNA was isolated from the liver (A) and spleen (B) of Balb/c mice (n = 5/group) treated by intravenous administration with 5 × 108 TU of the indicated vector. miRNA levels were quantitated by RT-qPCR using primers and probe specific for the indicated miRNAs. Results are presented as the mean plus or minus SEM.
In the liver, we found high levels of let-7a, miR-16, and miR-122, and lower levels of miR-142-3p, reflecting the lower contribution of hematopoietic cells in the tissue. In the spleen, miR-122a was expressed to very low levels, whereas the levels of let-7a remained similar to those found in the liver. Interestingly, levels of miR-16 were close to 5-fold higher in the spleen, suggesting some tissue bias for miR-16 expression. In accordance with its largely hematopoietic composition, in the spleen miR-142-3p levels were 50-fold higher than in the liver. This pattern is consistent with the expression profile of miR-142-3p–regulated vectors, in which high levels of transgene expression are observed in the liver, but not in the spleen. Crucially, we found no significant differences in the levels of miRNA expression between either of the 2 treatment groups. Thus, these findings indicate that introduction of the LV.ET.FIX.142-3pT vector does not affect the natural levels of miR-1423p in vivo.
miR-142-3p–regulated LV mediates stable hF.IX gene transfer and reduced clotting times in hemophilia B mice
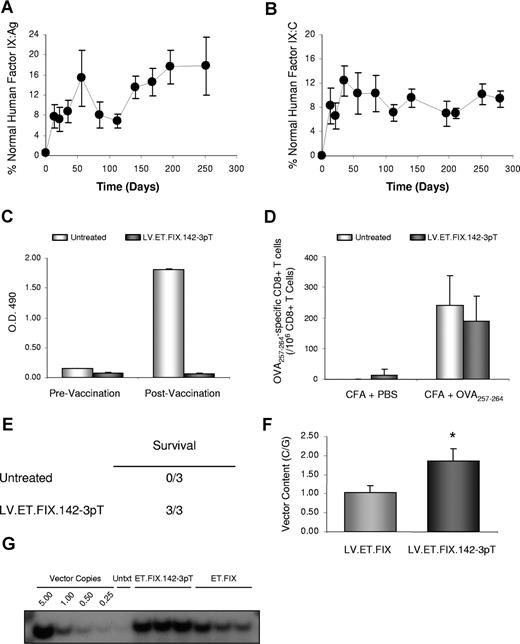
To evaluate the effectiveness of the miR-142-3p–regulated LV for F.IX gene transfer, we treated 6- to 8-week-old hemophilia B mice with 5 × 108 IU LV.ET.FIX.142-3pT (n = 7). In contrast to the LV.ET.F.IX vector, all mice treated with LV.ET.FIX.142-3pT had sustained F.IX expression for more than 280 days after injection (last time analyzed). Levels of hF.IX, as determined by hF.IX-specific ELISA, averaged 9.05% plus or minus 2.04% of normal human F.IX levels, which corresponds to approximately 452.5 ng/mL plus or minus 102 ng/mL of hF.IX (Figure 4A). Importantly, expression of hF.IX antigen directly correlated with an improvement in F.IX activity. As shown in Figure 4B, LV.ET.FIX.142-3pT–treated mice had up to 15% of human F.IX activity.
miR-142-3p–regulated LV mediates stable hF.IX gene expression and correction of hemophilia B mice. (A) Measurement of hF.IX:Ag in hemophilia B mice treated with 5 × 108 IU of LV.ET.FIX.142-3pT. Results are presented as the mean plus or minus SEM (n = 7/group). This includes evaluation of 2 different preparations of LV.ET.FIX.142-3pT and 3 different cohorts of hemophilia B mice. (B) Measurement of hF.IX:C by activated partial thromboplastin time (aPTT) in the same hemophilia B mice as shown in panel A. Results are presented as the mean plus or minus SEM (n = 7). (C) ELISA immunoassay of mouse plasma (1:100 dilution) to detect anti-hF.IX antibodies before (white bar) and after (gray bar) vaccination with hF.IX. Quantification was performed by analysis of absorbance at optical density (OD) 490 nm. Results are presented as the mean plus or minus SEM (n = 3/group). (D) Measurement of anti-OVA257–264 CD8+ T cells. Untreated ( ) and LV.PGK.FIX.142-3pT–treated (
) and LV.PGK.FIX.142-3pT–treated ( ) hemophilia B mice (n = 3/group) were immunized with the immunodominant epitope of OVA, OVA257-264, in complete Freurd adjuvant. The frequency of anti-OVA257-264 CD8+ T cells was measured by ELISPOT assay. Results are presented as the mean plus or minus SEM. (E) Tail transection was performed on LV.ET.FIX.142-3pT–treated and untreated hemophilia B mice (n = 3/group). Mice were monitored for 24 hours to assess survival. (F) Measurement of LV content in the liver of hemophilia B mice by qPCR at more than 280 days after injection (n = 7/group). Results are presented as the mean plus or minus SEM (*P < .05). (G) Measurement of LV content in the liver of hemophilia B mice by Southern blot analysis at more than 280 days after injection. Analysis was performed on the livers of 3 mice per treatment group. Untxt indicates untreated.
) hemophilia B mice (n = 3/group) were immunized with the immunodominant epitope of OVA, OVA257-264, in complete Freurd adjuvant. The frequency of anti-OVA257-264 CD8+ T cells was measured by ELISPOT assay. Results are presented as the mean plus or minus SEM. (E) Tail transection was performed on LV.ET.FIX.142-3pT–treated and untreated hemophilia B mice (n = 3/group). Mice were monitored for 24 hours to assess survival. (F) Measurement of LV content in the liver of hemophilia B mice by qPCR at more than 280 days after injection (n = 7/group). Results are presented as the mean plus or minus SEM (*P < .05). (G) Measurement of LV content in the liver of hemophilia B mice by Southern blot analysis at more than 280 days after injection. Analysis was performed on the livers of 3 mice per treatment group. Untxt indicates untreated.
miR-142-3p–regulated LV mediates stable hF.IX gene expression and correction of hemophilia B mice. (A) Measurement of hF.IX:Ag in hemophilia B mice treated with 5 × 108 IU of LV.ET.FIX.142-3pT. Results are presented as the mean plus or minus SEM (n = 7/group). This includes evaluation of 2 different preparations of LV.ET.FIX.142-3pT and 3 different cohorts of hemophilia B mice. (B) Measurement of hF.IX:C by activated partial thromboplastin time (aPTT) in the same hemophilia B mice as shown in panel A. Results are presented as the mean plus or minus SEM (n = 7). (C) ELISA immunoassay of mouse plasma (1:100 dilution) to detect anti-hF.IX antibodies before (white bar) and after (gray bar) vaccination with hF.IX. Quantification was performed by analysis of absorbance at optical density (OD) 490 nm. Results are presented as the mean plus or minus SEM (n = 3/group). (D) Measurement of anti-OVA257–264 CD8+ T cells. Untreated ( ) and LV.PGK.FIX.142-3pT–treated (
) and LV.PGK.FIX.142-3pT–treated ( ) hemophilia B mice (n = 3/group) were immunized with the immunodominant epitope of OVA, OVA257-264, in complete Freurd adjuvant. The frequency of anti-OVA257-264 CD8+ T cells was measured by ELISPOT assay. Results are presented as the mean plus or minus SEM. (E) Tail transection was performed on LV.ET.FIX.142-3pT–treated and untreated hemophilia B mice (n = 3/group). Mice were monitored for 24 hours to assess survival. (F) Measurement of LV content in the liver of hemophilia B mice by qPCR at more than 280 days after injection (n = 7/group). Results are presented as the mean plus or minus SEM (*P < .05). (G) Measurement of LV content in the liver of hemophilia B mice by Southern blot analysis at more than 280 days after injection. Analysis was performed on the livers of 3 mice per treatment group. Untxt indicates untreated.
) hemophilia B mice (n = 3/group) were immunized with the immunodominant epitope of OVA, OVA257-264, in complete Freurd adjuvant. The frequency of anti-OVA257-264 CD8+ T cells was measured by ELISPOT assay. Results are presented as the mean plus or minus SEM. (E) Tail transection was performed on LV.ET.FIX.142-3pT–treated and untreated hemophilia B mice (n = 3/group). Mice were monitored for 24 hours to assess survival. (F) Measurement of LV content in the liver of hemophilia B mice by qPCR at more than 280 days after injection (n = 7/group). Results are presented as the mean plus or minus SEM (*P < .05). (G) Measurement of LV content in the liver of hemophilia B mice by Southern blot analysis at more than 280 days after injection. Analysis was performed on the livers of 3 mice per treatment group. Untxt indicates untreated.
Mice were also monitored for anti-hF.IX antibodies and were found to be negative, indicating that treatment with LV.ET.FIX.142-3pT did not provoke an anti-hF.IX humoral immune response (Figure 4C). Since the hemophilia B mice are capable of mounting an immune response to hF.IX,38,39 we wanted to understand whether the absence of antibodies was due to a mechanism of immunologic ignorance resulting from miR-142-3p detargeting of transgene expression from APCs. Untreated and LV.ET.FIX.142-3pT-treated mice were immunized by subcutaneous injection of 4μg hF.IX in incomplete Freund adjuvant, and monitored for changes in hF.IX expression and anti-F.IX antibody formation. In untreated hemophilia B mice, high levels of anti-F.IX antibodies were detected within 2 weeks of vaccination. In contrast, none of the mice pretreated with LV.ET.FIX.142-3pT developed anti-hF.IX antibodies, and, moreover, all of these mice continued to stably express hF.IX (Figure 4C).
It is becoming increasingly clear that miRNAs may play a critical role in T-cell activation, and perhaps are even involved in autoimmune disorders.40,41 Although a role for miR-142-3p in immune function has not been reported, we wanted to address whether introduction of our vector, which expresses a transcript carrying a target sequence for miR-142-3p, would interfere with general immune function. To do this, we immunized LV.ET.FIX.142-3pT–treated mice with the immunodominant epitope of chicken ovalbumin, OVA257-264. As shown in Figure 4D, the anti-OVA257-264 T-cell response was comparable between LV.ET.FIX.142-3pT–treated and non-LV–treated hemophilia B mice, indicating that mice treated with a vector encoding the miR-142-3pT sequence are capable of responding to unrelated antigens. Thus, it appears that in addition to providing sustained hF.IX expression, LV-mediated gene transfer can confer immunologic tolerance to an encoded antigen.
To determine whether treatment corrected the bleeding diathesis, we subjected the mice to tail-clip challenge as previously described.42 The tails of treated and untreated hemophilia B mice (n = 3) were transected at a specific diameter, and mice were monitored for clotting and survival. Whereas untreated hemophilic mice did not form a stable clot, and eventually died, all LV.ET.FIX.142-3pT–treated mice formed a stable clot, and survived the tail-clipping (Figure 4E).
Finally, mice were killed, and vector content was measured in the liver by qPCR. An average of 1.86 C/Gs was found in LV.ET.FIX.142-3pT–treated mice (Figure 4F), whereas in LV.ET.FIX-treated mice there were almost 50% fewer vector C/Gs, indicating a significant difference (P = .022 by Student t test) in vector content between the 2 treatment groups. These results were corroborated by Southern blot analysis (Figure 4G), which also indicated that vector integration was widely distributed throughout the genome without indication of clonal outgrowth (data not shown). This confirms that LV.ET.FIX.142-3pT is stably maintained in treated mice, and that the miR-142-3pT sequence prevents vector clearance.
It is worth noting that we do expect to find vector genomes in LV.ET.FIX-treated mice, even in the absence of detectable F.IX in the plasma. This is because the vector is not expressed, or only poorly so, in nonparenchymal cells of the liver, such as Kupffer cells and endothelial cells, which are well transduced following systemic vector injection, and thus these cells will not be susceptible to immune-mediated clearance.
Overall, these data indicate that LV.ET.FIX.142–3pT can mediate efficient and sustained hF.IX gene transfer and expression, and rescue the phenotype of hemophilia B mice.
Discussion
Here we show that LV-mediated F.IX gene transfer can achieve successful correction of the hemophilia B phenotype in a mouse model of the disease. Following a single, intravenous injection of an LV encoding hF.IX, up to 15% of normal F.IX activity could be attained in hemophilia B mice for over 280 days after injection. These levels provided clear improvement to the phenotype of the animal, as indicated by their ability to survive a once lethal wound. Importantly, hF.IX expression was sustained long-term, and thus demonstrates that LVs can mediate stable transfer and expression of a transgene encoding a circulating neo-antigen without provoking an immune response.
To improve the levels of F.IX expression we employed an optimized, hepatocyte-specific promoter construct in our vector. Unfortunately, while we were able to achieve levels of hF.IX expression of greater than 1%, this expression was only transient, and induction of an anti-hF.IX T-cell response occurred. As we have shown here with the synthetic ET promoter, and previously with a native hepatocyte-specific promoter,31 aberrant transgene expression can arise from a tissue-specific promoter. This, in turn, can trigger an immune response, and result in clearance of cells expressing the transgene.11,31
Dysregulated expression from a promoter can occur due to a variety of factors. For one, cloning and reconstitution of the transcriptional control element of a tissue-specific gene may not be completely faithful. In an effort to reduce the size of a promoter, distant enhancer and repressor elements are often excluded during vector construction. This can result in aberrant regulation of the promoter. Achieving improved expression levels, by incorporating additional enhancer elements, may also reduce a promoter's specificity. The superior expression mediated by the ET promoter compared with the Apo-HCR/AAT element evaluated by VandenDriessche and colleagues14 may, conversely, have resulted in a higher frequency of off-target transgene expression.
To overcome these problems, we have developed a novel strategy that exploits endogenous miRNA activity to prevent vector expression in nontarget cells. This strategy may be akin to one of the natural functions of miRNAs, which has been proposed to serve as a means for preventing expression of genes transcribed in a previous cellular state or that arise due to leaky transcription.43 As we demonstrate here, by incorporating target sequences for the hematopoietic-specific miRNA, miR-142-3p, we prevented off-target expression of the ET promoter in hematopoietic lineage cells.
Importantly, LV.ET.FIX.142-3pT produced similar levels of circulating hF.IX in the hemophilia B mice as the LV.ET.FIX vector produced in normal mice. In our original description of the miR-142-3p–regulated vector system,31 we compared GFP expression from a vector with or without the miR-142-3p target sequence in nonhematopoietic cells in vitro and, more relevantly, in transgenic mice. We did not observe a reduction in expression of a transgene carrying the miR-142-3p target sequence in nonhematopoietic cells as measured by fluorescent microscopy, FACS analysis, or RT-qPCR. These findings, along with the data presented here, indicate that addition of the 142-3pT sequence does not result in an overall reduction in transgene expression, but instead likely serves to eliminate off-target transgene expression in the small subset of hematopoietic cells where dysregulated transcription of the vector's promoter occurs.
Inclusion of the miR-142-3p target sequences enabled us to achieve sustained gene transfer and long-term expression of hF.IX in the hemophilia B mouse model, which we were unable to achieve without the miRNA target sequences. Since both treatment groups received vector dose–matched injections, the differential in vector content (approximately 1:2) between LV.ET.FIX- and LV.ET.FIX.142-3pT–treated mice confirms that inclusion of the miR-142-3pT sequence prevents vector clearance. That LV.ET.FIX-treated mice still had significant vector content, despite their lack of hF.IX production, is explained by the fact that transgene expression is controlled by a hepatocyte-specific promoter. An anti-hF.IX–specific immune response would only be expected to eliminate cells expressing the transgene, which would be predominately hepatocytes, and the small fraction of hematopoietic cells where the transgene is aberrantly expressed. Thus, the differential in vector content should largely reflect the frequency of LV.ET.FIX.142-3pT–transduced hepatocytes, which were not cleared because the anti-hF.IX immune response was avoided.
We believe that the 142-3pT sequence is able to prevent the induction of an anti-transgene immune response because of miR-142-3p's effective ability to suppress expression of our transgene in APCs. By preventing endogenous transgene expression in APCs at the time of vector administration, when innate costimulatory signals are being elicited,26 effective T-cell priming is avoided, and immune-mediated clearance of transgene-expressing cells does not occur. This extends our previous findings, with an intracellular antigen, to a transgene encoding a circulating antigen, F.IX. This supports previous observations indicating that endogenous expression of an antigen in APCs is primarily responsible for initiating the antigen-specific immune response following gene transfer.34
This finding is somewhat unexpected, since endogenous expression is not necessary for APCs to access circulating antigens for uptake and presentation. Nonetheless, although APCs may be able to uptake circulating F.IX, in order to activate T cells APCs must be mature.44,45 Systemic administration of LV induces a transient innate response, which provides an environment for APC maturation.14,26 However, because circulation of vector-expressed hF.IX does not occur for several days after vector administration, when the innate response has subsided,26 most APCs uptaking h.FIX antigens in this period will not be mature, and thus are not capable of activating T cells. In contrast, transduced APCs are more likely to be mature as a result of the innate response to the vector. If these mature APCs express the transgene endogenously, they will present transgene-derived antigens, and can subsequently prime T cells and initiate an adaptive immune response. This helps to explain how the miRNA regulation strategy, which effectively detargets transgene expression from APCs, prevents vector clearance and enables long-term hF.IX expression.
Avoiding clearance and establishing transgene expression in transduced hepatocytes may have had a role in the induction of tolerance to hF.IX that we observed in LV.ET.FIX.142-3pT–treated mice. The liver has been shown to be a site of peripheral tolerance induction,46 and has recently been exploited for this end by AAV-mediated gene transfer.25,47 Although the mechanism or mechanisms responsible for this ability of the liver remains unknown, roles for Fas ligand as well as T-regulatory cells have been reported.25,46,47 By establishing transgene expression in the liver through gene transfer, hF.IX antigens may enter a natural physiologic pathway, which results in immunologic tolerance. Future studies will be required to define the mechanism of tolerance induction and maintenance induced by our vector system.
Of note, an attempt was made to use the miR-142-3pT sequences in a vector encoding hF.IX under transcriptional control of the ubiquitously expressed cytomegalovirus (CMV) promoter. Unfortunately, mice treated with this vector developed anti-hF.IX antibodies (data not shown). Analysis of vector preparations revealed that there were significant levels of hF.IX antigen contaminating the vector preparation (Table S1), and thus injection of the vector would have been accompanied by hF.IX protein. This is not entirely unexpected since the CMV promoter is active in 293T producer cells, and hF.IX is a secreted molecule. Instead, hF.IX was not detected in vectors prepared with the ET element, since this promoter is not highly active in producer cells. These findings support the notion that induction of antitransgene immunity is highly dependent on the context of APC presentation, since hF.IX antigens delivered with the vector would be uptaken and presented by APCs exposed to the transient innate response induced by the vector.26 Future strategies to prevent transgene expression during vector production, or improved vector purification procedures, may help to alleviate this problem. This should allow for the use of ubiquitous promoters with the miR-142-3p–regulation strategy for LV-mediated transfer of genes encoding secreted factors.
Analysis of miRNA expression in treated and untreated animals revealed that the 142-3p–regulated vector did not affect the natural levels of expression of miR-142-3p or other miRNAs in either the liver or spleen. Our previous studies have shown that increasing the concentration of perfectly complementary miR-142-3p target sites, even to high copy, does not saturate miR-142-3p regulation.31 The use of a liver-specific promoter in the vector further reduces concerns of interfering with the miR-142-3p regulatory pathway, since expression of the transgene containing the miR-142-3p target sites occurs predominately in hepatocytes, which do not express miR-142-3p. However, continued study will be required to evaluate the effects of increasing a miRNA substrate in a cell, and when targets for miR-142-3p are identified, it will be important to monitor their expression following delivery of the vector.
As noted, LV has a number of attractive properties that make it a promising system for hemophilia B gene therapy. Nonetheless, several issues must be addressed before clinical testing can be proposed. Although preexisting immunity to HIV-derived antigens is unlikely to be a problem, neutralization of the VSV.G envelope may thwart efficient delivery. To circumvent this problem, it has been shown that LVs can be pseudotyped with the gp64 glycoprotein envelope from the baculovirus Autographa californica multinuclear polyhedrosis virus, and molecules of CD55/decay-accelerating factor, a complement regulatory protein, can be incorporated into the vector particle.48 The use of the gp64 envelope may provide an additional benefit by improving the hepatotropism of the vector, and reducing the incidence of antitransgene immunity.49
An additional concern regarding the use of LVs is insertional mutagenesis and the possibility of tumorigenesis. This risk is likely to be low in mitotically quiescent tissue such as the liver,50 however, caution must still be maintained, in particular when using a strong promoter such as the ET construct used in this study. The use of integrase-defective LVs may provide a means to alleviate the risk of vector-mediated tumorigenesis,51 provided that these vectors can mediate sufficient and stable transgene expression over time. Further evaluation of this issue will be necessary in sensitive rodent models52 and large animals53 to properly assess the risk/benefit ratio of the proposed therapy.
The work presented here provides one of the first potential clinical applications to exploit the recently discovered miRNA regulatory pathway. By tagging an hF.IX transgene with a target sequence for miR-142-3p, long-term LV-mediated delivery was made possible in immunocompetent hemophilia B mice. Importantly, this strategy mediated clinically relevant levels of F.IX, and was able to rescue the phenotype of these mice. Of course, we recognize that there are limitations to evaluating gene therapies in mouse models,7 and thus further evaluation in a large animal model and with a species-specific transgene will be required. Nonetheless, based on our findings, we believe that LV-mediated gene transfer represents a promising new therapy for the treatment of hemophilia B, and may be similarly applied to other genetic diseases.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
Acknowledgments
The authors would like to thank Andrew D. Simmons for the ET promoter, Inder Verma for providing the hemophilia B mice, and Anna Zingale and Paola Corbella for technical assistance. We would also like to thank Antonia Follenzi, Christine Hough, David Lillicrap, and Maria Grazia Roncarolo for helpful discussions.
This work was supported by grants from Telethon (TIGET grant), the Italian Ministry of Scientific Research, and an EU RIGHT Project grant to L.N. B.D.B. is the recipient of a Natural Science and Engineering Research Council of Canada (NSERC) fellowship.
Authorship
Contribution: B.D.B. designed and performed research, analyzed data, and wrote the paper; A.C. and A.A. designed and performed research and analyzed data; L.S.S., A.L., and P.D.V. performed research; A.D. analyzed data and edited the paper; and L.N. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luigi Naldini, San Raffaele Telethon Institute for Gene Therapy, San Raffaele Scientific Institute, Milan, Italy; e-mail:naldini.luigi@hsr.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal