Human 8p11 stem cell leukemia/lymphoma syndrome usually presents as a myeloproliferative disorder (MPD) that evolves to acute myeloid leukemia and/or lymphoma. The syndrome associated with t(8;13)(p11;q12) results in expression of the ZNF198-FGFR1 fusion tyrosine kinase that plays a pathogenic role in hematopoietic transformation. We found that ZNF198-FGFR1 activated both the AKT and mitogen activated protein kinase (MAPK) prosurvival signaling pathways, resulting in elevated phosphorylation of the AKT target FOXO3a at T32 and BAD at S112, respectively. These phosphorylated residues subsequently sequestered the proapoptotic FOXO3a and BAD to 14–3-3 to prevent apoptosis. We used a peptide-based 14–3-3 competitive antagonist, R18, to disrupt 14–3-3–ligand association. Expression of R18 effectively induced apoptosis in hematopoietic Ba/F3 cells transformed by ZNF198-FGFR1 compared with control cells. Moreover, purified recombinant transactivator of transcription (TAT)-conjugated R18 proteins effectively transduced into human leukemia cells and induced significant apoptosis in KG-1a cells expressing FGFR1OP2-FGFR1 fusion tyrosine kinase but not in control HL-60 and Jurkat T cells. Surprisingly, R18 was only able to dissociate FOXO3a, but not BAD as previously proposed, from 14–3-3 binding and induced apoptosis partially through liberation and reactivation of FOXO3a. Our findings suggest that 14–3-3 integrates prosurvival signals in FGFR1 fusion-transformed hematopoietic cells. Disrupting 14–3-3–ligand association may represent an effective therapeutic strategy to treat 8p11 stem cell MPD.
Introduction
The 8p11 myeloproliferative syndrome (EMS) is an aggressive myeloproliferative disorder (MPD) caused by the fusion of diverse partner genes to FGFR1, which is associated with acute myeloid leukemia (AML) or stem cell MPD. Positional cloning of recurrent chromosomal translocations associated with the MPD has demonstrated frequent involvement of the FGFR1 gene on 8p11.2–11.1. Four major translocations have been described, including t(8;13)(p11;q12), t(8;9)(p11;q33), t(6;8)(q27;p11), and t(8;22)(p11;q11), that result in fusion of distinct partners to FGFR1, including ZNF198,1 CEP110,2 FOP,3 and BCR,4 respectively. In each case, an N-terminal partner containing self-association motif is fused to the C-terminal tyrosine kinase domain of FGFR1. Recently, 4 more chromosomal abnormalities associated with this syndrome have been cloned, including t(8;19) (p12;q13.3), ins(12;8)(p11;p11p22), t(7;8)(q34;p11), and t(8;17)(p11;q23), which result in the N-terminal portions of HERV-K, FGFR1OP2, TIF1, and MYO18A fused in frame to the C-terminal FGFR1 kinase domain, respectively.5,,–8 Although the transforming properties of these newly identified FGFR1 fusions have not been characterized, ZNF198-, BCR-, FOP-, and CEP110-FGFR1 fusion tyrosine kinases are constitutively activated,9,,–12 probably through aggregation by the self-association motif of the fusion partners, suggesting that these fusion tyrosine kinases play a pathogenic role in 8p11 EMS. We have demonstrated in a murine bone marrow transplantation (BMT) model that mice undergoing transplantation with bone marrow cells expressing ZNF198-FGFR1 developed a myeloproliferative disease.13 Similarly, Roumiantsev et al reported that ZNF198-FGFR1 may also confer a predilection for development of lymphoproliferative as well as myeloproliferative disease in a murine model.14 Expression of BCR-FGFR1 or FOP-FGFR1 in primary bone marrow cells induces a fatal myeloproliferative disease in mice similar to the one observed in our BMT assay.14,15
However, despite the cytogenetic advances in understanding the molecular basis of 8p11 EMS, there is no effective clinical treatment for this syndrome. The current empirically derived cytotoxic chemotherapy has been inadequate for treating this disease. Patients with MPD are characterized by myeloid hyperplasia with peripheral blood eosinophilia and B- or T-cell lymphoma. As reported, the myeloid hyperplasia clinically shows a short chronic phase before it aggressively progresses to AML within a year of the original diagnosis; and cure requires allogeneic stem cell transplantation.16,17 Therefore, there is a compelling need to develop effective molecularly targeted therapies in the treatment of 8p11 EMS. Tyrosine kinase fusions including BCR-ABL are well-validated therapeutic targets in human leukemias.18 Indeed, we first reported that a small molecule inhibitor, PKC412, effectively inhibits ZNF198-FGFR1 in cells and animal disease models and was active in the treatment of a stem cell MPD patient with t(8;13)(p11;q12) translocation.13
We and others have shown that expression of ZNF198-FGFR1 results in increased tyrosine phosphorylation of PI3K, PLCγ, STAT1, and STAT5 in Ba/F3 cells.11,13 Deletion of the proline-rich dimerization domain in the N-terminal ZNF portion of ZNF198-FGFR1, as well as treatment with the small-molecule inhibitor PKC412 that targets FGFR1, results in abolition of the ability of the fusion tyrosine kinase to activate these pathways in cells or induce the myeloproliferative disease in mice.13 In addition, the FOP-FGFR1 fusion induces cell survival by activating the PLCγ, MAPK/ERK, and PI3K/AKT/mTOR pathways,12 and BCR-FGFR1 activates STAT5 and MAPK pathways.4,19 These findings suggest that activation of FGFR1 fusions and downstream signaling pathways plays an essential role in the pathogenesis of diseases induced by distinct FGFR1 fusion proteins. Because the emergence of molecular resistance to tyrosine kinase inhibitors including imatinib poses a significant clinical problem,20,21 it is of interest to develop alternative and/or complementary therapeutic strategies to target critical signaling molecules that are activated by, in our case, FGFR1 fusion tyrosine kinases, which may broadly interfere with their transforming potential and overcome drug resistance.
Here we used ZNF198-FGFR1 as an example and show that this FGFR1 fusion tyrosine kinase activates the AKT and MAPK pathways in hematopoietic Ba/F3 cells, and 14–3-3 integrates prosurvival signals through sequestering proapoptotic FOXO3a and BAD. The 14–3-3 proteins are a family of conserved, phosphoserine/threonine-binding proteins with 7 isoforms. The 14–3-3 proteins regulate many cellular processes that are important in cancer biology, such as apoptosis and cell-cycle checkpoints. It functions through binding to a large group of regulatory proteins that are critical for cell survival and proliferation, such as FOXO3a, BAD, PI3K, apoptosis signal-regulating kinase 1 (ASK1), and Raf-1.22,,,,–27 Activated AKT has been shown to promote survival signaling by phosphorylating and inactivating proapoptotic proteins such as Bcl-2 family member BAD at Ser136 and forkhead family member FOXO3a at Thr32 and Ser253. Activation of the mitogen activated protein kinase (MAPK) pathway leads to phosphorylation and inactivation of BAD at Ser112. These phosphorylated residues provide binding sites for 14–3-3 proteins, which subsequently sequester proapoptotic FOXO3a and BAD (reviewed by Fu22 and Porter27 ). BAD proteins bind to and inhibit antiapoptotic Bcl-2, whereas binding of 14–3-3 to phosphorylated BAD traps BAD in an inactive form, which results in liberation of Bcl-2 to promote cell survival.24,28 Phosphorylation of the proapoptotic transcription factor FOXO3a leads to cytoplasmic sequestration of FOXO3a by binding 14–3-3 and consequently prevention of FOXO3a-dependent proapoptotic transcriptional regulation.25
In this report, we evaluated the effects of disrupting 14–3-3–ligand association on cell survival by using a peptide-based 14–3-3 competitive antagonist R18. R18 is a 20–amino acid peptide isolated from a phage display library screen for its ability to bind the 14–3-3τ isoform. R18 binds to all isoforms of 14–3-3 protein very specifically and competitively interferes with interactions of 14–3-3 with multiple ligands, including Raf, ASK1, and ExoS.29,30 Here we present the mechanism that R18 effectively induces apoptosis in hematopoietic cells transformed by ZNF198-FGFR1 in vitro and in vivo, partially through disrupting the interaction of 14–3-3–FOXO3a but not 14–3-3–BAD association.
Materials and methods
DNA constructs and mutagenesis
Distinct ZNF198-FGFR1 constructs have been described.13 The pEGFP-FOXO3a and EGFP-FOXO3aAAA plasmids were gifts from Dr James Griffin (Dana-Farber Cancer Institute, Boston, MA). Dr Williams Sellers (Novartis, Boston, MA) kindly provided pcDNA3-FOXO3a and the reporter plasmid 3XIRS-luciferase. The YFP-tagged R18 dimer and mutant expression vectors pSCM138 and pSCM174 were described previously.31 The pREV-TRE-Hyg was converted to a Gateway destination vector, and pREV-TRE-Hyg-ZNF198-FGFR1 or YFP-R18 was generated as described.32 The pECFP-R18 dimer and pECFP-R18 mutant were generated by cloning an ApaI and BspEI fragment of pSCM-138 or pSCM-174, respectively, into the ApaI and BspEI sites of pECFP (Clonetech, Palo Alto, CA).
Cell culture
TonBaF cells were kindly provided by Dr George Daley at Children's Hospital, Boston, MA. Ba/F3 cells were cultured in RPMI 1640 medium in the presence of 10% fetal bovine serum (FBS) and 1.0 ng/mL interleukin-3 (IL-3) (R&D Systems, Minneapolis, MN). U2–0S human osteosarcoma cells were cultured in McCoy 5A medium with 10% FBS. COS-7 monkey kidney cells and 293T cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% FBS. Retroviral stocks were generated and viral titers were determined as described previously.33,34 Doxycycline-inducible ZNF198-FGFR1, R18 dimer, R18 mutant, and FOXO3a- and FOXO3aAAA-expressing cell lines were generated by retroviral transduction of individual pREV-TRE-Hyg vectors encoding distinct constructs into TonBaF cells as described.35 For cell viability assays, 105 TonBaF cells inducibly expressing R18 with stable expression of 4ZF were cultured in 24-well plates with increasing concentrations of doxycycline in the absence of IL-3. The relative cell viability at each experimental time point was determined using the Celltiter96AQueous One solution proliferation kit (Promega, Madison, WI). For apoptosis assays, 106 TonBaF cells inducibly expressing R18 dimer or mutant with stable expression of 4ZF were treated with IL-3 withdrawal and increasing concentration of doxycycline for 24 hours. Cells were collected and stained using phycoerythrin (PE)-conjugated annexin V labeling reagent (BD Pharmingen, San Diego, CA), followed by fluorescence-activated cell sorter (FACS) analysis for the apoptotic cell population.
Immunoprecipitation and Western blot
To assay for the phosphorylation of various proteins, Ba/F3 cells were treated with serum starvation in plain RPMI 1640 for 6 hours before lysis. Cells (approximately 107) were lysed, and cell extracts were clarified by centrifugation and used for immunoprecipitation or immunoblotting as described.32 Applied antibodies included antibodies against BAD, phospho-BAD (S112), p44/42 ERK, phospho-p44/42 ERK (Thr202/Tyr204), AKT, and phospho-AKT PARP, caspase 3 p-PISK p85 (Cell Signaling, Beverly, MA); antibodies against FGFR1, P27, GFP, and 14–3-3β (Santa Cruz Biotechnology, Santa Cruz, CA); antibodies against Bim1 (Affinity Bio Reagents, Golden, CO); and antibodies against phosphotyrosine (clone 4G10), FOXO3a, and phospho-FOXo3a (Thr32) (Upstate, Lake Placid, NY); p-actin (Sigma, St Louis, MO).
Mice
Syngeneic BALB/c mice were injected with 106 TonBaF cells inducibly expressing R18 variants that were stably transduced with 4ZF. Recipient mice were given drinking water with or without doxycycline at 5 mg/mL in a solution of 5% sucrose ad libitum for 5 days before injection. The injected mice were maintained for 15 days with drinking water in the presence or absence of doxycycline before being killed and pathologic analysis. The doxycycline solution was changed every 2 to 3 days with protection from light by wrapping the drinking water bottles with autoclaved paper towels and aluminum foil. Affected animals were humanely killed for harvest of the spleens. An unpaired t test was used to assess statistical significance (P value) for differences in spleen sizes.
Luciferase assay
Luciferase assays were performed according to the manufacturer's instructions (Dual-Luciferase Reporter Assay System; Promega). Briefly, U2-OS cells (1.5 × 105 cells per well) were cultured in 6-well plates and transfected with a total of 2μg appropriate plasmid DNA (pcDNA-FOXO3a, pMSCV-ZNF198-FGFR1 4ZF, 3× IRS luciferase, CMV-Renilla, or pcDNA). Two hours after transfection, cells were cultured in serumfree DMEM for 40 hours. Luciferase and Renilla luciferase activities were determined for each sample, as described in the Dual-Luciferase Reporter Assay System instructions.
Purification of recombinant TAT-YFP-R18 fusion proteins
In brief, the expressed fusion protein was purified by sonication of high-expressing BL21(DE3)pLysS cells obtained from 50 mL of culture with isopropyl-beta-D-thiogalactopyranoside (IPTG) induction for 4 hours. Cellular lysates were resolved by centrifugation and loaded onto a Ni-NTA column in 20 mM imidazole. The TAT fusion proteins were eluted with 250 mM imidazole and desalted on a PD-10 column into PBS. Purification efficiency was examined by Coomassie blue staining.
Fluorescent confocal microscopy
COS-7 cells were plated onto glass coverslips at a density of 105 cells per well in 12-well plates. Cells were cotransfected with indicated plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Four hours after transfection, cells were cultured in serumfree DMEM for 16 hours. Transfected cells were fixed by 1% formaldehyde in methanol for 1 hour, followed by washing 3 times with PBS. The slides were mounted with ProLong Gold antifade reagent (Invitrogen). Confocal imaging was carried out on a Zeiss LSM 510 META confocal microscope (Thornwood, NY) equipped with an argon-ion laser, and CFP or EGFP signals were detected.
Results
Both AKT and MAPK are activated in hematopoietic Ba/F3 cells transformed by ZNF198-FGFR1, resulting in phosphorylation of 14–3-3–binding proteins FOXO3a at T32 and BAD at S112, respectively
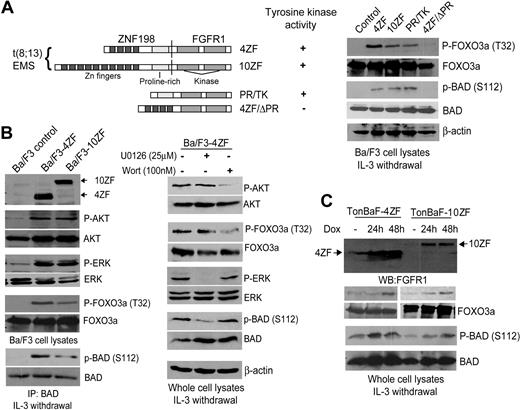
We previously evaluated the transforming properties of 2 ZNF198-FGFR1 variants cloned from stem cell MPD patients with t(8;13)(p11;q12) and related mutations in Ba/F3 cells.13 These included 2 isoforms of ZNF198-FGFR1 (4ZF and 10ZF, containing 4 and 10 N-terminal zinc fingers, respectively) as well as 2 truncation mutants, 4ZF/ΔPR and PR/TK (Figure 1A). 4ZF/ΔPR is a kinase dead mutant that has a deletion of the ZNF198 proline-rich domain from 4ZF, which is the oligomerization domain,10,13 whereas PR/TK is constitutively activated that lacks all ZNF198 zinc fingers but has retained the ZNF198 proline-rich domain (Figure 1A). Retroviral vectors encoding distinct ZNF198-FGFR1 variants were stably transduced into murine hematopoietic Ba/F3 cells. Ba/F3 cells require IL-3 for cell survival and proliferation, and our previous studies demonstrated that ZNF198-FGFR1 confers IL-3–independent proliferation to Ba/F3 cells.13 As shown in Figure 1B, in the absence of IL-3, expression of 4ZF and 10ZF resulted in phosphorylation and activation of AKT and ERK compared with control Ba/F3 cells. Activation of AKT by ZNF198-FGFR1 resulted in increased phosphorylation of the downstream proapoptotic substrate FOXO3a as assessed at Thr32, whereas activation of the ERK pathway resulted in phosphorylation of BAD at Ser112 (Figure 1A-B). In contrast, cells transduced with the kinase dead 4ZF/ΔPR mutant failed to induce phosphorylation of FOXO3a and BAD compared with cells expressing 4ZF, 10ZF, or PR/TK (Figure 1A). Treatment of Ba/F3 cells expressing 4ZF with MEK1 inhibitor U0126 and PI3K inhibitor wortmannin effectively decreased the phosphorylation level at BAD S112 and FOXO3a T32, respectively (Figure 1B). Phosphorylation at both FOXO3a T32 and BAD S112 negatively regulates these 2 proapoptotic protein factors by providing binding sites for 14–3-3 proteins, which consequently sequester FOXO3a and BAD,24,25 suggesting that 14–3-3 proteins integrate prosurvival signals of the AKT and ERK pathways in hematopoietic cells transformed by ZNF198-FGFR1.
Expression of ZNF198-FGFR1 activates both the AKT and ERK pathways, leading to phosphorylation at 14–3-3 binding sites of FOXO3a (T32) and BAD (S112), respectively. (A) (Left) Schematic diagram of ZNF198-FGFR1 fusion and truncation constructs. (Right) Expression of ZNF198-FGFR1 4ZF and 10ZF isoforms as well as activated truncation mutant PR/TK results in phosphorylation of FOXO3a (T32) and BAD (S112). Ba/F3 cells and cells expressing a kinase dead mutant 4ZF/ΔPR were included as negative controls. (B) Activation of the AKT and MAPK pathways in Ba/F3 cells stably expressing 4ZF and 10ZF isoforms resulted in phosphorylation of FOXO3a (T32) and BAD (S112), respectively. Cells stably expressing 4ZF were treated with U0126 and wortmannin at the indicated concentration for 90 minutes before immunoblotting. (C) Induced expression of 4ZF and 10ZF in TonBaF cells results in phosphorylation of FOXO3a (T32) and BAD (S112).
Expression of ZNF198-FGFR1 activates both the AKT and ERK pathways, leading to phosphorylation at 14–3-3 binding sites of FOXO3a (T32) and BAD (S112), respectively. (A) (Left) Schematic diagram of ZNF198-FGFR1 fusion and truncation constructs. (Right) Expression of ZNF198-FGFR1 4ZF and 10ZF isoforms as well as activated truncation mutant PR/TK results in phosphorylation of FOXO3a (T32) and BAD (S112). Ba/F3 cells and cells expressing a kinase dead mutant 4ZF/ΔPR were included as negative controls. (B) Activation of the AKT and MAPK pathways in Ba/F3 cells stably expressing 4ZF and 10ZF isoforms resulted in phosphorylation of FOXO3a (T32) and BAD (S112), respectively. Cells stably expressing 4ZF were treated with U0126 and wortmannin at the indicated concentration for 90 minutes before immunoblotting. (C) Induced expression of 4ZF and 10ZF in TonBaF cells results in phosphorylation of FOXO3a (T32) and BAD (S112).
Similar results were obtained in hematopoietic cells inducibly expressing 4ZF or 10ZF, which were generated by retroviral transduction of Ba/F3-derivative TonBaF cells (Figure 1C). TonBaF cells stably express the reverse Tet transactivator. Retroviruses containing the “Tet-on” pRev-TRE-Hyg-4ZF or 10ZF vectors that express 4ZF or 10ZF from the Tet-response elements were used to stably transform TonBaF cells. Immunoblotting results demonstrated doxycycline-inducible expression of 4ZF and 10ZF at 24 hours after the addition of doxycycline (Dox) (Figure 1C). The minimal level of 4ZF expression in the absence of doxycycline may be due to the basal level of doxycycline-independent activity of the Tet-response elements.
Inducible expression of the 14–3-3 antagonist, R18, attenuates 4ZF-induced transformation and induces apoptosis in TonBaF cells in vitro
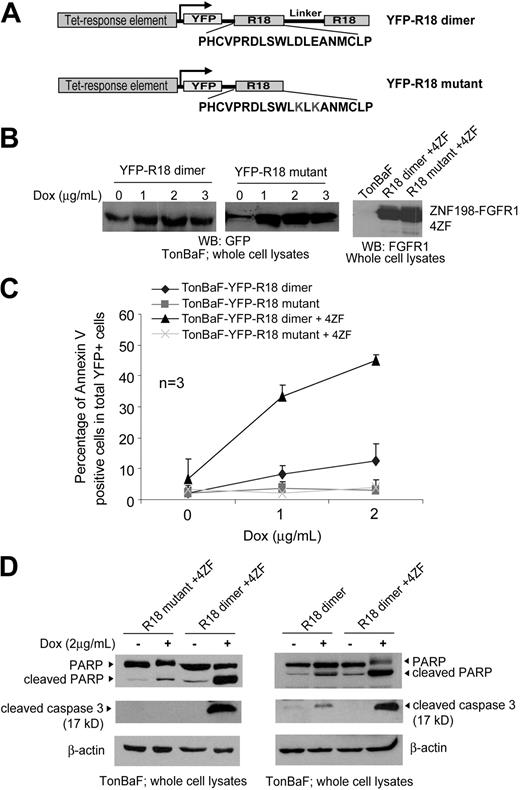
We next evaluated the inhibitory effects of a peptide-based 14–3-3 antagonist, R18, in FGFR1 fusion-transformed cells. We generated TonBaF cell lines inducibly expressing a dimeric version (R18 dimer; Figure 2A) combining 2 R18 peptides or a mutant form of R18 (R18 mutant; Figure 2A) that contains 1 R18 peptide with 2 substitutions, D12K and E14K, that abolish binding ability to 14–3-3. These cell lines were used to generate TonBaF cell lines that express either R18 dimer or R18 mutant in an inducible manner with stable expression of ZNF198-FGFR1 4ZF (Figure 2B).
Induced expression of a 14–3-3 antagonist R18 induces apoptosis in Ba/F3 cells transformed by ZNF198-FGFR1. (A) Schematic diagram of the structure and amino acid sequences of YFP-tagged R18 dimer and mutant. The R18 dimer construct contains 2 R18 motifs separated by a linker of 11 amino acids. (B) Inducible expression of YFP-tagged R18 dimer and mutant as well as stable expression of ZNF198-FGFR1 4ZF in the TonBaF cell lines. The GFP antibody applied cross-reacts with YFP. (C) TonBaF cells transformed by 4ZF are more sensitive to R18-induced apoptosis compared with control TonBaF-R18 cells. Cells were stained with PE-conjugated antiannexin V reagent and analyzed by FACS for the apoptotic population that is characterized as the fraction of annexin V/YFP double-positive cells in total YFP-positive cells (%). TonBaF cells inducibly expressing R18 cultured in the presence of IL-3 were included as controls. (D) R18 expression induced cleavage of PARP and caspase-3 in TonBaF R18 parental cells and 4ZF-transformed cells.
Induced expression of a 14–3-3 antagonist R18 induces apoptosis in Ba/F3 cells transformed by ZNF198-FGFR1. (A) Schematic diagram of the structure and amino acid sequences of YFP-tagged R18 dimer and mutant. The R18 dimer construct contains 2 R18 motifs separated by a linker of 11 amino acids. (B) Inducible expression of YFP-tagged R18 dimer and mutant as well as stable expression of ZNF198-FGFR1 4ZF in the TonBaF cell lines. The GFP antibody applied cross-reacts with YFP. (C) TonBaF cells transformed by 4ZF are more sensitive to R18-induced apoptosis compared with control TonBaF-R18 cells. Cells were stained with PE-conjugated antiannexin V reagent and analyzed by FACS for the apoptotic population that is characterized as the fraction of annexin V/YFP double-positive cells in total YFP-positive cells (%). TonBaF cells inducibly expressing R18 cultured in the presence of IL-3 were included as controls. (D) R18 expression induced cleavage of PARP and caspase-3 in TonBaF R18 parental cells and 4ZF-transformed cells.
The resulting cell lines were cultured in the presence or absence of doxycycline and assessed for R18-induced apoptosis. The cells were cultured in the absence or presence of IL-3 for 24 hours with doxycycline (Dox) at the indicated concentrations, followed by staining with PE-conjugated antiannexin V agents and flow cytometric analysis. Expression of YFP-R18 dimer induced significant apoptosis in cells stably transformed by 4ZF, which was assessed as the fraction of annexin V/YFP double-positive cells in a dose-dependent manner (Figure 2C) as well as cleavage of caspase-3 and PARP (Figure 2D, left). In contrast, R18 mutant failed to induce significant apoptosis in these cells. Moreover, in the presence of IL-3, R18 dimer induced much less apoptosis in the control parental TonBaF cells (Figure 2C-D), suggesting that the cells stably transformed by 4ZF are more sensitive to apoptosis induced by R18 expression.
Induction of R18 effectively inhibits 4ZF-induced transformation of Ba/F3 cells in vitro and attenuates disease development in vivo
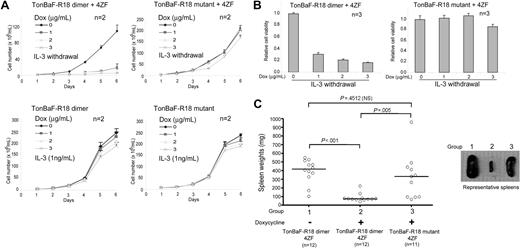
The TonBaF-R18 cell lines expressing 4ZF were cultured in the presence or absence of doxycycline and assessed for IL-3–independent proliferation as a surrogate for transformation. As shown in Figure 3A-B, stable expression of constitutively activated 4ZF conferred both TonBaF cell lines to IL-3 independence in the absence of doxycycline. However, doxycycline-induced expression of R18 dimer significantly attenuated IL-3–independent proliferation of 4ZF-transformed TonBaF cells, as assessed by proliferative rate as well as cell viability, whereas induced expression of R18 mutant failed to affect the proliferative rate and cell viability (Figure 3A-B). In contrast, induction of R18 dimer or mutant did not significantly affect the proliferative rate and cell viability of control TonBaF-R18 cells in the presence of IL-3 (Figure 3A-B). These data are in consonance with the results obtained from the apoptosis-based cell assay (Figure 2C), suggesting that TonBaF cells transformed by 4ZF are more sensitive to the inhibition of proliferation induced by R18 compared with control cells.
Induction of R18 effectively inhibits ZNF198-FGFR1–induced transformation in Ba/F3 cells in vitro and attenuates disease development in vivo. (A) R18 dimer inhibits 4ZF-conferred IL-3–independent proliferation of TonBaF cells. Cells were cultured with increasing dosages of Dox in the absence of IL-3 and counted daily. TonBaF cells inducibly expressing R18 dimer or mutant cultured in the presence of IL-3 were included as controls. (B) Inhibitory effects of R18 dimer on the IL-3 independence of 4ZF-transformed TonBaF cells. The relative cell viability was normalized to the viability of cells in the absence of Dox. (C) Inducible expression of R18 dimer attenuates tumor development in mice mediated by 4ZF-transformed TonBaF cells. (Left) Differential sizes of the spleens from each group of animals. Median values are represented by horizontal bars. Indicated P values are determined by an unpaired test. NS indicates not significant. (Right) Representative spleen examples from each group of animals.
Induction of R18 effectively inhibits ZNF198-FGFR1–induced transformation in Ba/F3 cells in vitro and attenuates disease development in vivo. (A) R18 dimer inhibits 4ZF-conferred IL-3–independent proliferation of TonBaF cells. Cells were cultured with increasing dosages of Dox in the absence of IL-3 and counted daily. TonBaF cells inducibly expressing R18 dimer or mutant cultured in the presence of IL-3 were included as controls. (B) Inhibitory effects of R18 dimer on the IL-3 independence of 4ZF-transformed TonBaF cells. The relative cell viability was normalized to the viability of cells in the absence of Dox. (C) Inducible expression of R18 dimer attenuates tumor development in mice mediated by 4ZF-transformed TonBaF cells. (Left) Differential sizes of the spleens from each group of animals. Median values are represented by horizontal bars. Indicated P values are determined by an unpaired test. NS indicates not significant. (Right) Representative spleen examples from each group of animals.
To evaluate the potentially therapeutic efficacy of targeting 14–3-3 in vivo, we used a murine mouse model in which 106 4ZF-transformed TonBaF cells inducibly expressing either R18 dimer or R18 mutant were injected into the tail veins of syngeneic BALB/c mice (Figure 3C). The injected mice were maintained on either normal drinking water or water containing 5 mg/mL doxycycline with 5% sucrose for 15 days. In the absence of doxycycline, the TonBaF-R18 dimer cells stably expressing 4ZF caused tumor development characterized by splenomegaly (median spleen size, 418 mg; Figure 3C). In contrast, doxycycline-induced expression of R18 dimer in the 4ZF-transformed cells resulted in a statistically significant decrease in spleen size (median, 73 mg; P < .001), whereas significant splenomegaly was observed in control animals injected with TonBaF-R18 mutant cells stably expressing 4ZF treated with doxycycline (median spleen size, 333 mg; P = .005). These results indicate that R18 markedly attenuates the efficacy of 4ZF-mediated tumorigenesis in this murine allograft model, whereas the R18 mutant has no significant inhibitory effects.
Protein transduction domain–conjugated TAT-YFP-R18 dimer effectively induces apoptosis in human leukemia KG-1a cells expressing 8p11 FGFR1 fusion
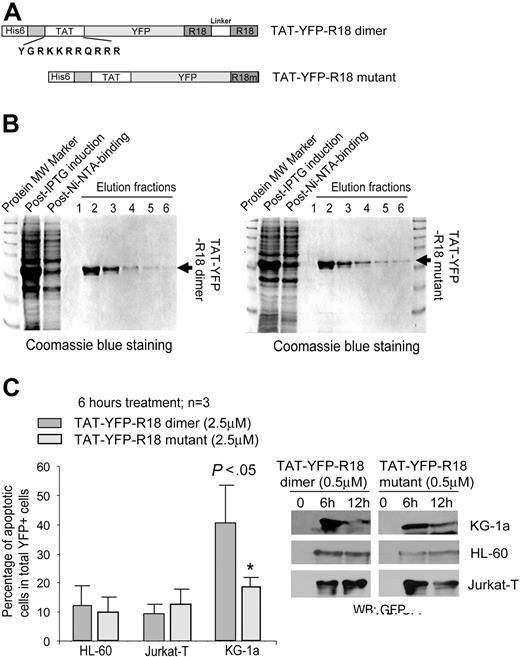
The HIV TAT-mediated protein transduction in a receptor-independent fashion was first discovered in 1988.36,37 We have generated bacterial expression plasmids to express fusion protein TAT-YFP-R18 containing an N-terminal 6-histidine leader followed by the 11–amino acid TAT protein transduction domain (YGRKKRRQRRR) and the YFP-tagged R18 dimer or mutant (Figure 4A). Purified recombinant R18 proteins were examined by Coomassie blue staining (Figure 4B).
TAT-YFP-R18 dimer is able to transduce into human leukemia KG-1a cells expressing FGFR1OP2-FGFR1 fusion and effectively induces apoptosis. (A) Schematic diagram of the structure of TAT-YFP-R18 dimer and -R18 mutant. Amino acid sequence of the TAT protein transduction domain is indicated. (B) Induced expression of recombinant TAT-YFP-R18 proteins in bacteria by IPTG. The fusion proteins were purified, and the purification efficiency was determined by Coomassie blue staining. (C) TAT-YFP-R18 dimer transduces into and induces significant apoptosis in KG-1a cells but not in control HL-60 and Jurkat T cells. The cells were incubated with TAT-YFP-R18 proteins for 6 hours, followed by staining with antiannexin V reagent and analysis by FACS for the apoptotic population that is characterized as the fraction of annexin V/YFP double-positive cells in total YFP-positive cells (%). Indicated P values were determined by the Student t test. Transduced TAT-YFP-R18 proteins were detected by Western blotting.
TAT-YFP-R18 dimer is able to transduce into human leukemia KG-1a cells expressing FGFR1OP2-FGFR1 fusion and effectively induces apoptosis. (A) Schematic diagram of the structure of TAT-YFP-R18 dimer and -R18 mutant. Amino acid sequence of the TAT protein transduction domain is indicated. (B) Induced expression of recombinant TAT-YFP-R18 proteins in bacteria by IPTG. The fusion proteins were purified, and the purification efficiency was determined by Coomassie blue staining. (C) TAT-YFP-R18 dimer transduces into and induces significant apoptosis in KG-1a cells but not in control HL-60 and Jurkat T cells. The cells were incubated with TAT-YFP-R18 proteins for 6 hours, followed by staining with antiannexin V reagent and analysis by FACS for the apoptotic population that is characterized as the fraction of annexin V/YFP double-positive cells in total YFP-positive cells (%). Indicated P values were determined by the Student t test. Transduced TAT-YFP-R18 proteins were detected by Western blotting.
Figure 4C shows that the purified TAT-YFP-R18 dimer and mutant effectively crossed the cell membrane and transduced into human leukemia cells, including KG-1a expressing the constitutively activated fusion tyrosine kinase FGFR1OP2-FGFR1 associated with ins(12,8)(p11;p11p22) stem cell MPD38 as well as HL-60 and Jurkat T. Moreover, treatment of TAT-YFP-R18 dimer effectively induced apoptosis in FGFR1OP2-FGFR1–transformed KG-1a cells but not in HL-60 and Jurkat T that are not transformed by any constitutively activated tyrosine kinase, which was assessed as the fraction of annexin V–positive/YFP-positive cells, compared with treatment of TAT-YFP-R18 mutant (Figure 4C).
These data suggest that the constitutively activated FGFR1OP2-FGFR1 fusion tyrosine kinase–transformed human leukemia cells are sensitive to R18-induced apoptosis, whereas R18 has minimal nonspecific cytotoxicity in human hematopoietic cells that are not transformed by leukemogenic fusion/mutant tyrosine kinases. These results are also consistent with the observation that Dox-induced expression of R18 effectively induces apoptosis in Ba/F3 cells transformed by 4ZF compared with control Ba/F3 cells (Figure 2C) and imply the therapeutic potential of R18 in treatment of human 8p11 MPD associated with different FGFR1 fusion tyrosine kinases.
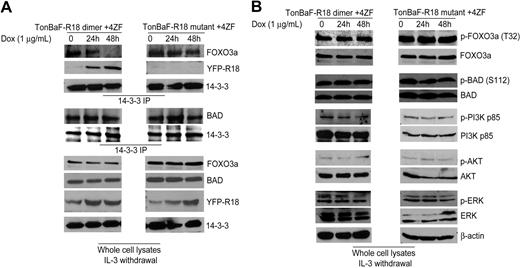
Induced R18 expression disrupts interaction between 14–3-3 and FOXO3a but not 14–3-3–BAD association in Ba/F3 cells transformed by 4ZF
To determine the molecular mechanism by which R18 induces apoptosis in 8p11 FGFR1 fusion-transformed hematopoietic cells, we performed coimmunoprecipitation experiments to examine whether R18 expression dissociates 14–3-3 from BAD and/or FOXO3a. Inducible TonBaF cell lines of R18 dimer or mutant with stable expression of 4ZF were treated with doxycycline for 24 and 48 hours. The 14–3-3 immunoprecipitates were obtained, and coimmunoprecipitated BAD or FOXO3a was detected by Western blot. As shown in Figure 5A (top), induced R18 dimer expression abolished 14–3-3-FOXO3a interaction and, at the same time, increased levels of R18 dimer were detected in the same 14–3-3 immunoprecipitates, whereas the FOXO3a–14–3-3 association was not affected by expression of R18 mutant. In contrast, the amount of BAD protein in the 14–3-3 immunoprecipitates was not altered by induction of either R18 dimer or R18 mutant (Figure 5A), suggesting R18 is only able to disrupt association of 14–3-3–FOXO3a but not 14–3-3–BAD.
R18 disrupts interaction between 14–3-3 and FOXO3a but not 14–3-3–BAD association in Ba/F3 cells transformed by ZNF198-FGFR1. (A) 4ZF-transformed TonBaF cells inducibly expressing YFP-tagged R18 dimer or mutant were cultured with Dox in the absence of IL-3 for 24 and 48 hours. Coimmunoprecipitated FOXO3a, BAD, and YFP-tagged R18 dimer or mutant in 14–3-3 immunoprecipitates were detected by specific antibodies. (B) Phosphorylation levels of FOXO3a, BAD, PI3K, AKT, and ERK in the cell lysates were probed by Western blotting as controls.
R18 disrupts interaction between 14–3-3 and FOXO3a but not 14–3-3–BAD association in Ba/F3 cells transformed by ZNF198-FGFR1. (A) 4ZF-transformed TonBaF cells inducibly expressing YFP-tagged R18 dimer or mutant were cultured with Dox in the absence of IL-3 for 24 and 48 hours. Coimmunoprecipitated FOXO3a, BAD, and YFP-tagged R18 dimer or mutant in 14–3-3 immunoprecipitates were detected by specific antibodies. (B) Phosphorylation levels of FOXO3a, BAD, PI3K, AKT, and ERK in the cell lysates were probed by Western blotting as controls.
The phosphorylation levels of both FOXO3a and BAD were not altered by expression of R18 (Figure 5B)—nor was the phosphorylation status of PI3K, AKT, ERK (Figure 5B), and 4ZF (data not shown). Thus, this difference might be due to the relatively high binding affinity of 14–3-3 for BAD such that R18 is able to compete with FOXO3a for 14–3-3 but insufficient to disrupt 14–3-3–BAD association.
R18 induces apoptosis partially through FOXO3a in TonBaF cells expressing 4ZF
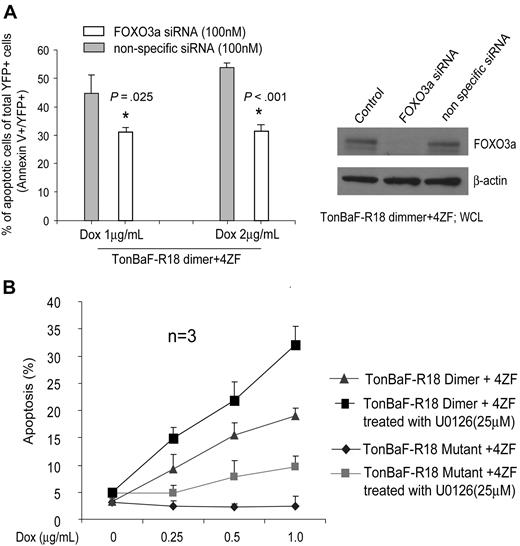
R18 has been shown to competitively interfere with the interaction of 14–3-3 with multiple ligands including Raf-1, ASK1, and ExoS.29,39 Therefore, we proposed that R18 may induce apoptosis by disrupting multiple interactions of 14–3-3 with its ligands and partially through liberation of FOXO3a from sequestration by 14–3-3. Indeed, targeted down-regulation of FOXO3a by specific siRNA significantly attenuated R18-inudced apoptosis in TonBaF cells stably expressing 4ZF, which, however, was insufficient to completely abrogate R18 proapoptotic effects compared with control nonspecific siRNA (Figure 6A). These data strongly support our hypothesis that FOXO3a is a critical signaling effector and contributes to R18-induced apoptosis.
R18 induces apoptosis in part through FOXO3a in cells expressing ZNF198-FGFR1. (A) Cells were transfected with siRNA for 24 hours before Dox induction. The apoptotic population was characterized as the fraction of annexin V/YFP double-positive cells in total YFP-positive cells (%). Indicated P values were determined by the Student t test. (B) MEK1 inhibitor U0126 treatment enhanced R18-induced apoptosis in the 4ZF-transformed TonBaF cells. Cells were treated with an increasing concentration of Dox in the presence or absence of U0126 for 24 hours, followed by annexin V staining and FACS analysis for the apoptotic population.
R18 induces apoptosis in part through FOXO3a in cells expressing ZNF198-FGFR1. (A) Cells were transfected with siRNA for 24 hours before Dox induction. The apoptotic population was characterized as the fraction of annexin V/YFP double-positive cells in total YFP-positive cells (%). Indicated P values were determined by the Student t test. (B) MEK1 inhibitor U0126 treatment enhanced R18-induced apoptosis in the 4ZF-transformed TonBaF cells. Cells were treated with an increasing concentration of Dox in the presence or absence of U0126 for 24 hours, followed by annexin V staining and FACS analysis for the apoptotic population.
Moreover, we observed that the MEK1 inhibitor U0126 treatment enhanced R18 dimer proapoptotic activity in TonBaF cells transformed by 4ZF compared with induction of R18 mutant (Figure 6B), which supports the proposed model that targeting 14–3-3 by R18 induces apoptosis in FGFR1 fusion-transformed cells partially through reactivation of FOXO3a phosphorylated by activated AKT but not BAD inhibited by activated ERK.
R18 induces apoptosis by rescuing the nuclear localization of FOXO3a in cells expressing 4ZF
To further elucidate the role of FOXO3a in R18-induced apoptosis, we first tested whether 4ZF-induced phosphorylation of FOXO3a negatively regulated its transcriptional activity by nuclear to cytoplasmic relocalization and sequestration, which might be reversed by the 14–3-3 antagonist, R18.
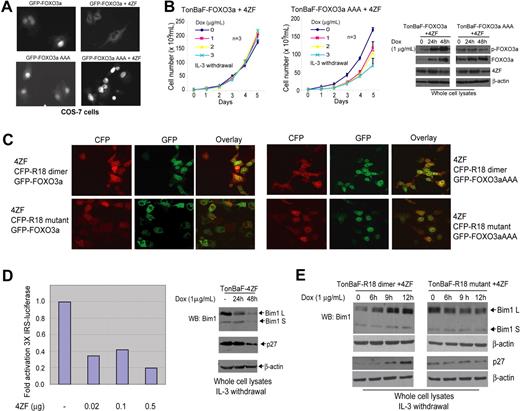
We transiently transfected COS-7 monkey kidney cells with expression vectors encoding EGFP-tagged FOXO3a or FOXO3aAAA triple mutants (T32A/S253A/S315A), in which each of 3 serine and threonine-phosphorylation sites regulated by AKT was mutated to alanine.25 Cells expressing EGFP-FOXO3a alone showed a nuclear pattern of EGFP signal distribution, whereas cells coexpressing both EGFP-FOXO3a and 4ZF demonstrated primary cytoplasmic localization of FOXO3a. In contrast, EGFP-FOXO3aAAA was predominantly localized in the nuclei in the absence or presence of 4ZF expression (Figure 7A). Moreover, doxycycline-induced expression of FOXO3aAAA significantly attenuated the IL-3–independent proliferation of TonBaF cells transformed by 4ZF, evidenced by a slower proliferative rate and decreased cell viability, which were due to increased apoptosis and decreased mitotic rate (data not shown), compared with cells without doxycycline treatment. Induced expression of wild-type FOXO3a, however, did not alter the proliferative rate and cell viability in cells transformed by 4ZF (Figure 7B). These data suggest that mutations at the AKT phosphorylation sites and consequent 14–3-3–binding sites of FOXO3a overcome 4ZF-dependent inhibition, and nuclear relocation of FOXO3aAAA attenuates 4ZF-induced transformation as R18 does (Figure 2C).
R18 induces apoptosis by rescuing the nuclear localization of FOXO3a in cells expressing ZNF198-FGFR1. (A) Expression of constitutively activated 4ZF fusion tyrosine kinase results in cytoplasmic localization of FOXO3a. (B) Induced expression of nuclear-localized FOXO3aAAA mutant attenuates IL-3 independence of TonBaF cells transformed by 4ZF. (C) Expression of R18 dimer inhibits cytoplasmic sequestration of FOXO3a induced by 4ZF and rescues nuclear localization of FOXO3a. COS7 cells were cotransfected with 4ZF, CFP-tagged R18 dimer or mutant, and EGFP-FOXO3a or EGFP-FOXO3aAAA mutant. The molar ratio of DNA amount in the cotransfection experiment was 2:1:1 (4ZF/R18/FOXO3a) to ensure successful 4ZF transfection in cells that are both CFP and EGFP positive. (D) Expression of 4ZF abrogates FOXO3a-dependent transcription regulation. (Left) U2-OS cells were transiently cotransfected with the 3× IRS reporter plasmid encoding luciferase, pCMV-Renilla, and an increasing amount of plasmid encoding 4ZF. The luciferase activity was measured and normalized to Renilla luciferase activity. The fold of luciferase activity was normalized to the activity of cells in the absence of 4ZF transfection. Representative data are shown from repeated experiments. (Right) Decreased expression of Bim1 and p27kip1 in TonBaF cells inducibly expressing 4ZF. Two isoforms of Bim1, L (long) and S (short), were detected. (E) Induced expression of R18 dimer rescued FOXO3a-dependent expression of Bim1 and p27kip1 in TonBaF cells expressing 4ZF. The samples were analyzed by using a Zeiss LSM 510 META microscope with a 40×/1.30 objective lens. The pictures were taken by using a photo multipler tube (PMT) and analyzed by Adobe Photoshop 6.0 (Adobe, San Jose, CA).
R18 induces apoptosis by rescuing the nuclear localization of FOXO3a in cells expressing ZNF198-FGFR1. (A) Expression of constitutively activated 4ZF fusion tyrosine kinase results in cytoplasmic localization of FOXO3a. (B) Induced expression of nuclear-localized FOXO3aAAA mutant attenuates IL-3 independence of TonBaF cells transformed by 4ZF. (C) Expression of R18 dimer inhibits cytoplasmic sequestration of FOXO3a induced by 4ZF and rescues nuclear localization of FOXO3a. COS7 cells were cotransfected with 4ZF, CFP-tagged R18 dimer or mutant, and EGFP-FOXO3a or EGFP-FOXO3aAAA mutant. The molar ratio of DNA amount in the cotransfection experiment was 2:1:1 (4ZF/R18/FOXO3a) to ensure successful 4ZF transfection in cells that are both CFP and EGFP positive. (D) Expression of 4ZF abrogates FOXO3a-dependent transcription regulation. (Left) U2-OS cells were transiently cotransfected with the 3× IRS reporter plasmid encoding luciferase, pCMV-Renilla, and an increasing amount of plasmid encoding 4ZF. The luciferase activity was measured and normalized to Renilla luciferase activity. The fold of luciferase activity was normalized to the activity of cells in the absence of 4ZF transfection. Representative data are shown from repeated experiments. (Right) Decreased expression of Bim1 and p27kip1 in TonBaF cells inducibly expressing 4ZF. Two isoforms of Bim1, L (long) and S (short), were detected. (E) Induced expression of R18 dimer rescued FOXO3a-dependent expression of Bim1 and p27kip1 in TonBaF cells expressing 4ZF. The samples were analyzed by using a Zeiss LSM 510 META microscope with a 40×/1.30 objective lens. The pictures were taken by using a photo multipler tube (PMT) and analyzed by Adobe Photoshop 6.0 (Adobe, San Jose, CA).
We next investigated whether R18 functions similarly by rescuing FOXO3a nuclear localization. COS-7 cells were transiently transfected with 4ZF, EGFP-tagged FOXO3a or FOXO3aAAA triple mutant, and CFP-tagged R18 dimer or mutant (Figure 7C). The patterns of CFP and EGFP signal distribution indicated the expression and localization of R18 and FOXO3a, respectively. Consistent with previous observations, in the presence of 4ZF, cells coexpressing both EGFP-FOXO3a and R18 mutant primarily demonstrated cytoplasmic localization of FOXO3a (Figure 7C, left 6 panels). In contrast, most cells coexpressing both EGFP-FOXO3a and R18 dimer showed a nuclear pattern of EGFP signal distribution. The control EGFP-FOXO3aAAA was predominantly localized in the nucleus in the presence of 4ZF expression despite expression of R18 dimer or mutant (Figure 7C, right 6 panels). Similar results were observed in NIH3T3 cells (data not shown). Together, these data indicate that 4ZF-induced cytoplasmic sequestration of phospho-FOXO3a by 14–3-3 binding is disrupted by R18 expression, which results in nuclear relocalization of FOXO3a and contributes to R18-induced apoptosis.
Rescued nuclear relocation of FOXO3a by R18 up-regulates FOXO3a transcription targets Bim1 and p27kip1
To define the molecular mechanism by which R18 attenuates transformation and induces apoptosis in cells transformed by 4ZF, we directed our attention to the transcriptional targets of FOXO3a. First we tested the effect of 4ZF expression on FOXO3a-mediated transactivation in a transient transfection assay. We coexpressed 4ZF and FOXO3a in U2–0S human osteosarcoma cells and assayed for transactivation of the FOXO3a-responsive promoter element IRS coupled to a luciferase reporter (3× IRS-luc).35 As shown in Figure 7D (left), 4ZF-induced phosphorylation of FOXO3a resulted in a dose-dependent decrement in IRS-mediated transactivation of the luciferase reporter in U2–0S cells.
In addition, we observed that regulation of a spectrum of FOXO3a downstream targets was altered by induced expression of 4ZF. The proapoptotic Bcl-2 family member Bim1 and cyclin-dependent kinase (CDK) inhibitor p27kip1 have been identified as downstream transcription targets of FOXO3a and implicated in regulating cell survival and proliferation.40,–42 We observed a time-dependent decrease in protein levels of Bim1 S (short isoform of Bim1) and Bim1 L (long isoform) as well as p27kip1 after the induction of 4ZF expression with doxycycline (Figure 7D, right). These data showed that 4ZF expression confers a cell survival and proliferative advantage by down-regulating Bim-1 and p27kip1, which is transcriptionally up-regulated by FOXO3a.
Consistent with previous observations, we found that induction of R18 dimer expression significantly increased protein expression levels of both Bim1 and p27kip1 in TonBaF cells stably transformed by 4ZF (Figure 7E). These data support the hypothesis that R18-induced inhibition of cell survival and proliferation in 4ZF-transformed hematopoietic cells functions partially through reactivation of FOXO3a-mediated apoptosis and growth arrest by up-regulating Bim1 and p27kip1.
Discussion
Our data suggest that 14–3-3 integrates AKT- and ERK-mediated prosurvival signals in ZNF198-FGFR1 fusion tyrosine kinase–induced hematopoietic transformation. Targeting 14–3-3 by the competitive 14–3-3 inhibitor R18 induces apoptosis in FGFR1 fusion-transformed cells partially through reactivation of the proapoptotic activity of FOXO3a but not BAD. The 14–3-3 proteins may function as a general prosurvival signal integrator in the aberrant signaling induced by FGFR1 fusion tyrosine kinases in hematopoietic cells. This is in consonance with several lines of evidence that suggest a critical role of 14–3-3 proteins in promoting tumorigenesis and resistance to therapies.43,–45 One exception is that epigenetic inactivation of 14–3-3σ isoform has been identified in several epithelial tumors including carcinomas of breast, skin, and prostate. However, 14–3-3σ is expressed primarily in epithelial cells.46,47
The results that the R18 mutant lacks binding ability to 14–3-3 and fails to induce significant apoptosis compared with the R18 dimer strongly argue that the phenotype of R18-induced apoptosis requires 14–3-3 binding. Moreover, compared with control TonBaF cells, cells stably transformed by ZNF198-FGFR1 are more sensitive to apoptosis induced by R18 expression (Figure 2C-D). In consonance with these data, the protein transduction domain (PTD)–conjugated R18 dimer effectively induces apoptosis in human leukemia KG-1a cells, which are transformed by constitutively activated FGFR1OP2-FGFR1 fusion, compared with the control human leukemia cell lines HL-60 and Jurkat T that are not transformed by constitutively activated tyrosine kinases. These studies together provide “proof of principle” that 14–3-3–ligand association may represent a potential target in inhibition of hematologic transformation induced by 8p11 FGFR1 fusion tyrosine kinases.
Previous studies have implied that R18 may function by causing BAD to dissociate from 14–3-3 because both Bcl-2 and Bcl-XL overcome R18-induced apoptosis.31,48 However, we found that only FOXO3a, but not BAD, was released to induce apoptosis in response to R18 induction in cells stably expressing ZNF198-FGFR1 4ZF. This release was accompanied by restoration of FOXO3a nuclear localization as well as up-regulation of FOXO3a transcription targets including Bim1 and p27kip1. Bim1 is a proapoptotic Bcl-2 family member, and thus Bcl-2 and Bcl-XL may inhibit R18-induced apoptosis by antagonizing the FOXO3a downstream transcription target Bim1 instead of BAD.
The 14–3-3 proteins are essential for cell survival and suppress the apoptosis process by multiple interactions with proteins of the core mitochondria machinery including BAD and BAX, proapoptotic transcription factors such as FOXO3a, as well as upstream proapoptotic signaling pathways. R18 is able to disrupt the interaction of 14–3-3 with multiple ligands.29,39 Therefore, although R18 may be insufficient to compete with BAD for 14–3-3 binding, R18 may induce apoptosis by competitively interfering with multiple interactions of 14–3-3 with its ligands including FOXO3a. In fact, we have observed that induced expression of a FOXO3aAAA mutant (Figure 7B) is less potent in attenuating the IL-3–independent proliferation of TonBaF cells transformed by 4ZF compared with cells inducibly expressing R18 (Figure 3A). This difference is probably due to the possibility that R18 may simultaneously interfere with multiple 14–3-3–ligand interactions besides 14–3-3–FOXO3a association. These data are consistent with the observations that targeted down-regulation of FOXO3a by specific siRNA significantly attenuated but failed to completely abrogate R18 proapoptotic activity (Figure 6A).
Therefore, in contrast to the conventional strategies targeting oncogenic fusion/mutant tyrosine kinases—in our case 8p11 FGFR1 fusions—our data provide proof of concept that development of 14–3-3 antagonists to inhibit multiple or a whole class of 14–3-3–ligand interactions may provide a novel strategy to induce apoptosis by simultaneously abrogating multiple signaling pathways.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We gratefully acknowledge the critical reading of the manuscript by Drs Benjamin Lee, Brian Huntly, Ifor Williams, and Edmund Waller. Dr Benjamin Lee kindly helped us with statistical analysis. We greatly appreciate the generous suggestion of Drs Songli Xu and Maureen Powers with confocal microscopy.
This work was supported in part by National Institutes of Health (NIH) grant CA120272 (J.C.), the Leukemia and Lymphoma Society (J.C.), and the Golfer Against Cancer Foundation (J.C., S.L.). J. Chen is a Fellow of the Leukemia and Lymphoma Society and a Georgia Cancer Coalition Distinguished Cancer Scholar.
National Institutes of Health (NIH)
Authorship
Contribution: S.D., S. Kang, and J.C. designed research, performed research, analyzed data, and wrote the paper; S. Kardar performed research; T.G., H.F., S.L., H.J.K., and F.K. provided reagents and materials.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jing Chen, Winship Cancer Institute, Emory University School of Medicine, 1365-C Clifton Rd NE, C-3002, Atlanta, GA 30322; e-mail: jchen@emory.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal