Abstract
Iron (Fe) plays an important role in proliferation, and Fe deficiency results in G1/S arrest. Despite this, the precise role of Fe in cell-cycle control remains unclear. Cyclin D1 plays a critical function in G1 progression by interacting with cyclin-dependent kinases. Previously, we examined the effect of Fe depletion on the expression of cell-cycle control molecules and identified a marked decrease in cyclin D1 protein, although the mechanism involved was unknown. In this study, we showed that cyclin D1 was regulated posttranscriptionally by Fe depletion. Iron chelation of cells in culture using desferrioxamine (DFO) or 2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone (311) decreased cyclin D1 protein levels after 14 hours and was rescued by the addition of Fe. Cyclin D1 half-life in control cells was 80 ± 15 minutes (n = 5), while in chelator-treated cells it was significantly (P < .008) decreased to 38 ± 3 minutes (n = 5). Proteasomal inhibitors rescued the Fe chelator–mediated decrease in cyclin D1 protein, suggesting the role of the proteasome. In Fe-replete cells, cyclin D1 was degraded in an ubiquitin-dependent manner, while Fe depletion induced a ubiquitin-independent pathway. This is the first report linking Fe depletion–mediated growth suppression at G1/S to a mechanism inducing cyclin D1 proteolysis.
Introduction
Proliferation is dependent upon iron (Fe).1–4 The highest demand for Fe occurs during the late G1 and S phases of the cell cycle,5,6 due in part to the activity of the Fe-requiring enzyme of DNA synthesis, ribonucleotide reductase (RR).5,6 Iron-deprivation inhibits RR activity and results in G1/S arrest.7 However, little is known at the molecular level concerning the role of Fe in cell-cycle progression.
Cell-cycle progression is controlled in part by the cyclins, cyclin-dependent kinases (cdk's), and cyclin-dependent kinase inhibitors (cdki's).7,8 Cyclins interact with cdk's to form catalytically active heterodimers.8 Fe depletion decreases cyclin D1 (CD1) protein expression.9,10 However, the mechanism responsible remains unknown. CD1 assembles with cdk-4 or cdk-6, generating an active holoenzyme that catalyzes a rate-limiting step in G1/S progression.7,8 This complex phosphorylates the retinoblastoma protein which regulates S-phase entrance.8 Overexpression of CD1 releases cells from their normal controls and acts as an oncogene.11,12 In fact, pharmacologic targeting of CD1 may lead to novel antitumor agents.13
CD1 is regulated by changes in transcription, protein stability, mRNA turnover, and nucleocytoplasmic transport of the transcript and protein.14–19 CD1 protein expression can be regulated by degradation via the ubiquitin-dependent20 or ubiquitin-independent21 proteasome pathway. Phosphorylation of CD1 at Thr-286 by glycogen synthase kinase-3β (GSK-3β) can result in ubiquitination and proteasomal degradation.14,16 However, a GSK-3β–independent pathway of proteasomal CD1 degradation has been reported.17 CD1 can also be targeted by the von Hippel–Lindau protein (pVHL) that is a substrate recognition component of the E3-ubiquitin ligase that targets proteins for proteasomal degradation.22–24
Apart from ubiquitin-dependent degradation, ubiquitin-independent processes have been identified involving association of proteins with antizyme21,25,26 and/or NAD(P)H-quinone oxidoreductase (NQO1).27,28 Association of a protein with antizyme leads to proteasomal degradation,26 while NQO1 binding leads to stabilization against proteasomal degradation.28
Considering the mechanism of G1/S arrest after Fe depletion and the role of CD1, this study examines alterations in CD1 expression after Fe chelation. CD1 mRNA levels showed no change after Fe depletion, while its protein expression was markedly decreased. The reduction in CD1 was dependent on proteasomal activity, and our results indicate that G1/S arrest after Fe depletion is mediated, in part, by the decrease in CD1 via proteasomal degradation.
Materials and methods
Cell treatments
2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone (311) was synthesized as reported.29 Desferrioxamine (DFO) was from Novartis (Basel, Switzerland).
Cell culture
Renal carcinoma cells (RCCs; 786-O) and 786-O cells transfected with VHL (WT-8) or vector alone (PRC-3) were from Dr W. Kaelin (Harvard Research Centre, Cambridge, MA). A31N-ts20 cells30 were from Dr H. Ozer (University of Medicine and Dentistry of New Jersey, Newark [UMDNJ]–New Jersey Medical School). All other cell types were from the American Type Culture Collection (Manassas, VA) and were cultured as described.31
RNA isolation and semiquantitative RT-PCR
RNA was isolated using TRIzol (Invitrogen, Melbourne, VIC, Australia). Reverse transcription–polymerase chain reaction (RT-PCR) was done via standard methods32 using the primers listed in Table 1RT-PCR was shown to be semiquantitative by an optimization protocol that demonstrated it was in the log-phase of amplification. β-actin was used as an RNA-loading control.
Primers for amplification of human CD1, TfRI, and β-actin
| Pair no. . | Primer name . | Genotype/accession no. . | Oligonucleotides (5′-3′) . | Prod size, 1 . | |
|---|---|---|---|---|---|
| Forward . | Reverse . | ||||
| 1 | Cyclin D1 | M 73554 | CTGGATGCTGGAGGTCTGCGAGGA | TGAACTTCACATCTGTGGCACAGA | 400 |
| 2 | TfR-1 | NM_003234 | TCAGGTCAAAGACAGCGCTCAAAACTC | AGTCTCCTTCCATATTCCCAAACAGCTTTT | 358 |
| 3 | β-actin | NM_001101 | CCCGCCGCCAGCTCACCATGG | AAGGTCTCAAACATGATCTGGGTC | 397 |
| Pair no. . | Primer name . | Genotype/accession no. . | Oligonucleotides (5′-3′) . | Prod size, 1 . | |
|---|---|---|---|---|---|
| Forward . | Reverse . | ||||
| 1 | Cyclin D1 | M 73554 | CTGGATGCTGGAGGTCTGCGAGGA | TGAACTTCACATCTGTGGCACAGA | 400 |
| 2 | TfR-1 | NM_003234 | TCAGGTCAAAGACAGCGCTCAAAACTC | AGTCTCCTTCCATATTCCCAAACAGCTTTT | 358 |
| 3 | β-actin | NM_001101 | CCCGCCGCCAGCTCACCATGG | AAGGTCTCAAACATGATCTGGGTC | 397 |
Western analysis and immunoprecipitation
Western analysis was performed as described.10,31 Lysate protein concentrations were assayed using the BCA protein assay (Pierce, Rockford, IL). Optimization of primary and secondary antibody conditions was performed using a range of antibody concentrations. Membranes were then washed and developed using the ECL Plus Western blot detection reagent (Amersham Biosciences, Piscataway, NJ). Development of X-ray film was over a range of exposure times to ensure optimal sensitivity. Quantitation of Western results was performed by scanning X-ray film, and the picture was analyzed using Quantity One software (Biorad, Hercules, CA). To ensure equal loading of proteins, membranes were probed for β-actin. These experimental conditions were implemented to ensure linearity of the substrate reaction and best film sensitivity.
Immunoprecipitation was achieved using standard methods.33 All antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA) or Sigma-Aldrich (St Louis, MO).
20S and 26S proteasome activity
To assay effects of chelators on purified 20S proteasomes, the 20S Proteasome Activity Kit (Alexis, San Diego, CA) was used.
Cytoplasmic extracts were used to measure 26S proteasome activity.17 Briefly, MCF-7 cells were incubated for 24 hours at 37°C with chelators and/or proteasomal inhibitors and lysed. Cytoplasmic proteins from MCF-7 cells were incubated for 30 minutes at 30°C with the fluorogenic substrate Suc-Leu-Leu-Val-Tyr-AMC (Calbiochem-Merck, Darmstadt, Germany), and the fluorescent product was measured using a VICTOR3 Perkin Elmer counter (Shelton, CT).
MCF-7 cell synchronization and cycle analysis
In some experiments, cultures were synchronized by serum starvation.17 Briefly, cells were washed with PBS and serum-starved for 24 hours at 37°C. Starved cells were restimulated with 10% FCS for 16 hours at 37°C in the presence or absence of 250 μM DFO or 25 μM 311. Cells were harvested and the cell cycle was assayed using FACSCalibur flow cytometry (BD Biosciences, Franklin Lakes, NJ) and CELLQUEST software.34
Statistical analysis
Data were compared using the Student t test. Results are presented as mean ± SD (number of experiments) and were considered statistically significant when the P value was less than .05.
Results
Effect of Fe depletion on CD1 mRNA and protein levels
Incubation with DFO or 311 results in cellular Fe depletion,29,34–37 and causes a marked decrease in CD1 protein.10 Since CD1 is regulated at the transcriptional and posttranscriptional levels,14–17 we examined whether these processes were involved in the response to Fe depletion.
MCF-7 cells were incubated with DFO (250 μM) or 311 (25 μM) for 24 hours at 37°C. Both chelators can inhibit tumor growth in vitro and in vivo, with 311 being most effective.3,38 We used a 10-fold greater concentration of DFO than 311 because of the low permeability and decreased chelation efficacy of DFO.34,35 These chelator concentrations prevent transferrin-Fe uptake and effect cellular Fe mobilization, inducing Fe depletion.31,35,39 This leads to up-regulation of Fe-responsive genes, such as transferrin receptor-1 (TFR1).31,35,39
In this investigation, chelators and other agents were added with serum to examine the effect of Fe depletion on CD1 expression. The effect of the Fe chelators on CD1 expression were compared with actinomycin D (AD; 9 nM), which at this low concentration acts as a DNA-damaging agent, but does not inhibit transcription.31,40,41 This was done to assess the ability of Fe chelators to induce DNA damage (eg, through RR inhibition)6,42,43 compared with their effect on other Fe-requiring pathways. Neither DFO nor 311 had no significant effect on CD1 mRNA expression compared with the control (Figure 1A), while its protein level was markedly (P < .001) reduced (Figure 1B). On the other hand, AD had no effect on CD1 mRNA or protein expression compared with the relevant control. As a positive control for Fe depletion, TfR1 mRNA and protein expression were assessed. These studies demonstrated up-regulation of TfR1 mRNA (Figure 1C) and protein (Figure 1D) in chelator-treated cells compared with the control. Apart from MCF-7 cells, the effect of chelators on decreasing CD1 was observed in SK-N-MC neuroepithelioma cells and SK-Mel-2 melanoma and SK-Mel-28 melanoma cells (data not shown). Hence, the chelator-mediated effect on CD1 was not specific for 1 cell type.
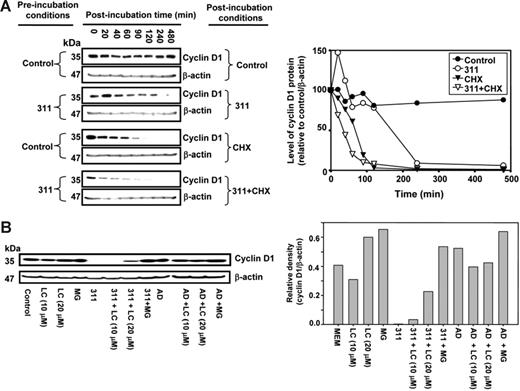
Iron chelators down-regulate cyclin D1 protein but not mRNA expression, an effect that can be reversed by Fe supplementation. Cellular Fe depletion after a 24-hour incubation with DFO (250 μM) or 311 (25 μM) either (A) has no effect on cyclin D1 mRNA; (B) decreases cyclin D1 protein levels; (C) markedly increases both TfR1 mRNA; and (D) markedly increases both TfR1 protein levels. actinomycin D (AD; 9 nM) was used as a DNA-damage control (A-D) and had no effect on cyclin D1 or TfR1 expression. (Ei) Kinetic study over 24 hours with Fe chelators showing that cyclin D1 mRNA levels were not significantly altered compared with cells incubated with control medium, while (Eii) cyclin D1 protein levels are decreased after 14 hours during incubation with chelators. MCF-7 cells were incubated with control medium or medium containing 311 (25 μM) or DFO (250 μM) for up to 24 hours at 37°C. Cells were harvested and RT-PCR and Western analysis performed. (Fi-iv) Reincubation of 311-treated cells with the Fe source FAC restores cyclin D1 protein levels. Cells were preincubated with either medium alone or medium containing 311 (25 μM) for 24 hours at 37°C and then reincubated with FAC (100 μg/mL) in the presence or absence of medium containing 311 (25 μM) for 4 to 24 hours at 37°C. Cells were harvested for Western analysis. Results are a representative experiment from 4 performed.
Iron chelators down-regulate cyclin D1 protein but not mRNA expression, an effect that can be reversed by Fe supplementation. Cellular Fe depletion after a 24-hour incubation with DFO (250 μM) or 311 (25 μM) either (A) has no effect on cyclin D1 mRNA; (B) decreases cyclin D1 protein levels; (C) markedly increases both TfR1 mRNA; and (D) markedly increases both TfR1 protein levels. actinomycin D (AD; 9 nM) was used as a DNA-damage control (A-D) and had no effect on cyclin D1 or TfR1 expression. (Ei) Kinetic study over 24 hours with Fe chelators showing that cyclin D1 mRNA levels were not significantly altered compared with cells incubated with control medium, while (Eii) cyclin D1 protein levels are decreased after 14 hours during incubation with chelators. MCF-7 cells were incubated with control medium or medium containing 311 (25 μM) or DFO (250 μM) for up to 24 hours at 37°C. Cells were harvested and RT-PCR and Western analysis performed. (Fi-iv) Reincubation of 311-treated cells with the Fe source FAC restores cyclin D1 protein levels. Cells were preincubated with either medium alone or medium containing 311 (25 μM) for 24 hours at 37°C and then reincubated with FAC (100 μg/mL) in the presence or absence of medium containing 311 (25 μM) for 4 to 24 hours at 37°C. Cells were harvested for Western analysis. Results are a representative experiment from 4 performed.
Effect of incubation time with chelators on CD1 mRNA and protein expression
To examine the effect of incubation time with Fe chelators on CD1 mRNA expression, MCF-7 cells were incubated with control medium or medium containing 311 (25 μM) for 2 to 24 hours at 37°C, and RT-PCR was performed. CD1 mRNA levels in treated cells were not affected compared with their respective controls at each time point (Figure 1Ei). CD1 protein was not significantly affected by Fe chelation compared with the relevant control up to 10 hours (Figure 1Eii). However, after 14 hours of incubation with DFO or 311, CD1 protein was decreased (P < .001) to 60% ± 4% (n = 4) of the relevant control. This decreased CD1 expression after Fe depletion became more marked at 18 and 24 hours (Figure 1Eii).
The decrease in CD1 protein mediated by chelators is Fe dependent
To assess whether decreased CD1 expression in the presence of chelators was Fe dependent, MCF-7 cells were preincubated with medium alone (control) or medium containing 311 (25 μM) for 24 hours. This medium was removed and the cells were incubated (after incubation) for 4 to 24 hours with either control medium or medium containing ferric ammonium citrate (FAC; 100 μg/mL), 311 (25 μM), or 311 (25 μM) and FAC (100 μg/mL) (Figure 1F). FAC was used as an Fe source as it donates Fe to many cell types, including MCF-7.31,44
Pre- and postincubation of cells with medium alone led to little alteration in CD1 protein (Figure 1Fi). Similarly, preincubation of cells with control medium followed by postincubation with FAC led to no change in CD1 (Figure 1Fii). Pre- and postincubation of MCF-7 cells with 311 led to ablation of CD1 (Figure 1Fiii). However, preincubation with 311 (25 μM) followed by postincubation with 311 (25 μM) in the presence of a large excess of Fe as FAC (100 μg/mL; [Fe] = 280 μM), led to CD1 recovery (Figure 1Fiv). In fact, CD1 began to increase after a postincubation with FAC of 8 hours. Hence, in chelator-treated cells, donation of Fe using FAC resulted in CD1 reconstitution. Incubation of cells with DFO or 311 as Fe complexes had no effect on CD1 (data not shown), again suggesting the importance of cellular Fe chelation.
Cell synchronization and cell-cycle regulation after Fe chelation
CD1 protein expression oscillates during the cell cycle, increasing at mid-G1, complexing to cdk-4 and cdk-6 and producing peak activation near the G1/S transition.8,45–48 In cycling cells, CD1 is expressed at high levels in the G1 and G2 phases and at lower levels in S phase.49 Here, we examined cell-cycle status and CD1 expression of MCF-7 cells before and after chelation to determine their relationship.
In control cells growing in 10% FCS (unsynchronized), the percentage of cells between G1, G2, and S phases were 36% ± 3% (n = 3), 24% ± 1% (n = 3), and 30% ± 2% (n = 3), respectively (Figure 2A). Upon DFO treatment (250 μM), accumulation of cells occurred in G1, the percentages in G1, G2, and S being 46% ± 3% (3), 16% ± 1% (n = 3), and 28% ± 3% (n = 3), respectively (Figure 2A). Similar results were found in cells treated with 311 (25 μM; Figure 2A). These data were consistent with the G1/S arrest found upon Fe depletion.34,50,51 Western blotting in parallel with flow cytometry demonstrated that CD1 was markedly (P < .001) decreased in the presence of the chelators (Figure 2A).
Cellular Fe depletion decreases cyclin D1 expression and induces a G1/S arrest. (A) Fe chelators induce a G1/S arrest and ablate cyclin D1 protein expression. MCF-7 cells were treated with 10% FCS medium alone or medium containing 10% FCS and the Fe chelators (DFO [250 μM] or 311 [25 μM]) for 24 hours at 37°C. (B) Serum starvation induces a G1/S arrest but does not affect cyclin D1 expression. In addition, G1/S arrest induced by serum starvation is reversible upon addition of serum but does not markedly affect cyclin D1 expression. Cells were washed twice with PBS and then incubated with either serum-free media or media containing 10% FCS for 24 hours at 37°C. The serum-starved cells were then released into media containing 10% FCS for 16 hours at 37°C. (C) G1/S arrest induced by serum starvation is not reversible upon addition of 10% FCS and 250 μM DFO for 16 hours. Cells were washed with PBS and then incubated with either serum-free media or media containing 10% FCS for 24 hours at 37°C. The serum-starved cells were then released into media containing 10% FCS and 250 μM DFO for 16 hours at 37°C. Cells were harvested after each step of the synchronization process for flow cytometric and Western blot analysis. The same results were found for 311. Results shown are from a representative experiment from 3 performed.
Cellular Fe depletion decreases cyclin D1 expression and induces a G1/S arrest. (A) Fe chelators induce a G1/S arrest and ablate cyclin D1 protein expression. MCF-7 cells were treated with 10% FCS medium alone or medium containing 10% FCS and the Fe chelators (DFO [250 μM] or 311 [25 μM]) for 24 hours at 37°C. (B) Serum starvation induces a G1/S arrest but does not affect cyclin D1 expression. In addition, G1/S arrest induced by serum starvation is reversible upon addition of serum but does not markedly affect cyclin D1 expression. Cells were washed twice with PBS and then incubated with either serum-free media or media containing 10% FCS for 24 hours at 37°C. The serum-starved cells were then released into media containing 10% FCS for 16 hours at 37°C. (C) G1/S arrest induced by serum starvation is not reversible upon addition of 10% FCS and 250 μM DFO for 16 hours. Cells were washed with PBS and then incubated with either serum-free media or media containing 10% FCS for 24 hours at 37°C. The serum-starved cells were then released into media containing 10% FCS and 250 μM DFO for 16 hours at 37°C. Cells were harvested after each step of the synchronization process for flow cytometric and Western blot analysis. The same results were found for 311. Results shown are from a representative experiment from 3 performed.
Serum starvation to induce synchronization and G1/S arrest17,52 resulted in a pronounced increase in the percentage of cells in G1 (65% ± 5% [n = 3]; Figure 2B), as found with chelators in the presence of serum (Figure 2A). However, in contrast to CD1 ablation after chelator treatment, CD1 levels after serum starvation remained similar to the control (Figure 2B). Synchronization of cells using serum starvation followed by incubation in media containing 10% FCS resulted in release from the G1 arrest, with cells cascading into the other phases (Figure 2B). In contrast, release of serum-starved cells into media containing 10% FCS and DFO (250 μM) did not result in any significant change in phase distribution, while CD1 decreased markedly (Figure 2C). Similar observations were found when serum-starved cells were released into 10% FCS and 311 (25 μM; data not shown). These results suggested that the decrease in CD1 which occurred by chelator-mediated Fe depletion can be independent of G1 arrest and was not simply because the chelators induced G1 arrest.
Half-life of CD1 protein is decreased after Fe chelator treatment
To investigate the decrease in CD1 protein after Fe depletion, we examined CD1 degradation using the protein synthesis inhibitor cycloheximide (CHX)53 (Figure 3Ai-iv). MCF-7 cells were preincubated with media alone or media containing 311 for 12 hours followed by postincubation (0-480 minutes) with CHX (10 μg/mL)37,53,54 in the presence or absence of 311. With CHX alone, CD1 half-life was 80 ± 15 minutes (n = 5) (Figure 3Aiii). However, in the presence of 311 and CHX, the half-life of CD1 was decreased (P < .008) to 38 ± 3 minutes (n = 5) (Figure 3Aiv).
Incubation with 311 decreases cyclin D1 protein half-life and this is mediated by proteasomal activity. (A) 311 decreases cyclin D1 half-life. MCF-7 cells were preincubated with either medium or medium containing 311 (25 μM) for 12 hours at 37°C and then postincubated for 20 to 480 minutes at 37°C with either media alone (control), 311 (25 μM), the protein synthesis inhibitor CHX (10 μg/mL), or CHX (10 μg/mL) and 311 (25 μM). Cells were harvested, and Western blot analysis was performed. (B) The proteasomal inhibitors LC and MG prevented the chelator-mediated decrease in cyclin D1 protein levels, while the DNA-damaging agent Act D (AD) had no effect. MCF-7 cells were incubated for 24 hours at 37°C with either LC, MG, 311, 311 and LC, 311 and MG, AD, AD and LC, AD and LC, or AD and MG. Cells were harvested, and Western blot analysis was performed. Results in panels A and B are representative of 5 and 3 experiments, respectively.
Incubation with 311 decreases cyclin D1 protein half-life and this is mediated by proteasomal activity. (A) 311 decreases cyclin D1 half-life. MCF-7 cells were preincubated with either medium or medium containing 311 (25 μM) for 12 hours at 37°C and then postincubated for 20 to 480 minutes at 37°C with either media alone (control), 311 (25 μM), the protein synthesis inhibitor CHX (10 μg/mL), or CHX (10 μg/mL) and 311 (25 μM). Cells were harvested, and Western blot analysis was performed. (B) The proteasomal inhibitors LC and MG prevented the chelator-mediated decrease in cyclin D1 protein levels, while the DNA-damaging agent Act D (AD) had no effect. MCF-7 cells were incubated for 24 hours at 37°C with either LC, MG, 311, 311 and LC, 311 and MG, AD, AD and LC, AD and LC, or AD and MG. Cells were harvested, and Western blot analysis was performed. Results in panels A and B are representative of 5 and 3 experiments, respectively.
Role of the proteasome in the Fe depletion–mediated decrease in CD1
The lack of alteration of CD1 mRNA levels (Figure 1Ei) and decrease in CD1 protein half-life (Figure 3A) after Fe chelation indicated posttranscriptional regulation was responsible for decreased CD1. Further, it is known that CD1 is regulated by proteasomal degradation.16 To examine the role of proteasomal degradation of CD1 after Fe chelation, we assessed the effect of a 24-hour incubation with the proteasomal inhibitors MG-132 (MG) and lactacystin (LC)17,55 (Figure 3B).
Incubation of control MCF-7 cells with LC (10 μM) had no significant effect on CD1, while LC or MG at 20 μM slightly increased CD1 (Figure 3B). Incubation with 311 (25 μM) ablated CD1 protein (Figure 3B). The addition of LC (20 μM) with 311 (25 μM) increased CD1 by 95-fold compared with 311 alone. Incubation of cells with 311 and MG (20 μM) rescued CD1 (Figure 3B). The ability of the proteasome inhibitors to rescue CD1 in the presence of chelator suggested Fe depletion induces proteasome-mediated CD1 degradation.
The ability of proteasomal inhibitors to prevent the chelator-mediated decrease in CD1 potentially suggests these inhibitors were rescuing cells from G1/S arrest and thereby were reducing the ability of chelators to inhibit proliferation. However, flow cytometry demonstrated that the G1/S arrest was still observed with MG and the chelator, despite CD1 reconstitution (data not shown). In fact, after a 24-hour incubation with MG (20 μM) and 311 (25 μM), the proportion of cells in G1 was 45% ± 4% (n = 3). This was not significantly different from the proportion in G1 in the presence of 311 alone (45% ± 1% [n = 3]). Moreover, MG did not prevent the inhibition of proliferation observed with 311, despite its ability to rescue CD1 (data not shown). This is probably because proteasomal inhibitors prevent proliferation themselves and are being examined as potent antitumor agents.56,57
Chelator-induced CD1 proteolysis is independent of GSK-3β
Under some conditions, GSK-3β may be involved in proteasomal degradation of CD1.14,16,17 This enzyme phosphorylates CD1 at Thr-286, which induces ubiquitin-dependent proteasomal degradation.14,16 To assess the role of GSK-3β in proteasomal-mediated degradation of CD1 after Fe chelation, we investigated the effect of lithium chloride (LiCl) which inhibits this enzyme17,58 (data not shown). We used β-catenin as a positive control, as it is a downstream target of GSK-3β and is degraded via its action.17,58 Moreover, LiCl prevents β-catenin degradation by this mechanism.17 Iron-replete cells incubated with control medium and increasing LiCl (1-30 mM) resulted in no significant change in CD1 or β-catenin expression (data not shown). Hence, these results suggested GSK-3β was not involved in proteasomal degradation of these proteins when cells were Fe replete. The effect of LiCl was then examined in cells incubated with 311 (25 μM). Incubation with this chelator ablated CD1 protein expression, and this was not rescued by LiCl. In contrast, in the presence of 311 (25 μM), LiCl significantly (P < .01) increased β-catenin expression, but only at 30 mM, indicating inhibition of GSK-3β activity. These studies suggested decreased CD1 expression after Fe chelation was independent of GSK-3β.
CD1 protein degradation after Fe depletion is proteasome dependent
To further elucidate the mechanism of CD1 protein degradation and to rule out the effect of protein synthesis on the Fe chelator–mediated decrease in CD1, the protein synthesis inhibitor CHX (10 μg/mL) was used (Figure 4). Incubation of MCF-7 cells with control medium for up to 24 hour had no effect on CD1 expression, while 311 (25 μM) markedly decreased CD1 only after 10 hours (Figure 4A). In contrast, CHX alone or CHX and 311 inhibited CD1 after 2 hours (Figure 4B). Incubation of cells with medium containing MG (20 μM) alone did not change CD1, while addition of MG and 311 rescued CD1 (Figure 4C) when compared to cells treated with 311 alone (Figure 4A). When control cells were incubated with MG and CHX to inhibit proteasomal degradation and protein synthesis, there was only a slight decrease in protein with time, which was not significant over 5 experiments (Figure 4D). This observation demonstrated that the CHX and MG concentrations appropriately inhibited protein synthesis and proteasomal degradation, as the protein level was constant up to 24 hours (Figure 4D). In contrast, the addition of MG and CHX in the presence of 311 resulted in a decrease (P < .001) of CD1 after 2 hours (Figure 4D). Hence, when protein synthesis and proteasomal degradation were blocked in 311-treated cells, CD1 continued to decrease. This suggested another degradation pathway apart from that mediated by the proteasome in the chelator-mediated decrease in CD1 expression. The identity of the nonproteasomal mechanism remains unknown and was not pursued considering the significance of the proteasome in CD1 degradation (Figure 3B).
Inhibition of both protein synthesis and proteasomal degradation results in the identification of another mechanism of Fe depletion–mediated cyclin D1 degradation. (A-D) Effect of 311 on cyclin D1 expression in MCF-7 cells in the presence of the proteasomal inhibitor MG, and the protein synthesis inhibitor CHX. MCF-7 cells were incubated for 0.5 hours to 24 hours at 37°C with either control medium or this medium containing 311 (25 μM) in the presence of either or both MG (20 μM) and CHX (10 μg/mL). Cells were harvested, and Western blot analysis was performed. Results shown are from a representative experiment of 3 performed.
Inhibition of both protein synthesis and proteasomal degradation results in the identification of another mechanism of Fe depletion–mediated cyclin D1 degradation. (A-D) Effect of 311 on cyclin D1 expression in MCF-7 cells in the presence of the proteasomal inhibitor MG, and the protein synthesis inhibitor CHX. MCF-7 cells were incubated for 0.5 hours to 24 hours at 37°C with either control medium or this medium containing 311 (25 μM) in the presence of either or both MG (20 μM) and CHX (10 μg/mL). Cells were harvested, and Western blot analysis was performed. Results shown are from a representative experiment of 3 performed.
CD1 degradation is independent of pVHL expression at low cellular density
Considering proteasomal degradation of CD1 after Fe depletion, it was relevant to examine the role of pVHL.59,60 When cells are Fe replete and under normal oxygen tension, hydroxylation of Pro564 of hypoxia-inducible factor-1α (HIF-1α) enables pVHL binding. pVHL activates the ubiquitin E3 ligase that ubiquitinates HIF-1α for proteasomal degradation.59,60 In confluent cells, CD1 is also a target of pVHL, which serves as a substrate recognition component for polyubiquitination and proteasomal degradation.24 This may relate to the tumor suppressor function of pVHL and its role in the contact inhibition of cellular growth.24 Hence, it was important to examine if pVHL could regulate CD1 in response to Fe levels.
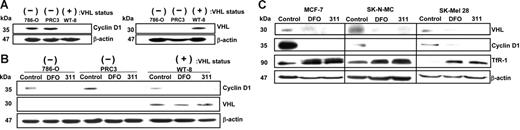
To assess the role of pVHL in CD1 expression, we examined CD1 protein in parent RCCs (786-O) that do not express VHL and compared the results with 786-O cells transfected with pVHL (WT-8) or vector alone (PRC3 cells).24 Using confluent cultures, CD1 was down-regulated in pVHL+ WT-8 cells compared with pVHL− cells (ie, 786-O and PRC3; Figure 5A). These results confirm CD1 is regulated by pVHL at high cellular density.24
Cyclin D1 is down-regulated in confluent RCCs expressing pVHL, and both cyclin D1 and pVHL are down-regulated after Fe chelation. (A) Cyclin D1 is down-regulated in confluent cultures of human RCCs expressing pVHL. Western blot analysis of cyclin D1 and pVHL expression using confluent cultures of human parental RCCs (786-O); these cells either transfected with the VHL hyperexpression vector (WT-8) or this vector without the VHL insert (PRC3). (B) DFO and 311 down-regulate cyclin D1 protein regardless of pVHL status. The 786-O, WT-8, and PRC3 cells under partially confluent culture conditions were incubated with media alone (control) or media containing DFO (250 μM) or 311 (25 μM) for 24 hours. Western blot analysis was performed to detect cyclin D1 and pVHL expression. (C) DFO and 311 down-regulate pVHL and cyclin D1 but up-regulate TfR1 in a variety of cell types. MCF-7 breast cancer cells, SK-N-MC neuroepithelioma cells, and SK-Mel-28 melanoma cells were incubated with either media alone or media containing 311 (25 μM) or DFO (250 μM) over 24 hours, and Western analysis was performed. Results shown are a typical experiment from 3 performed.
Cyclin D1 is down-regulated in confluent RCCs expressing pVHL, and both cyclin D1 and pVHL are down-regulated after Fe chelation. (A) Cyclin D1 is down-regulated in confluent cultures of human RCCs expressing pVHL. Western blot analysis of cyclin D1 and pVHL expression using confluent cultures of human parental RCCs (786-O); these cells either transfected with the VHL hyperexpression vector (WT-8) or this vector without the VHL insert (PRC3). (B) DFO and 311 down-regulate cyclin D1 protein regardless of pVHL status. The 786-O, WT-8, and PRC3 cells under partially confluent culture conditions were incubated with media alone (control) or media containing DFO (250 μM) or 311 (25 μM) for 24 hours. Western blot analysis was performed to detect cyclin D1 and pVHL expression. (C) DFO and 311 down-regulate pVHL and cyclin D1 but up-regulate TfR1 in a variety of cell types. MCF-7 breast cancer cells, SK-N-MC neuroepithelioma cells, and SK-Mel-28 melanoma cells were incubated with either media alone or media containing 311 (25 μM) or DFO (250 μM) over 24 hours, and Western analysis was performed. Results shown are a typical experiment from 3 performed.
Studies were initiated to assess the effect of chelators on CD1 in the presence and absence of pVHL under partial confluence (Figure 5B), as these conditions were used to examine CD1 expression in this investigation. In the absence of chelators, CD1 expression in pVHL+ WT-8 cells was similar to that in pVHL− cell types (786-O and PRC3; Figure 5B). Hence, under semiconfluent conditions, these results were different to when cells were confluent (Figure 5A). Regardless of pVHL status, in all cell types, DFO and 311 markedly down-regulated CD1 (Figure 5B). Hence, since most experiments in this study were under semiconfluent conditions, our results suggest pVHL was not involved in CD1 degradation after Fe chelation.
Interestingly, both pVHL and CD1 expression in MCF-7 cells were observed to be down-regulated after Fe depletion compared with the control (Figure 5C). Similar results were also found for SK-N-MC and SK-Mel-28 cells. As a positive control, TfR1 protein was examined and was up-regulated by chelators in all cell types (Figure 5C). These results suggest that pVHL is regulated by Fe status. Notably, Fe chelation did not regulate pVHL in the pVHL+ WT-8 cells (Figure 5B) because its expression is mediated by a transfected vector. The effect of Fe depletion on pVHL expression has not been previously reported, but could facilitate the increase in HIF-1α after Fe depletion.59,60 Since pVHL expression is down-regulated by Fe chelation, it would probably not mediate the decrease in CD1 via its function as a substrate recognition component for proteasomal degradation.
General polyubiquitination of proteins is decreased in 311-treated cells, while proteasomal activity remains constant
To investigate how Fe depletion increases proteasome-mediated CD1 degradation, we examined general protein polyubiquitination [(Ub)n] using antiubiquitin and Western analysis (Figure 6A). This was performed after a 24-hour incubation of cells with 311 (25 μM) or 311 (25 μM) in the presence of MG (20 μM) or LC (10 or 20 μM; Figure 6A). MG and LC were positive controls and markedly (P < .001) increased polyubiquitination, demonstrating inhibited proteasomal activity (Figure 6A; lanes 3, 5, and 7). In contrast, when cells were incubated with 311 alone (Figure 6A; lane 2), it only slightly reduced polyubiquitination compared with the control (Figure 6A; lane 1). When 311 was combined with MG or LC and incubated with cells, there was a marked (P < .001) decrease in polyubiquitination when compared with LC or MG alone (Figure 6A; lanes 3 [MG] and 4 [MG + 311]).
Iron chelators decrease general (Ub)n and cyclin D1 polyubiquination when proteasome activity is inhibited. (A) General protein polyubiquitination is decreased when cells are incubated with 311 in the presence of the proteasomal inhibitors MG or LC. MCF-7 cells were incubated with media alone or medium containing 311 (25 μM) in the presence and absence of MG (20 μM) or LC (10 and 20 μM) for 24 hours at 37°C. Total protein was extracted from cells and Western analysis was performed. (B) 311 decreases cyclin D1 protein expression and also decreases polyubiquitination of cyclin D1 in the presence of MG. MCF-7 cells were incubated with either media alone or media containing 311 (25 μM) in the presence and absence of MG (20 μM) for 8, 18, and 24 hours. Immunoprecipitation was performed using anti–cyclin D1 antibody. Complexes were then separated by SDS-PAGE and probed with anti–cyclin D1 or antiubiquitin antibody using Western analysis. Results shown are typical experiments from 3 performed. The horizontal broken line in the densitometric analysis represents a value of 100%.
Iron chelators decrease general (Ub)n and cyclin D1 polyubiquination when proteasome activity is inhibited. (A) General protein polyubiquitination is decreased when cells are incubated with 311 in the presence of the proteasomal inhibitors MG or LC. MCF-7 cells were incubated with media alone or medium containing 311 (25 μM) in the presence and absence of MG (20 μM) or LC (10 and 20 μM) for 24 hours at 37°C. Total protein was extracted from cells and Western analysis was performed. (B) 311 decreases cyclin D1 protein expression and also decreases polyubiquitination of cyclin D1 in the presence of MG. MCF-7 cells were incubated with either media alone or media containing 311 (25 μM) in the presence and absence of MG (20 μM) for 8, 18, and 24 hours. Immunoprecipitation was performed using anti–cyclin D1 antibody. Complexes were then separated by SDS-PAGE and probed with anti–cyclin D1 or antiubiquitin antibody using Western analysis. Results shown are typical experiments from 3 performed. The horizontal broken line in the densitometric analysis represents a value of 100%.
These data may suggest chelators increase proteasomal activity, which decreases ubiquitinated protein. Considering this, direct measurement of 26S proteasomal activity was assessed in cells incubated with DFO (250 μM) or 311 (25 μM) for 24 hours or when isolated 20S proteasomes were incubated with DFO (250 μM) or 311 (25 μM) for 4 hours. These studies demonstrated no effect of chelators on proteasomal activity. In contrast, the positive control MG significantly (P < .001) reduced proteasomal activity to 35% to 40% when either cells or purified proteasomes were incubated with this agent. Further, addition of DFO (250 μM) or 311 (25 μM) to MG had no effect on proteasomal activity when compared with MG alone (data not shown). These results suggest that proteasomal degradation of CD1 after Fe chelation was not due to a direct increase in proteasomal activity.
Polyubiquitination of CD1 is decreased in 311-treated cells when proteasomal activity is inhibited
To determine the effect of chelation on CD1 polyubiquitination, CD1 was immunoprecipitated with anti-CD1 and Western analysis was performed with this antibody or antiubiquitin (Figure 6B). Cells were incubated for 8, 18, or 24 hours with medium alone or medium containing 311 (25 μM) in the presence or absence of MG (20 μM). These time points were chosen since there was little effect of chelators on CD1 protein at 8 hours, while a pronounced decrease occurred at 18 and 24 hours (Figure 1Eii).
Immunoprecipitation and Western blotting with anti-CD1 demonstrated that compared with the control after 8 hours there was no effect on CD1 of incubating cells with 311 (Figure 6B). However, Fe chelation caused near ablation of CD1 (P < .001) compared with the relative controls at 18 and 24 hours (Figure 6B). Incubation with 311 alone did not have any marked effect on CD1 ubiquitination compared with the control at each time point, despite near ablation of CD1 at 18 and 24 hours (Figure 6B). Incubation with MG or MG and 311 increased CD1 relative to the respective control at all time points.
Immunoprecipitation with anti-CD1 and Western blotting with antiubiquitin demonstrated that compared with the control after 8 hours, there was a slight increase in CD1 polyubiquitination in the presence of MG or MG and 311 (Figure 6B). CD1 polyubiquitination resulted in high molecular weight species that were above the immunoglobulin G (IgG) heavy chain (at 50 kDa; not shown), in agreement with previous work.61 After 18 or 24 hours, MG markedly (P < .001) increased CD1 ubiquitination compared with the controls (Figure 6B) due to its ability to inhibit CD1 degradation.61 Hence, in the absence of chelators, CD1 is ubiquitinated prior to proteasomal degradation.
In the presence of 311 and MG, there was a marked (P < .001) decrease in CD1 ubiquitination compared with MG alone at 18 and 24 hours (Figure 6B). The decrease in CD1 ubiquitination in the presence of chelator is in agreement with the general decrease in protein polyubiquitination in Figure 6A. Hence, decreased CD1 ubiquitination mediated by Fe depletion would not favor proteasomal degradation of the protein via this pathway. Further studies then examined the role of ubiquitin-independent proteasomal degradation of CD1 after Fe depletion.
Chelators induce ubiquitin-independent CD1 degradation
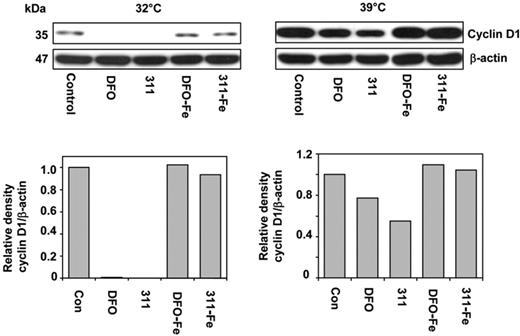
To determine the role of ubiquitin-independent CD1 degradation after Fe chelation, we used the well-characterized A31N-ts20 cell line that has a temperature-sensitive E1 ubiquitin–activating enzyme.27,30 This molecule is inactivated at 39°C, preventing ubiquitin-dependent proteasomal degradation, but it is active at 32°C.27,30 Cells were grown for 24 hours at 32°C or 39°C prior to the experiment to ensure maximum activation or inactivation of E1, respectively.27 This media was then removed and the cells incubated for 24 hours at either temperature with control media, DFO (250 μM), 311 (25 μM), or their Fe(III) complexes at these concentrations.
Examining the effect of temperature on CD1 expression in control cells showed that a shift from 32°C to 39°C resulted in a 10-fold increase in CD1 (Figure 7). This demonstrated that at 39°C CD1 ubiquitination was decreased, leading to reduced degradation and CD1 accumulation. It also indicates that upon incubation with control medium, proteasomal degradation of CD1 occurs via a ubiquitin-dependent process, as suggested by our MG studies using MCF-7 cells (Figure 6B).
Iron chelators induce ubiquitin-independent degradation of CD1. A31N-ts20 cells were used because they have a temperature-sensitive E1 ubiquitin–activating enzyme which is active at 32°C, but is inactivated at 39°C, preventing ubiquitin-dependent proteasomal degradation. Cells were grown at 32°C or 39°C for 24 hours prior to the experiment. The medium was then removed and the cells were incubated for 24 hours at either temperature with control media or media containing DFO (250 μM), 311 (25 μM), or their Fe(III) complexes, namely DFO-Fe (250 μM) or 311-Fe (25 μM); Western analysis was then performed. Results shown are from a representative experiment from 3 performed.
Iron chelators induce ubiquitin-independent degradation of CD1. A31N-ts20 cells were used because they have a temperature-sensitive E1 ubiquitin–activating enzyme which is active at 32°C, but is inactivated at 39°C, preventing ubiquitin-dependent proteasomal degradation. Cells were grown at 32°C or 39°C for 24 hours prior to the experiment. The medium was then removed and the cells were incubated for 24 hours at either temperature with control media or media containing DFO (250 μM), 311 (25 μM), or their Fe(III) complexes, namely DFO-Fe (250 μM) or 311-Fe (25 μM); Western analysis was then performed. Results shown are from a representative experiment from 3 performed.
Incubation of A31N-ts20 cells at 39°C (which inhibits ubiquitin-dependent proteasomal degradation) in the presence of DFO or 311 reduced CD1 expression compared with the control, 311 being more effective due to its greater permeability35 (Figure 7). These observations were consistent with the hypothesis that chelators induce a ubiquitin-independent process to degrade CD1 by the proteasome. However, the decrease in CD1 after incubation with chelators at 39°C was less marked than at 32°C. This was probably because of the large CD1 accumulation at 39°C that occurs during the 24-hour preincubation prior to the addition of chelator. The Fe complexes of 311 and DFO had no effect on CD1, suggesting Fe chelation was responsible for the decrease observed (Figure 7).
Discussion
CD1 plays a rate-limiting role in G1/S progression and is a known oncogene and target for cancer treatment.12,62–64 We investigated the mechanism involved in the Fe chelation–mediated decrease in CD1 protein and showed it was predominantly due to proteasomal degradation. These observations are of interest, since this is the first report to link Fe depletion–mediated growth suppression at G1/S to CD1 proteolysis.
Our investigation demonstrated that Fe depletion decreases CD1 protein expression, but had no effect on its mRNA level (Figure 1E). Potentially, this could suggest regulation by iron regulatory protein (IRP)–1 and IRP-2 by a mechanism similar to that found for ferritin.3 However, analysis of the 5′ untranslated region of CD1 mRNA demonstrated that there was no iron-responsive element (IRE) that could mediate regulation of expression via the IRPs. Further studies showed that CD1 protein half-life was significantly reduced after incubation with chelators, indicating increased CD1 degradation. Proteasomal inhibitors prevented decreased CD1 during incubation with chelators, suggesting a role of the proteasome. The effect of Fe depletion in decreasing CD1 protein by the proteasome was reminiscent of the effects of all-trans retinoic acid20 and Coxsackievirus17 on reducing CD1 expression and inducing G1 arrest. Thus, diverse signals that induce G1/S arrest may use proteasome-mediated CD1 degradation as a common regulator.
Considering the rate-limiting role that CD1 plays in G1/S progression,8 its regulation by cellular Fe levels appears to be important for preventing entrance into the S phase, where Fe is essential for ribonucleotide reductase activity, and thus, DNA synthesis.1–3,50 It is known that chelators perturb multiple molecular targets to inhibit cellular growth,9,10,65 and the antiproliferative response observed is due to this combination effect. At present, the relative contributions of the inhibition of ribonucleotide reductase6,42 and CD1 degradation to the observed growth arrest after Fe chelation are not known.
In the ubiquitin-proteasome degradation pathway, proteins are conjugated to ubiquitin in a reaction involving ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3).66 The polyubiquitinated substrate is then degraded.66,67 Interestingly, pVHL serves as a substrate recognition component of an E3 ubiquitin ligase, targeting CD1 for proteasomal degradation.22–24 Therefore, increased CD1 degradation after Fe depletion could be mediated by mechanisms affecting proteasomal activity, and these are discussed below.
First, phosphorylation of Thr286 on CD1 by GSK-3β is required for ubiquitination and proteasomal degradation.14,16 We showed that CD1 ubiquitination was important for its degradation in Fe-replete cells, given the marked increase in CD1 and its ubiquitinated products when proteasomal activity was inhibited (Figure 6B). Incubation of cells with 311 and the GSK-3β inhibitor LiCl (30 mM) resulted in an accumulation of a substrate of this enzyme, namely β-catenin, but it did not block CD1 decrease upon Fe depletion. This indicates GSK-3β is not involved in CD1 regulation during Fe depletion. Luo et al17 reported similar findings using a viral model of G1/S arrest where the decrease of CD1 in host cells was GSK-3β independent.
A second mechanism of increased CD1 degradation after chelation could be mediated by enhanced proteasomal activity. However, there was no alteration in proteasomal activity in the presence of chelator. A third mechanism of CD1 degradation could involve alterations in ubiquitin conjugation. Since pVHL targets CD1 for proteasomal degradation via activation of E3 ubiquitin ligase, it may be involved. However, in semiconfluent RCCs, the chelator-mediated decrease in CD1 was independent of pVHL expression. Moreover, Fe depletion decreased both pVHL and CD1 (Figure 5C), suggesting pVHL does not mediate decreased CD1 expression. Immunoprecipitation and Western blotting showed that Fe depletion inhibited CD1 ubiquitination (Figure 6B). This would prevent proteasomal degradation and could not explain decreased CD1 during Fe depletion. Hence, ubiquitin-independent proteasomal degradation21,27,68 of CD1 after Fe chelation may be involved.
To examine the role of a ubiquitin-independent process in CD1 degradation, we used A31N-ts20 cells that have a temperature-sensitive E1 ubiquitin–activating enzyme.27,30 This protein is inactive at 39°C, preventing ubiquitin-dependent proteasomal degradation, but is active at 32°C.27,30 We demonstrated that Fe chelators decreased CD1 in A31-ts20 cells at 39°C, suggesting an ubiquitin-independent mechanism was induced by Fe depletion. This was not found for the Fe complexes, indicating Fe chelation mediates CD1 degradation.
The nature of the ubiquitin-independent pathway of CD1 degradation identified in this investigation remains unknown, but it may be mediated by NQO127,28 or antizyme.21 NQO1 binds and stabilizes proteins, while NQO1 inhibitors such as curcumin disrupt this association and stimulate proteasomal degradation.27 Interestingly, curcumin decreases CD1 expression69 and acts as a Fe chelator.70 These observations suggest a link between Fe-depletion, NQO1, and CD1 degradation. However, further studies are clearly required to investigate this hypothesis.
The decreased CD1 expression after incubation with chelators could be suggested to be unrelated to Fe depletion and simply be the consequence of G1 arrest followed by rapid CD1 degradation. However, a number of points of evidence from this study indicated that decreased CD1 expression was directly due to cellular Fe chelation. First, Fe supplementation with a Fe salt rescued CD1 degradation (Figure 1F), and the Fe complexes of DFO and 311 had no effect on CD1 expression (Figure 7). Second, Fe chelation induced a G1/S arrest and totally abrogated CD1 expression, while serum starvation resulted in a pronounced G1/S arrest, but did not markedly reduce CD1 levels (Figure 2). This indicated that decreased CD1 expression and cell-cycle arrest can be independent under some conditions. Third, the decrease in CD1 expression via the proteasome in Fe-replete cells was mediated by a ubiquitin-dependent process, while in contrast, Fe depletion induced ubiquitin-independent degradation of CD1 (Figures 6B, 7).
In conclusion, the Fe chelator–mediated down-regulation of CD1 occurs via a ubiquitin-independent pathway through the proteasome. In contrast, in Fe-replete cells, ubiquitin-dependent degradation was involved. These observations are relevant, as increased CD1 could be important for proliferation and tumorigenesis.12,62,71 Iron depletion using chelators can inhibit tumor growth,1–3,32 and targeting CD1 degradation may represent a new chemotherapeutic approach.
Authorship
Author contributions: D.R. designed the study, obtained grant funding, and wrote the manuscript. E.N.-T. designed studies, wrote the paper, and performed experiments. D.F., and J.P. designed studies, wrote the paper, prepared figures, and performed experiments.
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: D. R. Richardson, Department of Pathology, Blackburn Bldg D06, Western Avenue, University of Sydney, New South Wales, 2006 Australia; e-mail: d.richardson@med.usyd.edu.au.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Dr David Lovejoy, Dr Robert Sutak, Dr Jon Howard, Ms Megan Whitnall, and Ms Danuta Kalinowski (Iron Metabolism and Chelation Program, Department of Pathology, University of Sydney) are thanked for their kind help in reviewing the paper prior to submission. Special acknowledgment is given to Mr Yohan Suryo Rahmanto for computational assistance, data analyses, and critical assessment of the manuscript.
Supported by a fellowship and grants from the National Health and Medical Research Council of Australia and Australian Research Council to D.R.R.


![Figure 2. Cellular Fe depletion decreases cyclin D1 expression and induces a G1/S arrest. (A) Fe chelators induce a G1/S arrest and ablate cyclin D1 protein expression. MCF-7 cells were treated with 10% FCS medium alone or medium containing 10% FCS and the Fe chelators (DFO [250 μM] or 311 [25 μM]) for 24 hours at 37°C. (B) Serum starvation induces a G1/S arrest but does not affect cyclin D1 expression. In addition, G1/S arrest induced by serum starvation is reversible upon addition of serum but does not markedly affect cyclin D1 expression. Cells were washed twice with PBS and then incubated with either serum-free media or media containing 10% FCS for 24 hours at 37°C. The serum-starved cells were then released into media containing 10% FCS for 16 hours at 37°C. (C) G1/S arrest induced by serum starvation is not reversible upon addition of 10% FCS and 250 μM DFO for 16 hours. Cells were washed with PBS and then incubated with either serum-free media or media containing 10% FCS for 24 hours at 37°C. The serum-starved cells were then released into media containing 10% FCS and 250 μM DFO for 16 hours at 37°C. Cells were harvested after each step of the synchronization process for flow cytometric and Western blot analysis. The same results were found for 311. Results shown are from a representative experiment from 3 performed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/9/10.1182_blood-2006-10-047753/4/m_zh80090700150002.jpeg?Expires=1764958619&Signature=huauAQ99mmnpYx1FrpVmvDhvZ95YsjvH0sT1Lzx~vcYaIBYvc9iCsuz5vm9NwKLA48d8MJ0zWACL19rdON3TJHPwRVmJCFOJ1wshe8UlKjzL5l0w~ncIQ3cV9sq1dvSy4OClY8we4nLtxYVXqXZxiUiSzSYU71pE1pO03h3Pb2NAisb0WaMPyylXr4KH6OedHcIHFwCoB7PccMjKIhR5-kWzrnH2cXyFo3monFjIPCx~XSR~fu1AMID6WEU~BaMJ4Vh2hc5kn-E-Vcx0NGwmyYXb6MIPbtL6N7EcMVzANsrUAYQ4XCQw7P4vFStQMHs36KVktJF7erG1iOuoyeYDYA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal