Abstract
Aberrant DNA methylation is the most frequent molecular alteration in acute myeloid leukemia (AML). To identify methylation-silenced genes in AML, we performed microarray analyses in U937 cells exposed to the demethylating agent 5-aza-deoxy-cytidine. Overall, 274 transcripts were significantly induced. Interestingly, C/EBPδ expression was significantly induced (more than 10-fold) by demethylation whereas expression of all other C/EBP family members remained unchanged. The C/EBPδ promoter was strongly methylated in different leukemic cell lines and showed signs of a repressed chromatin state. Analyses of the promoter regions of the entire C/EBP family (α, β, γ, δ, ϵ, ζ) in bone marrow samples from AML patients (n = 80) and controls (n = 15) by mass spectrometry revealed that C/EBPδ is the most commonly hypermethylated C/EBP gene in AML. Hypermethylation occurred in more than 35% of AML patients at primary diagnosis. A significant correlation (P = .016) was observed between hypermethylation of the C/EBPδ promoter and low expression of C/EBPδ in AML patients. C/EBPδ promoter activity was strongly repressed by methylation in vitro, and transcriptional repression partially depended on MeCP2 activity. C/EBPδ exhibited growth-inhibitory properties in primary progenitor cells as well as in Flt3-ITD–transformed cells. Taken together, C/EBPδ is a novel tumor suppressor gene in AML that is silenced by promoter methylation.
Introduction
The CCAAT/enhancer binding proteins (C/EBPs) constitute a family of transcription factors involved in tissue differentiation, metabolism, and immune responses.1,2 C/EBP proteins play important roles in growth regulation of hematopoietic as well as nonhematopoietic tissues
In the hematopoietic system, all members of the C/EBP family (α, β, δ, γ, ϵ, ζ) are expressed during myeloid development.3–5 C/EBPα, C/EBPβ, and C/EBPδ are predominantly expressed in granulocytes, monocytes, and eosinophils while C/EBPϵ is found at later stages of differentiation of granulocytes and T cells.6–8 Overexpression of either C/EBPα, C/EBPβ, C/EBPδ, or C/EBPϵ induces granulocytic differentiation in myeloblastic cell lines.9–13 Neutrophilic differentiation of BCR-ABL–positive CML cell lines was induced by ectopic expression of C/EBPδ.14
Outside of the hematopoietic system, C/EBPα, C/EBPβ, and C/EBPδ are key proteins mediating the differentiation of preadipocytes to adipocytes and playing an important role in normal functions of the liver.15 C/EBPβ regulates the growth of ovarian granulosa cells, mammary epithelial cells, and keratinocytes.2 C/EBPδ appears to play a role in the growth and differentiation of lung epithelial cells16–18 and mammary epithelial cells19–21 as shown by overexpression studies. A growth inhibitory role in prostate cancer has been demonstrated.22 Huang et al recently showed that loss of C/EBPδ leads to chromosomal instability, suggesting an involvement of C/EBPδ in tumor suppression.23 Nonetheless, no mutations in C/EBPδ have been reported, raising doubts as to its potential role as a tumor suppressor.24
In the pathogenesis of leukemias and other cancers, gene silencing by aberrant DNA methylation is a frequent event.25,26 Promoter hypermethylation of bona fide tumor suppressor genes has been identified in different types of cancers including leukemias.26–28 In hematopoietic malignancies, hypermethylation of genes including E-cadherin, DAP kinase, estrogen receptor (ER) alpha, and the cell cycle regulators p15INK4B and p16INK4A are associated with gene inactivation.29–32
Here, we used a genomewide screening to identify methylation-silenced genes. Our findings demonstrate that C/EBPδ is frequently silenced by methylation in acute myeloid leukemia (AML). Its unique methylation pattern among all members of the C/EBP family suggests a role in AML pathogenesis. Strong growth inhibitory effects in primary bone marrow cells and Flt3–internal tandem duplication (Flt3-ITD)–transformed cells provide evidence that C/EBPδ acts as a growth suppressor.
Materials and methods
Cell culture
The U937 cell line was cultured in RPMI media containing 10% heat-inactivated fetal calf serum (FCS) and the 32Dcl3 cells in RPMI medium supplemented with 10% WEHI as a source of interleukin-3 and 10% FCS. Media for all cells were supplemented with 100 U/mL penicillin, 100 mg/mL streptomycin, and 20 mM glutamine. Cells were cultured in 5% CO2 at 37°C in a humified atmosphere. For aza-deoxy-cytidine (Aza) treatment, U937 cells were seeded at a density of 5 × 105/mL with 5-aza-deoxy-cytidine (Sigma, St Louis, MO) at 100 nM concentration. Cells were harvested after 2, 4, and 6 days of treatment for RNA preparation.
Microarray analysis
Total RNA was extracted from U937 cells using RNAeasy kit (Qiagen, Hilden, Germany). Ten micrograms of total RNA were reverse transcribed into cDNA with an oligo(dT)–T7 polymerase primer. Subsequently, T7 polymerase was used for in vitro transcription. During transcription, cRNA was labeled with biotinylated oligonucleotides. The resulting labeled cRNAs were fragmented and hybridized to human U133A oligonucleotide microarrays containing probes for approximately 22 000 independent transcripts (Affymetrix, Santa Clara, CA) as described previously.33 The microarray hybridizations were performed according to the MAIME (minimum information about a microarray experiment) standards.34 Three independent arrays were hybridized for each sample, and analyses were performed comparing results from 3 untreated control versus 3 Aza-treated arrays. Differentially expressed genes were identified using the significance analysis of microarrays (SAM) analysis with a false discovery rate set at 20%. Genes that scored positive according to the SAM algorithm and were induced at least 2-fold were considered to be significant.
Quantitative real-time RT-PCR
Real-time reverse transcriptase–polymerase chain reactions (RT-PCRs) were performed as described.35 Primer sequences used for expression analyses are provided in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). For C/EBPδ expression analysis, specific primers were used in combination with a probe labeled with 5′-FAM and 3′-TAMRA in a real-time PCR master mix (Eurogentec, San Diego, CA). Relative gene expression levels were calculated using standard curves generated by serial dilutions of U937 cDNA (before and after Aza treatment) for human cell lines samples. The expression level of GAPDH was used as an internal standard. Statistical analyses were done with SPSS 12.0 (Chicago, IL), and the nonparametric Mann-Whitney U test was used to compare the expression levels of C/EBPδ between AML and controls.
Western blot analyses
Cell lysates were prepared as described.36 Antibodies against C/EBPδ (Active Motif Europe, Rixensart, Belgium), G-CSFR, c-myc (Santa Cruz Biotechnology, Santa Cruz, CA), or actin (Sigma) were used, and Western blots were performed as described.37 Densitometric analysis was performed by using a high-sensitive charged-coupled device camera (INTAS, Göttingen, Germany), and the bands were quantified using the GelPro Analyzer software (INTAS), according to the manufacturer's instructions.
In vitro methylation and luciferase reporter assay
The C/EBPδ-pGL3 (generous gift from Dr J. De Wille), C/EBPα,38 (kind gift from Dr G. Behre), cyclin E, and c-myc35 promoter luciferase reporter constructs were in vitro methylated by SssI methylase (New England Biolabs, Ipswich, MA) treatment following manufacturer's instructions. The 32D cells were transfected by electroporation with 15 μg of methylated or mock-methylated pGL3 promoter constructs together with Renilla vector pRLSV40 (300 ng). S2 Drosophila cells were transfected as described.39 Luciferase activity was analyzed after 24 hours using a dual reporter assay (Promega, Madison, WI). Luciferase experiments were independently repeated 3 times.
Transduction of mouse hematopoietic progenitors cells and colony assays
Bone marrow cells for transduction were obtained from the long bones of C3H/j mice as described.35 Cells were transduced with retroviral supernatants collected from Phoenix packaging cells transfected with ER-inducible pBabe-puro-C/EBPδ (kind gift from Dr Alan D. Friedman) or pBabe-puro empty vector control. Transduced cells were selected with 1 μg/mL puromycin for 2 days and seeded in IL-3, IL-6, and SCF-supplemented methylcellulose dishes in the presence of puromycin (1 μg/mL). β-estradiol (Sigma) was added for C/EBPδ activation. Ethanol was used as solvent control. After 10 days, colonies were collected from methylcellulose and replated (5000 cells per dish) on fresh methylcellulose medium with supplements. The remaining cells from the colony assays were used for Western blot analysis. The morphology of C/EBPδ cells cultured in the presence or absence of β-estradiol was analyzed by Wright-Giemsa staining.
Generation of stable 32D cell lines
Stable cell lines were generated as described.40 Briefly, 32D-Flt3-ITD cells were electroporated with 10 μg pBabe-puro-C/EBPδ ER-inducible construct or empty pBabe-puro vector as control. Cells were selected with puromycin (1 mg/mL). Blasticidin was added to the cultures to ensure Flt3-ITD expression. Selected cells were analyzed for C/EBPδ expression by Western blotting.
Growth curve and colony assay
Stable 32D-Flt3-ITD cells expressing C/EBPδ ER-inducible or empty vector were seeded at a density of 1 × 105/mL in growth medium (RPMI and 10% FCS) without WEHI or IL-3. A total of 1 μM β-estradiol was added to the medium, and ethanol was used as solvent control. Cell growth was analyzed by counting viable cells using trypan blue dye exclusion method for 4 consecutive days. For colony assays, cells with ER-inducible C/EBPδ or empty vector were seeded at a density of 103 cells per dish in triplicate in 35 × 10-mm dishes containing Iscove modified Dulbecco medium (IMDM; Life Technologies, Grand Island, NY), 1% methylcellulose, 20% FCS, 20 ng/mL G-CSF, and 1 μM β-estradiol or ethanol as solvent control. Colonies (more than 50 cells) were counted on day 12. Experiments were performed independently 3 times. Photographs of the colony assays were taken with an Olympus C5050 digital camera attached to an Olympus CKX1 inverted microscope with an Olympus Plan 2×/0.05 NA objective lens (Olympus, Hamburg, Germany).
For analysis of morphology, the C/EBPδ-overexpressing 32D-Flt3-ITD cells or the empty vector cells were treated with β-estradiol or ethanol for 7 days. Cytospins were prepared and stained with Wright-Giemsa. Photographs were taken with a digital camera from Diagnostic Instruments (model 2.3.1; Vision System, Puchheim, Germany) attached to a Zeiss fluorescent microscope equipped with a Zeiss 100×/1.30 NA oil objective lens (Zeiss, Jena, Germany).
Detection of CpG methylation in cell lines
Genomic DNA was extracted using DNAzol (Gibco Life Technology, Invitrogen, Carlsbad, CA). The DNA was treated with bisulfite using the CpG genome kit (Chemicon R, Temecula, CA) following manufacturer's instructions. A nested PCR was performed using primers designed by MethPrimer program (UCLA, San Francisco, CA).41 Primers sequences were TTTTTTAYGGGTTTTTATTTTTYGT (outer-F), CCTTTTCTAACCCCRACTAACRTA (outer-R), GATTTTATTTTTAATTTYGAGGAGY (nested, forward), and CCTTTTCTAACCCCRACTAACRTA (nested, reverse). The PCR product (356 bp) was blunt-end cloned into pST1-blue vector (Novagen, EMD Biosciences, Beeston, Nottingham, United Kingdom). At least 10 clones were sequenced for each cell line using T7 sequencing primer.
Chromatin immunoprecipitations
The chromatin immunoprecipitations were performed as described with minor modifications.42 Briefly, U937 cells were treated with Aza for 6 days. Cells were crosslinked with 1% formaldehyde for 10 minutes. After washing with ice-cold PBS, the cell pellets were resuspended in lysis buffer (150 mM NaCl/25 mM Tris HCl [pH 7.5], 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate) and sonicated for 90 seconds at 30% amplitude. The lysates were incubated either with 10 μL anti-K4 methylated histone H3 antibody or antiacetylated histone H3 antibody (Upstate Biotechnology, Lake Placid, NY) at 4°C overnight. Isotype control IgG antibody was used as a negative control. Protein A/G plus agarose beads (Santa Cruz Biotechnology) were added for 1 hour at 4°C. After washing, the immunoprecipitates were eluted by incubating with elution buffer (0.1 M NaHCO3, 1% SDS). Samples were treated with RNaseA (50 μg/mL) and incubated overnight at 65°C to reverse the crosslinks. Proteins were removed by treatment with EDTA, 1 M Tris Cl (pH 6.5), and proteinase K for 1 hour at 42°C. DNA was extracted using a PCR purification kit (Qiagen) and eluted in 100 μL ddH2O. C/EBPδ promoter sequences were amplified in the immunoprecipitates by real-time PCR using primers GGCCAAGTCCTGGTTTTGATT (forward), GCCTTCTCTTCTTCCTGTTTGTG (reverse), and probe FAM-CCCGAGGAGCGAGGAGGTTCCA-TAMRA.
Methylation analyses in AML patients
Genomic DNA was isolated by means of standard procedures from bone marrow samples obtained for diagnostic reasons from patients with acute myeloid leukemia or nonmalignant diseases for control purposes. The more sequential and detailed clinical information about all AML patients and controls are provided in Table S2. Informed consent was obtained from all patients and controls.
The target regions were amplified using bisulfite-modified DNA and the primer pairs described in Table 1The PCR reactions were carried out in a total volume of 5 μL using 1 pmol of each primer, 40 μM dNTP, 0.1 unit of Hot Star Taq DNA polymerase (Qiagen), 1.5 mM MgCl2, and buffer supplied with the enzyme (final concentration 1 ×). The reaction mix was preactivated for 15 minutes at 95°C. The reactions were amplified in 45 cycles of 95°C for 20 seconds, 62°C for 30 seconds, and 72°C for 30 seconds followed by 72°C for 3 minutes. Unincorporated dNTPs were dephosphorylated by adding 1.7 μL H2O and 0.3 units of shrimp alkaline phosphatase (SAP; Sequenom, San Diego, CA). The reaction was incubated at 37°C for 20 minutes, and SAP was then heat inactivated for 10 minutes at 85°C.
Regions of CEBP family members analyzed by MALDI-TOF MS
| Name . | Chromosome: regions analyzed . | Analyzed regions relative to transcriptional start site . | Length . | No. of analyzed CpGs . | Left primer . | Right primer . |
|---|---|---|---|---|---|---|
| CEBPA_1 | chr19: 38485156-38485692 | +4 to −531 | 536 | 75 | TTTTATGGGGGAGTTAGAGTTTTTT | CCCCAACAACTCAAAACCAAAAC |
| CEBPA_2 | chr19: 38485997-38486426 | −836 to −1265 | 429 | 45 | GGGTTGGAAAATTTTTTTTATAATTATTTT | CACTCAAAAAACCCCAAAACCTAAC |
| CEBPA_3 | chr19: 38484095-38484522 | +639 to +1066 | 427 | 51 | TTGTTTATGGTTTTGATTAAGGAGTTTTTT | ACCAAACCACCATACACCTACAAC |
| CEBPB_1 | chr20: 48240467-48241013 | −317 to +229 | 546 | 66 | TTAGGATAGTTTTTAGTTTAGAGGGG | CATACTAAATCCCAAACCACCAAAC |
| CEBPB_2 | chr20: 48239913-48240493 | −291 to −871 | 580 | 45 | ATTGTAGTTGGGAGAAGTGAAGTGATTTAT | CCCCTCTAAACTAAAAACTATCCTAAAAAA |
| CEBPB_3 | chr20: 48241915-48242141 | +1131 to +1357 | 226 | 27 | GGTTGTAGAAGAAGGTGGAGTAGTTGT | CTACCAAATACCCCAATACCAAAA |
| CEBPD_1 | chr8: 48813003-48813574 | −321 to +250 | 571 | 73 | GTGGTATAGTTTTAGGGTGGGTA | CAAAAAAAATATCATTCCCAACAAC |
| CEBPD_2 | chr8: 48813596-48814045 | +272 to +721 | 449 | 52 | TTTTTTTGTTTGTGGGTTTGGAATT | CTCCCCCATCTACTCTACTTTTAAC |
| CEBPD_3 | chr8: 48812469-48813026 | −298 to −855 | 557 | 72 | GTTGGGTAGTTGTTTGAAGAATTGT | TACCCACCCTAAAACTATACCAC |
| CEBPD_4 | chr8: 48814020-48814282 | +696 to +958 | 262 | 26 | GTTAAAAGTAGAGTAGATGGGGGAGA | CAACTACTTTAAATTCAAAACACTTTTT |
| CEBPG_1 | chr19: 38555928-38556250 | −322 to −645 | 322 | 26 | TTAATGTTAATTTTATGTGTGAAATATTTG | CATTTTCTAAAATAATTTTATAAACCACC |
| CEBPZ_1 | chr2: 37311753-37312199 | +46 to +492 | 447 | 33 | AAGGTTAGGATTAAGAAAAGTTATTTTT | AAACCTTTAAAATTCCATACCAAAC |
| CEBPZ_2 | chr2: 37312183-37312383 | +62 to −138 | 201 | 13 | TGGAATTTTAAAGGTTTTTTGATTG | ACCCCAAACCTAACCTCAACAATAC |
| CEBPZ_3 | chr2: 37312183-37312602 | +62 to −357 | 420 | 32 | TGGAATTTTAAAGGTTTTTTGATTG | CAAAAACAACACCCACTTACCCAC |
| CEBPZ_4 | chr2: 37312359-37312781 | −114 to −536 | 423 | 32 | GTATTGTTGAGGTTAGGTTTGGGGT | ATATTTCCCTCTCCAAATAAAAAATA |
| CEBPE_1 | chr14: 22658012-22658410 | −304 to +94 | 399 | 11 | GGAGAGGTCAATGGAGGCCTCATGCTCACA | CTCCCTGAGTCACCCCAAGGGGAG |
| Name . | Chromosome: regions analyzed . | Analyzed regions relative to transcriptional start site . | Length . | No. of analyzed CpGs . | Left primer . | Right primer . |
|---|---|---|---|---|---|---|
| CEBPA_1 | chr19: 38485156-38485692 | +4 to −531 | 536 | 75 | TTTTATGGGGGAGTTAGAGTTTTTT | CCCCAACAACTCAAAACCAAAAC |
| CEBPA_2 | chr19: 38485997-38486426 | −836 to −1265 | 429 | 45 | GGGTTGGAAAATTTTTTTTATAATTATTTT | CACTCAAAAAACCCCAAAACCTAAC |
| CEBPA_3 | chr19: 38484095-38484522 | +639 to +1066 | 427 | 51 | TTGTTTATGGTTTTGATTAAGGAGTTTTTT | ACCAAACCACCATACACCTACAAC |
| CEBPB_1 | chr20: 48240467-48241013 | −317 to +229 | 546 | 66 | TTAGGATAGTTTTTAGTTTAGAGGGG | CATACTAAATCCCAAACCACCAAAC |
| CEBPB_2 | chr20: 48239913-48240493 | −291 to −871 | 580 | 45 | ATTGTAGTTGGGAGAAGTGAAGTGATTTAT | CCCCTCTAAACTAAAAACTATCCTAAAAAA |
| CEBPB_3 | chr20: 48241915-48242141 | +1131 to +1357 | 226 | 27 | GGTTGTAGAAGAAGGTGGAGTAGTTGT | CTACCAAATACCCCAATACCAAAA |
| CEBPD_1 | chr8: 48813003-48813574 | −321 to +250 | 571 | 73 | GTGGTATAGTTTTAGGGTGGGTA | CAAAAAAAATATCATTCCCAACAAC |
| CEBPD_2 | chr8: 48813596-48814045 | +272 to +721 | 449 | 52 | TTTTTTTGTTTGTGGGTTTGGAATT | CTCCCCCATCTACTCTACTTTTAAC |
| CEBPD_3 | chr8: 48812469-48813026 | −298 to −855 | 557 | 72 | GTTGGGTAGTTGTTTGAAGAATTGT | TACCCACCCTAAAACTATACCAC |
| CEBPD_4 | chr8: 48814020-48814282 | +696 to +958 | 262 | 26 | GTTAAAAGTAGAGTAGATGGGGGAGA | CAACTACTTTAAATTCAAAACACTTTTT |
| CEBPG_1 | chr19: 38555928-38556250 | −322 to −645 | 322 | 26 | TTAATGTTAATTTTATGTGTGAAATATTTG | CATTTTCTAAAATAATTTTATAAACCACC |
| CEBPZ_1 | chr2: 37311753-37312199 | +46 to +492 | 447 | 33 | AAGGTTAGGATTAAGAAAAGTTATTTTT | AAACCTTTAAAATTCCATACCAAAC |
| CEBPZ_2 | chr2: 37312183-37312383 | +62 to −138 | 201 | 13 | TGGAATTTTAAAGGTTTTTTGATTG | ACCCCAAACCTAACCTCAACAATAC |
| CEBPZ_3 | chr2: 37312183-37312602 | +62 to −357 | 420 | 32 | TGGAATTTTAAAGGTTTTTTGATTG | CAAAAACAACACCCACTTACCCAC |
| CEBPZ_4 | chr2: 37312359-37312781 | −114 to −536 | 423 | 32 | GTATTGTTGAGGTTAGGTTTGGGGT | ATATTTCCCTCTCCAAATAAAAAATA |
| CEBPE_1 | chr14: 22658012-22658410 | −304 to +94 | 399 | 11 | GGAGAGGTCAATGGAGGCCTCATGCTCACA | CTCCCTGAGTCACCCCAAGGGGAG |
Two microliters of the PCR reaction were directly used as template in a 6.5 μL transcription reaction. Twenty units of T7 DNA polymerase (Epicentre, Madison, WI) were used to incorporate either dCTP or dTTP in the transcripts. Ribonucleotides were used at 1 mM and the dNTP substrate at 2.5 mM; other components in the reaction were as recommended by the supplier. In the same step, RNase A (Sequenom) was added to cleave the in vitro transcript. The mixture was then further diluted with H2O to a final volume of 27 μL. Conditioning of the phosphate backbone prior to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was achieved by the addition of 6 mg CLEAN Resin (Sequenom). Further experimental details have been described elsewhere.43
Mass spectrometry measurements
Fifteen microliters of the cleavage reactions were robotically dispensed onto silicon chips preloaded with matrix (SpectroCHIP; Sequenom). Mass spectra were collected using a MassARRAY mass spectrometer (Bruker-Sequenom, Billerica, MA). Spectra were analyzed using proprietary peak picking and spectra interpretation tools.
Analyses of the data
To analyze methylation status of CEBP genes in bone marrow samples from AML or controls, the mean fraction of methylation for each CpG of the controls was analyzed. The number of hypermethylated CpGs (higher than mean plus 2 times the standard deviation in controls) was counted for each gene and each sample. Statistical analyses were done with SPSS 12.0. Nonparametric tests were used to compare differences in methylation levels.
Results
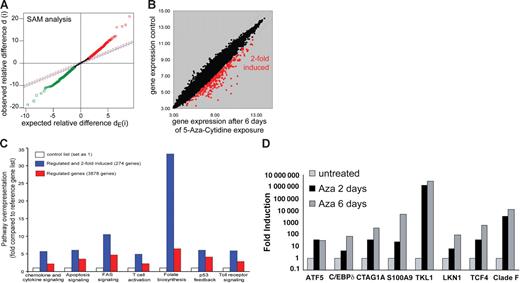
Aberrant DNA hypermethylation has emerged as a major contributing factor in tumor suppressor silencing. We performed a genomewide expression analysis to identify methylation-silenced genes in leukemia using an Aza–based demethylation assay. Global gene expression profiles of U937 cells were obtained before and after 6 days of Aza treatment. There was no significant cell death observed by Aza treatment (11.2% ± 1.58% after 6 days of treatment). The resulting expression data set was normalized with BRB Array Tools, and significance analysis of microarrays (SAM) was performed to identify genes differentially expressed between Aza-exposed (3 arrays) and control cells (3 arrays). Genes scored as positive by SAM and with induction levels more than 2-fold were regarded as significant (Figure 1A-B). Overall, 276 genes were consistently induced by demethylation (Table S3). Pathway analysis using the PANTHER (Protein Analysis Through Evolutionary Relationships) Classification System44,45 revealed several pathways with an increased number of induced transcripts (Figure 1C). Among these were genes involved in folic acid metabolism. Folic acid metabolism is closely associated with methyltransferase activity, and induction of this pathway might represent a cellular response mechanism to Aza treatment.46–48 To confirm the microarray results, we performed real-time PCR analyses for expression of several genes that were induced by demethylation. For all tested genes (n = 8), significant mRNA induction was observed in response to Aza treatment compared with nonexposed controls (Figure 1D).
Microarray analysis of Aza-treated U937 cells reveals multiple differentially regulated genes. (A) SAM of a paired analysis between Aza-exposed or nonexposed U937 cells to identify genes that are induced by Aza treatment. The scatter plot indicates the expected as well as the observed variability. Induced and repressed genes are indicated in red and green, respectively. (B) The scatter plot depicts the induced expression of genes (2-fold) by 6 days of Aza treatment of U937 cells. Red dots indicate 274 genes that were significantly induced after 6 days of Aza treatment. (C) Pathway analysis by PANTHER Classification System indicates biochemical pathways to be regulated by Aza treatment of U937 cells for all differentially expressed genes or for genes that were 2-fold induced (n = 274). (D) Relative levels of mRNA expression of transcripts identified from microarray analysis. Real-time RT-PCR was performed using mRNA from U937 cells treated with Aza for the indicated time. Expression levels for each gene were normalized to GAPDH levels.
Microarray analysis of Aza-treated U937 cells reveals multiple differentially regulated genes. (A) SAM of a paired analysis between Aza-exposed or nonexposed U937 cells to identify genes that are induced by Aza treatment. The scatter plot indicates the expected as well as the observed variability. Induced and repressed genes are indicated in red and green, respectively. (B) The scatter plot depicts the induced expression of genes (2-fold) by 6 days of Aza treatment of U937 cells. Red dots indicate 274 genes that were significantly induced after 6 days of Aza treatment. (C) Pathway analysis by PANTHER Classification System indicates biochemical pathways to be regulated by Aza treatment of U937 cells for all differentially expressed genes or for genes that were 2-fold induced (n = 274). (D) Relative levels of mRNA expression of transcripts identified from microarray analysis. Real-time RT-PCR was performed using mRNA from U937 cells treated with Aza for the indicated time. Expression levels for each gene were normalized to GAPDH levels.
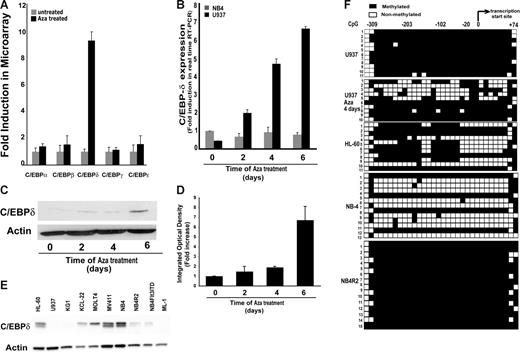
Among these genes, C/EBPδ was highly induced (more than 10-fold) after 6 days of Aza treatment (Figure 1D). The expression of other C/EBP proteins (α, β, γ, ϵ) remained unchanged after Aza treatment whereas C/EBPδ mRNA was induced by several folds (Figure 2A). The discovery of C/EBPδ as a methylation-silenced gene was intriguing because it is a member of the C/EBP family of transcription factors, which are known to play an important role in lineage commitment and differentiation.2 The induction of C/EBPδ expression in response to Aza treatment was confirmed on RNA as well as on the protein level (Figure 2B-D). Treatment with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) did not influence C/EBPδ expression (data not shown).
Aza treatment induced the expression of C/EBPδ. (A) Relative levels of mRNA expression of C/EBP family members' transcripts in Aza-cytidine–exposed and nonexposed U937 cells. Indicated are means and standard deviations. (B) Relative levels of C/EBPδ mRNA in Aza-treated or untreated U937 and NB4 cells by real-time RT-PCR. Expression levels were normalized to GAPDH levels. Results shown are mean ± SD of 3 independent experiments. (C) Western blot analysis of C/EBPδ expression in lysates from Aza-treated U937 cells. (D) Densitometry of the Western blot shown in panel C normalized to actin verified induction of C/EBPδ protein after Aza treatment of U937 cells. (E) Western blot showing expression of C/EBPδ in different leukemic cell lines. Total protein lysate was subjected to Western blotting and probed with anti-C/EBPδ and antiactin antibodies. (F) Bisulfite sequencing for C/EBPδ promoter methylation analysis in leukemic cell lines. Filled squares indicate methylated and open squares nonmethylated CpG dinucleotides. The transcriptional start site and the location of the analyzed CpGs (n = 32) are indicated. Individual clones were sequenced.
Aza treatment induced the expression of C/EBPδ. (A) Relative levels of mRNA expression of C/EBP family members' transcripts in Aza-cytidine–exposed and nonexposed U937 cells. Indicated are means and standard deviations. (B) Relative levels of C/EBPδ mRNA in Aza-treated or untreated U937 and NB4 cells by real-time RT-PCR. Expression levels were normalized to GAPDH levels. Results shown are mean ± SD of 3 independent experiments. (C) Western blot analysis of C/EBPδ expression in lysates from Aza-treated U937 cells. (D) Densitometry of the Western blot shown in panel C normalized to actin verified induction of C/EBPδ protein after Aza treatment of U937 cells. (E) Western blot showing expression of C/EBPδ in different leukemic cell lines. Total protein lysate was subjected to Western blotting and probed with anti-C/EBPδ and antiactin antibodies. (F) Bisulfite sequencing for C/EBPδ promoter methylation analysis in leukemic cell lines. Filled squares indicate methylated and open squares nonmethylated CpG dinucleotides. The transcriptional start site and the location of the analyzed CpGs (n = 32) are indicated. Individual clones were sequenced.
C/EBPδ expression is negatively regulated by methylation in leukemic cell lines
We analyzed C/EBPδ expression in several leukemic cell lines including U937 (Figure 2E). C/EBPδ was expressed in HL-60, NB4, KCL22, MOLT-4, and MV411 cells, but several other leukemic cells such as KG1, ML-1, and U937 did not show detectable expression of C/EBPδ. To evaluate promoter methylation we analyzed the C/EBPδ upstream region for CpG islands. The 5′ upstream region of C/EBPδ promoter contains a CpG island with high GC content (75%). We performed bisulfite sequencing of the region between −408 to +24 relative to the transcription start site, which contains 32 CpG dinucleotides. Four cell lines (U937, HL-60, NB4, and NB4R2) with different levels of C/EBPδ expression were chosen for the analysis. We found a high degree of methylation for U937 and NB4R2, moderate levels in HL-60, and low methylation levels in NB4 cells (Figure 2F). In U937 cells, methylation decreased after Aza treatment (Figure 2F). This finding is in line with the induced expression of C/EBPδ after Aza exposure in U937 cells (Figure 2B-C). Also, in other cell lines a close association between methylation and loss of expression was observed. The NB4 (PML-RARα fusion protein positive) and NB4R2 (ATRA-resistant PML-RARα fusion protein positive) cells differed in C/EBPδ promoter methylation status and expression (Figure 2E-F). These data indicated that C/EBPδ expression is negatively correlated with promoter methylation in leukemic cell lines.
DNA methylation suppresses C/EBPδ promoter activity
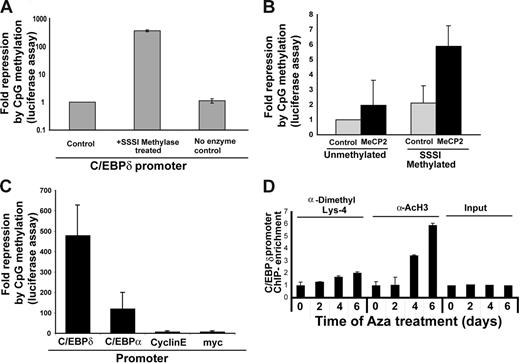
To analyze the effects of DNA methylation on promoter activity, we used a C/EBPα promoter luciferase construct (−393) treated with or without SssI methylase enzyme to methylate the promoter in vitro. In vitro–methylated or mock-methylated C/EBPα promoter constructs were transfected into 32D cells along with a Renilla expression plasmid. C/EBPα promoter activity was strongly repressed by methylation (Figure 3A). Methyl CpG binding protein (MeCP2), a member of the MBD family of proteins, is known to induce transcriptional repression of methylated CpG promoters by recruiting histone deacetylases (HDACs).49 MeCP2 is thought to be active in the presence of multiple methylated CpG dinucleotides such as the CpG-dense C/EBPα promoter. Therefore, we analyzed whether repression of the methylated C/EBPα promoter was MeCP2 dependent. Because MeCP2 does not have a homolog in Drosophila, we cotransfected S2 Drosophila cells with in vitro–methylated or mock-methylated −393 C/EBPα promoter constructs in the presence or absence of MeCP2 expression construct. Activity of the methylated promoter was further repressed 4-fold in the presence of MeCP2, which indicates that MeCP2 might play a role in suppression of the transcriptional activity of the methylated C/EBPα promoter (Figure 3B).
C/EBPδ promoter activity is suppressed by methylation. (A) Luciferase assays showing C/EBPδ promoter repression in response to methylation. Methylated (SssI) or mock-methylated C/EBPδ promoter reporter constructs were transfected into 32D cells. The fold repression was calculated as 1/activity (normalized to Renilla values) with the control set as 1. The results are shown as the mean ± SD of 3 independent experiments. (B) Luciferase assay in S2 Drosophila cells, indicating that C/EBPδ promoter repression is dependent on MeCP2 activity. Methylated or mock-methylated reporter constructs were transfected into S2 Drosophila cells along with an Sp1 expression plasmid and either an empty vector or a MeCP2 expression vector. Fold repression was calculated as 1/relative activity, with the control set as 1. The results are shown as the mean ± SD of 3 independent experiments. (C) Luciferase assay showing fold repression in promoter activity of C/EBPδ, C/EBPα, cyclin E, and c-myc promoters after SssI methylase treatment. Methylated (SssI) or mock-methylated C/EBPδ promoter reporter constructs were transfected into 32D cells. The results are shown as the mean ± SD of 3 independent experiments. (D) Chromatin modification of histone H3-K4 methylation and histone H3 acetylation. Chromatin immunoprecipitations were performed with Aza-exposed U937 cells. Antibodies against histone H3-methylated K4 (K4-Me), acetylated histone H3, or IgG isotype control were used. Real-time PCR was performed using C/EBPδ promoter–specific primers and probe. The results shown are mean ± SD of 2 independent experiments.
C/EBPδ promoter activity is suppressed by methylation. (A) Luciferase assays showing C/EBPδ promoter repression in response to methylation. Methylated (SssI) or mock-methylated C/EBPδ promoter reporter constructs were transfected into 32D cells. The fold repression was calculated as 1/activity (normalized to Renilla values) with the control set as 1. The results are shown as the mean ± SD of 3 independent experiments. (B) Luciferase assay in S2 Drosophila cells, indicating that C/EBPδ promoter repression is dependent on MeCP2 activity. Methylated or mock-methylated reporter constructs were transfected into S2 Drosophila cells along with an Sp1 expression plasmid and either an empty vector or a MeCP2 expression vector. Fold repression was calculated as 1/relative activity, with the control set as 1. The results are shown as the mean ± SD of 3 independent experiments. (C) Luciferase assay showing fold repression in promoter activity of C/EBPδ, C/EBPα, cyclin E, and c-myc promoters after SssI methylase treatment. Methylated (SssI) or mock-methylated C/EBPδ promoter reporter constructs were transfected into 32D cells. The results are shown as the mean ± SD of 3 independent experiments. (D) Chromatin modification of histone H3-K4 methylation and histone H3 acetylation. Chromatin immunoprecipitations were performed with Aza-exposed U937 cells. Antibodies against histone H3-methylated K4 (K4-Me), acetylated histone H3, or IgG isotype control were used. Real-time PCR was performed using C/EBPδ promoter–specific primers and probe. The results shown are mean ± SD of 2 independent experiments.
We compared the effects of C/EBPα promoter methylation with the effects of methylation on several other promoters: C/EBPα, cyclin E, and c-myc. Interestingly, C/EBP promoters (α and δ) were strongly repressed by SssI treatment compared with cyclin E and c-myc. These results suggest that the C/EBPδ promoter is by far the most sensitive promoter with regard to methylation (Figure 3C). Nonetheless, these in vitro analyses cannot be directly extended to the in vivo situations. Therefore, we analyzed the association between DNA methylation and chromatin formation of the C/EBPδ promoter in vivo.
Chromatin modifications by demethylation of the C/EBPδ promoter
Epigenetic repression by DNA methylation is associated with corresponding histone modifications that contribute to gene repression and heterochromatin formation. To determine whether the reexpression of C/EBPδ induced by Aza treatment altered the chromatin structure, we performed chromatin immunoprecipitation for histone modifications using antibodies against methylated lysine-4 histone H3 and acetylated histone H3. Lysine-4 dimethylation of histone H3 and especially histone acetylation were significantly increased in response to Aza treatment of U937 cells (Figure 3D).
Methylation analysis of the C/EBP gene family in AML patient samples
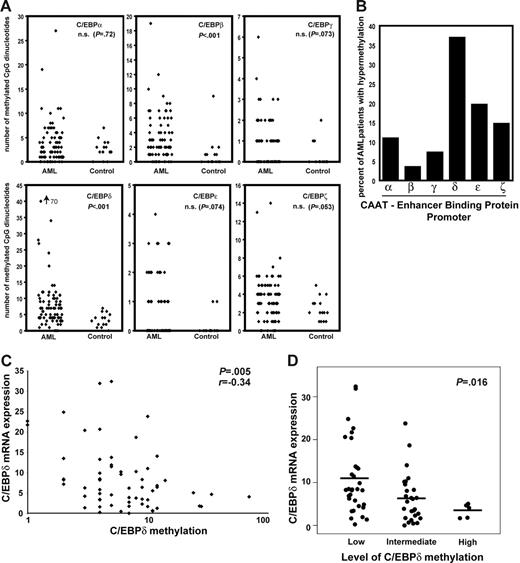
C/EBP genes play important roles in hematopoietic differentiation. Due to functional redundancy, the role of individual C/EBP proteins is difficult to assess. For C/EBPδ to be considered a tumor suppressor, it would be important to elucidate its methylation status in comparison with other C/EBP family members in AML patients. Genomic DNA from AML patients (n = 81) and healthy controls (n = 15) was bisulfite treated and subsequently PCR amplified with primers that bound irrespective of methylation. Quantitative methylation analysis was performed using a specialized MALDI-TOF assay. This method allows the quantitative analysis of a large number of CpG dinucleotides.43,50 Background methylation levels were calculated for each CpG using 15 control samples. Hypermethylation of a CpG was defined as the mean methylation level in controls plus 2 times the standard deviation. Subsequently, the number of hypermethylated CpGs was calculated for each sample and each gene (Figure 4A). Significant differences in the number of methylated CpG dinucleotides between AML and control samples were found for C/EBPβ and C/EBPδ, whereas no significant differences were found for the other C/EBP members. These analyses included the region of the C/EBPα promoter (−837 to −1266) that has been shown to be methylated in lung cancer.51 Importantly, more than 35% of all AML samples showed higher levels of C/EBPδ methylation than all controls. In contrast, none of the other C/EBP family members were methylated in more than 20% of the patient samples (Figure 4B). No association between C/EBP methylation and age of the patients or stage of disease was found. These data demonstrate the occurrence of aberrant promoter methylation of C/EBPδ in primary leukemic samples. These findings indicate that the C/EBPδ promoter is the most abundantly methylated C/EBP gene in AML.
The C/EBPδ promoter is hypermethylated in acute myeloid leukemia. (A) The number of hypermethylated CpGs in the promoter region of the C/EBP family members was analyzed in AML patients and controls by MALDI-TOF. Hypermethylation was defined as methylation above the mean methylation levels in controls plus 2 times the controls' standard deviation. The number of hypermethylated CpGs was analyzed for each sample and each gene. (B) The C/EBPδ promoter was methylated at a high frequency in AML. The bars indicate the percent of AML patients' samples hypermethylated for the different C/EBP genes as assessed by MALDI-TOF. (C-D) Correlation between C/EBPδ promoter methylation and mRNA expression. C/EBPδ promoter methylation was associated with decreased C/EBPδ expression levels (P = .005). The scatter plot indicates the relative mRNA expression levels normalized to GAPDH with regard to a low (up to 5 hypermethylated CpGs), intermediate (6 to 15 hypermethylated CpGs), and high (more than 150 methylated CpGs) degree of C/EBPδ promoter methylation (P = .016, nonparametric test). The median level of expression is indicated for each group with a horizontal line.
The C/EBPδ promoter is hypermethylated in acute myeloid leukemia. (A) The number of hypermethylated CpGs in the promoter region of the C/EBP family members was analyzed in AML patients and controls by MALDI-TOF. Hypermethylation was defined as methylation above the mean methylation levels in controls plus 2 times the controls' standard deviation. The number of hypermethylated CpGs was analyzed for each sample and each gene. (B) The C/EBPδ promoter was methylated at a high frequency in AML. The bars indicate the percent of AML patients' samples hypermethylated for the different C/EBP genes as assessed by MALDI-TOF. (C-D) Correlation between C/EBPδ promoter methylation and mRNA expression. C/EBPδ promoter methylation was associated with decreased C/EBPδ expression levels (P = .005). The scatter plot indicates the relative mRNA expression levels normalized to GAPDH with regard to a low (up to 5 hypermethylated CpGs), intermediate (6 to 15 hypermethylated CpGs), and high (more than 150 methylated CpGs) degree of C/EBPδ promoter methylation (P = .016, nonparametric test). The median level of expression is indicated for each group with a horizontal line.
Promoter methylation is associated with C/EBPδ silencing in AML
Next, we analyzed whether methylation of the promoter influences the expression of C/EBPδ in AML patients. C/EBPδ is usually expressed in human CD34+ primary progenitor cells at low levels with induction upon granulocytic differentiation (real-time RT-PCR data not shown).
Real-time RT-PCR was performed on cDNA samples obtained from the patients' samples used for methylation analysis. We found a close correlation between a higher number of methylated CpGs and low expression of C/EBPδ mRNA (P = .005) (Figure 4C). We divided all samples into 3 groups based on the number of methylated CpGs (low, up to 5 methylated CpGs; intermediate, 6 to 15 methylated CpGs; high, more than 15 methylated CpGs). Samples with high CpG methylation levels showed significantly lower C/EBPδ mRNA expression compared with samples containing low numbers of methylated CpGs (mean, 3.45 versus 10.92; P = .016). Samples grouped as intermediate showed mean relative expression levels of 6.32 (Figure 4D).
C/EBPδ possesses growth suppressive functions in primary hematopoietic progenitors
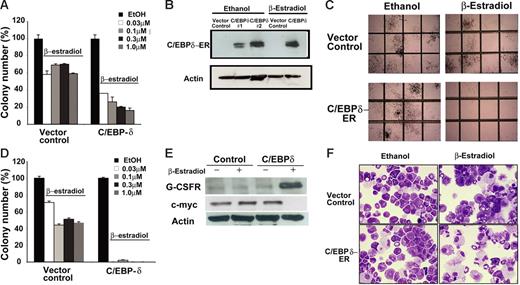
A role of C/EBPδ in inducing granulopoiesis has been suggested in cell lines.12 We analyzed whether C/EBPδ has growth inhibitory effects on primary bone marrow cells. C/EBPδ-ER–inducible or empty vector (kind gift from Dr A. Friedman) were transduced by retroviral transduction into primary cells obtained from murine bone marrow. Transduced cells were selected with puromycin for 2 days and then cultured in methylcellulose with growth factors in the presence or absence of β-estradiol to induce C/EBPδ activity. We used 4 different concentrations of β-estradiol (0.03 μM, 0.1 μM, 0.3 μM, 1 μM) to find the lowest concentration that did not affect the growth of control cells. Addition of β-estradiol inhibited colony growth of control cells to some extent, but in C/EBPδ-overexpressing cells we observed a dose-dependent decrease on colony growth (Figure 5A). C/EBPδ expression markedly reduced the number of granulocyte macrophage colony-forming unit (CFU-GM), macrophage CFU (CFU-M), and granulocyte, erythroid, macrophage, megakaryocyte CFU (CFU-GEMM) colonies compared with control (Figure 5A). Expression of C/EBPδ in the colonies was confirmed by Western blotting (Figure 5B).
C/EBPδ suppresses growth of primary hematopoietic progenitors. (A) Colony assays showing the growth inhibitory effect of C/EBPδ on hematopoietic progenitor cells. Murine bone marrow cells were transduced with an ER-inducible C/EBPδ construct or empty vector as a control. After 48 hours of transduction, cells were puromycin selected for 2 days. Colony assays were performed in the presence of β-estradiol to activate C/EBPδ. The results are shown as the mean ± SD of 3 independent experiments. (B) Western blot analysis of the colonies obtained in panel A. Blots were probed with anti-C/EBPδ and antiactin antibodies to confirm ectopic expression of C/EBPδ in the colonies. (C) C/EBPδ-transduced primary bone marrow cells in colony assay after replating. C/EBPδ expression completely abolished the colony growth of primary cells on replating. Photograph of representative areas of the plates demonstrates the lack of colony growth after replating in C/EBPδ-overexpressing cells in the presence of β-estradiol. (D) The bars indicate the inhibitory effect of C/EBPδ on colony formation in replating experiments. Results show mean ± SD of triplicates of 2 independent experiments. (E) Western blot analysis with G-CSFR and c-myc antibodies in total protein lysate from C/EBPδ or empty vector control–transduced bone marrow cells. The blot was stripped and rehybridized with actin antibodies. (F) Wright-Giemsa–stained cytospin slides under light microscopy. C/EBPδ or the empty vector–transduced primary bone marrow cells were incubated with β-estradiol for induction of C/EBPδ. Representative morphologic changes seen at day 7 of β-estradiol treatment are shown.
C/EBPδ suppresses growth of primary hematopoietic progenitors. (A) Colony assays showing the growth inhibitory effect of C/EBPδ on hematopoietic progenitor cells. Murine bone marrow cells were transduced with an ER-inducible C/EBPδ construct or empty vector as a control. After 48 hours of transduction, cells were puromycin selected for 2 days. Colony assays were performed in the presence of β-estradiol to activate C/EBPδ. The results are shown as the mean ± SD of 3 independent experiments. (B) Western blot analysis of the colonies obtained in panel A. Blots were probed with anti-C/EBPδ and antiactin antibodies to confirm ectopic expression of C/EBPδ in the colonies. (C) C/EBPδ-transduced primary bone marrow cells in colony assay after replating. C/EBPδ expression completely abolished the colony growth of primary cells on replating. Photograph of representative areas of the plates demonstrates the lack of colony growth after replating in C/EBPδ-overexpressing cells in the presence of β-estradiol. (D) The bars indicate the inhibitory effect of C/EBPδ on colony formation in replating experiments. Results show mean ± SD of triplicates of 2 independent experiments. (E) Western blot analysis with G-CSFR and c-myc antibodies in total protein lysate from C/EBPδ or empty vector control–transduced bone marrow cells. The blot was stripped and rehybridized with actin antibodies. (F) Wright-Giemsa–stained cytospin slides under light microscopy. C/EBPδ or the empty vector–transduced primary bone marrow cells were incubated with β-estradiol for induction of C/EBPδ. Representative morphologic changes seen at day 7 of β-estradiol treatment are shown.
The replating potential of CFU-GMs is an indicator of self-renewal capacity of progenitor cells. We performed replating experiments by pooling the colonies from C/EBPδ-transduced colonies in the absence of β-estradiol. Cells were replated in methylcellulose in the presence or absence of β-estradiol (0.03 μM, 0.1 μM, 0.3 μM, 1 μM). Colonies from empty vector control in ethanol were also picked and replated in the presence of β-estradiol for control purposes. C/EBPδ induction completely abrogated the colony growth of replated cells in all 4 concentrations of β-estradiol while empty vector control–transduced cells showed a slight reduction in colony number in the presence of β-estradiol (Figure 5C-D). These findings confirmed the growth inhibitory role of C/EBPδ in primary myeloid progenitor cells. We next analyzed the expression of G-CSFR and c-myc in response to C/EBPδ induction in the primary cells from colony assay. Western blot analysis showed that G-CSFR protein expression increased and c-myc expression decreased in the presence of β-estradiol in C/EBPδ-overexpressing cells compared with control (Figure 5E).
Cell morphology was examined by cytospin and staining with Wright-Giemsa. Within 7 days of β-estradiol treatment, many of the C/EBPδ-overexpressing bone marrow cells underwent myeloid maturation with reduction in the nuclear-cytoplasmic ratio (Figure 5F). Control vector–transduced cells contained more promyelocytes compared with C/EBPδ-overexpressing cells, whereas the number of completely differentiated granulocytes was higher in C/EBPδ-expressing bone marrow (Figure 5F).
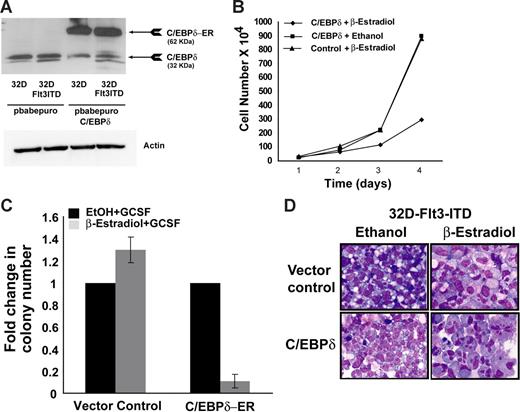
C/EBPδ inhibits the growth of 32D cells transformed by Flt3 mutations
Based on the finding that C/EBPδ is a growth inhibitor of myeloid progenitor cells, we analyzed the potential of C/EBPδ to act as a suppressor of leukemogenic transformation. We overexpressed the estrogen-inducible C/EBPδ construct in 32D cells, which were transformed by oncogenic Flt3 receptor with ITDs.40,52 These mutations are among the most frequent in AML. Ectopic expression of C/EBPδ-ER fusion protein was verified by Western blot analysis (Figure 6A). Cells were seeded in IL-3–free medium in the presence or absence of β-estradiol and were counted every 24 hours. A significant decrease in the cell number was observed in C/EBPδ-overexpressing cells in the presence of β-estradiol compared with vector control or C/EBPδ cells in ethanol (Figure 6B). We next assessed the ability of these cells to form colonies in methylcellulose. Cells were seeded in methylcellulose containing G-CSF in the presence of β-estradiol or ethanol. There was a 10-fold decrease in the colony number in the presence of β-estradiol compared with ethanol in C/EBPδ-overexpressing cells. No decrease in colony numbers was observed in empty vector cells in the presence of β-estradiol (Figure 6C). These findings suggest that C/EBPδ inhibits growth and colony-forming ability of Flt3-ITD–transformed cells. Overexpression of C/EBPδ in normal 32D cells induced granulocytic differentiation (data not shown), which is in line with previous findings 12 . To analyze the effect of C/EBPδ on differentiation of 32D-Flt3-ITD cells, we analyzed the change in cell morphology by cytospin and staining with Wright-Giemsa. The activation of C/EBPδ induces morphologic changes toward granulocytic differentiation after 7 days of β-estradiol treatment (Figure 6D). Most of the cells had a reduction in the nuclear-cytoplasmic ratio with segmented nuclei, a typical characteristic of mature granulocytes. In contrast, the morphology of empty vector control cells did not show any signs of granulocytic differentiation (Figure 6D). The expression of secondary granule proteins was also analyzed after 4 days of β-estradiol treatment. The G-CSFR and lysozyme expression was significantly induced in C/EBPδ-overexpressing cells in the presence of β-estradiol compared with control cells (Figure S1). No induction was observed in MPO and neutrophil elastase RNA after C/EBPδ activation (Figure S1).
C/EBP inhibits the growth of Flt3-ITD–transformed cells. (A) Western blot analysis of transfected 32D-Flt3-ITD cells. The 32D-Flt3-ITD stable cells were transfected with an ER-inducible C/EBPδ construct or empty vector. Cells were puromycin selected. Lysates were probed with anti-C/EBPδ and antiactin antibody. (B) Growth curve of C/EBPδ-expressing 32D-Flt3-ITD cells. 32D-Flt3-ITD cells with C/EBPδ or control cells were seeded in suspension cultures at the density of 2 × 105/mL in the presence of β-estradiol or ethanol. Cells were counted every 24 hours by trypan blue exclusion method. The results are shown as the mean ± SD of 2 independent experiments. (C) Colony assays were performed with 32D-Flt3-ITD cells expressing the C/EBPδ ER construct. A total of 1000 cells were seeded per dish with either β-estradiol or ethanol. Colonies were analyzed on day 8. Each bar represents the mean ± SD of a representative triplicate experiment. (D) Wright-Giemsa–stained cytospin slides under light microscopy. C/EBPδ or the empty vector–transduced 32D-Flt3-ITD cells were incubated with β-estradiol for induction of C/EBPδ. Representative morphologic changes seen at day 7 of β-estradiol treatment are shown.
C/EBP inhibits the growth of Flt3-ITD–transformed cells. (A) Western blot analysis of transfected 32D-Flt3-ITD cells. The 32D-Flt3-ITD stable cells were transfected with an ER-inducible C/EBPδ construct or empty vector. Cells were puromycin selected. Lysates were probed with anti-C/EBPδ and antiactin antibody. (B) Growth curve of C/EBPδ-expressing 32D-Flt3-ITD cells. 32D-Flt3-ITD cells with C/EBPδ or control cells were seeded in suspension cultures at the density of 2 × 105/mL in the presence of β-estradiol or ethanol. Cells were counted every 24 hours by trypan blue exclusion method. The results are shown as the mean ± SD of 2 independent experiments. (C) Colony assays were performed with 32D-Flt3-ITD cells expressing the C/EBPδ ER construct. A total of 1000 cells were seeded per dish with either β-estradiol or ethanol. Colonies were analyzed on day 8. Each bar represents the mean ± SD of a representative triplicate experiment. (D) Wright-Giemsa–stained cytospin slides under light microscopy. C/EBPδ or the empty vector–transduced 32D-Flt3-ITD cells were incubated with β-estradiol for induction of C/EBPδ. Representative morphologic changes seen at day 7 of β-estradiol treatment are shown.
Discussion
C/EBP genes are believed to be critically involved in hematopoietic differentiation and leukemogenesis.1,53 So far, especially C/EBPα has been implicated in leukemia pathogenesis, most strikingly due to the occurrence of mutations in AML with myeloblastic differentiation.54
Here, we demonstrate that C/EBPδ is specifically and frequently inactivated by promoter hypermethylation in acute myeloid leukemia. Methylation of C/EBP genes in AML most frequently occurs at the C/EBPδ promoter. Our findings indicate that C/EBPδ inactivation is likely to play a role in the pathogenesis of a significant number of AMLs.
We used a genomewide screen to identify methylation-silenced genes in leukemia. The microarray data were verified by real-time PCR. Genes involved in several pathways were significantly induced as indicated by gene distribution in the PANTHER Classification System.44,45 Alterations in these pathways are not necessarily the result of tumor suppressor gene reactivation. For example, induction of the folate biosynthesis pathway is more likely to represent the cellular response to an overall decrease in DNA methylation.46,55 Thus, identification of genes induced by demethylating agents needs to be complemented by direct DNA methylation analysis. In the current study, we chose to concentrate on C/EBPδ that was induced more than 10-fold in real-time RT-PCR analyses, and its 5′ upstream region contains a CpG island with a dense population of CpG dinucleotides. Our further experiments indicated that this CpG island is methylated in several leukemia cell lines that do not express C/EBPδ, whereas absence of methylation was associated with gene expression. In primary patient samples, more than 35% exhibited significant levels of C/EBPδ methylation, which again was associated with reduced expression levels. In vitro analyses confirmed that C/EBPδ promoter methylation was associated with strong transcriptional repression partially induced by MeCP2 and posttranslational histone modifications. Reversal of DNA methylation increases C/EBPδ histone acetylation, which suggests that DNA methylation might play a role in the hypoacetylated state of the promoter. These findings conclusively establish the association between C/EBPδ promoter methylation and loss of expression.
C/EBPδ promoter methylation has not been reported in leukemia. A recent paper described infrequent and weak C/EBPδ methylation in breast cancer specimens.56 Similar to C/EBPα mutations and inactivation that occur not only in leukemia but also in solid tumors,57 C/EBPδ inactivation might be more widespread in hematologic and solid tumor malignancies.
Two important questions arose from our findings of C/EBPδ methylation in acute myeloid leukemia. First, how specific is C/EBPδ methylation in leukemogenesis? To analyze this in detail we used an innovative MALDI-TOF–based technique to quantitate methylation of a huge number of CpG dinucleotides of many C/EBP family members. Importantly, C/EBPδ and C/EBPβ were the only genes with increased methylation levels in AML samples compared with normal bone marrow samples. Importantly, neither C/EBPα, C/EBPγ, C/EBPϵ, nor C/EBPζ were significantly methylated. Because the MALDI-TOF method is the most sensitive and quantitative method to be used for methylation analysis of a huge number of CpG dinucleotides43,50 (the number of analyzed CpGs in this study was 260), these findings are conclusive. Aberrant methylation commonly occurred in 2 genes whose 5′ upstream regions contained a CpG island (β and δ) but not in C/EBPα, which also contains a CpG island.
C/EBPα methylation has recently been reported in lung cancer.51 Our analyses that included the region methylated in lung cancer indicate that a few AML patient samples may have increased C/EBPα methylation as well but that C/EBPα methylation is a rather rare event in AML. This is in line with previous findings.58
DNA hypermethylation of CpG islands regulating tumor suppressor gene expression is now recognized as an important feature of human neoplasia. Especially in leukemia, inactivation of several tumor suppressor genes by promoter hypermethylation has been a topic of considerable interest in the last few years. Acute myeloid leukemia blasts show frequent hypermethylation of ER, p15, MyoD, PITX2, GPR37, and SDC4 genes.55,59,60 Our findings concerning C/EBPδ methylation raise the question of C/EBPδ tumor suppressor functions. Growth inhibitory effects of C/EBPδ have been shown in human breast cancer61 and prostate cancer.22 In hematopoietic cells, involvement of C/EBPδ as an inducer of granulocytic differentiation has been demonstrated in BCR-ABL–positive CML cell lines and the 32D cell line.12,14 Importantly, loss of C/EBPδ can promote genomic instability.23 These data provide evidence that C/EBPδ has growth inhibitory properties in cell lines, but analyses of the effects on primary cells has been lacking. In the current study, we used an ER-inducible C/EBPδ to show that C/EBPδ has a growth inhibitory effect on primary bone marrow cells, which was enhanced on replating. These findings may indicate a decreased self-renewal capacity of C/EBPδ-expressing hematopoietic progenitor cells. C/EBPα has also been implicated in regulating self-renewal.62 Growth inhibitory effects of C/EBPδ had so far not been reported for AML-specific oncogenes. In the current study we used 32D cells transformed by the mutant form of the fms-like tyrosine kinase Flt3.40 Again, C/EBPδ suppressed proliferation of Flt3-ITD–transformed cells. These results provide further evidence that C/EBPδ is a tumor suppressor gene that is frequently silenced by methylation in acute myeloid leukemia.
Taken together, a genomewide screening approach identified C/EBPδ as a frequently methylated tumor suppressor gene in acute myeloid leukemia. These findings strengthen the hypothesis that C/EBP proteins are important tumor suppressors in acute myeloid leukemia, and strategies to reinduce these factors in AML cells may prove useful for antileukemic therapy in the future.
Authorship
Contribution: S.A., H.S., and C.M.-T. designed the research, analyzed the data, and wrote the manuscript; S.A. performed most of the experiments; W.K.H. performed microarray hybridizations; M.E. and D.v.d.B. performed MALDI-TOF MS experiments; N.T., S.K., W.E.B., H.S., and C.M.-T. provided analytical tools and analyzed the data; and all authors checked the final version of the manuscript.
Conflict-of-interest disclosure: M.E. and D.v.d.B. are employed by Sequenom and hold Sequenom stocks. The other authors declare no competing financial interests.
Correspondence: Carsten Müller-Tidow, Department of Medicine A, Hematology and Oncology, University of Münster, Domagkstr.3, 48129 Münster, Germany; e-mail: muellerc@uni-muenster.de.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by a Heisenberg grant from the Deutsche Forschungsgemeinschaft (C.M.-T.), the Nationales Genomforschungsnetz (NGFN)–2 LeukemiaNet, the José-Carreras Leukämiestiftung, and the Deutsche Krebshilfe. We are grateful to Sandra Doths, Barabara Mlody, Linda Kamp, and Annette Westermann for excellent technical assistance. We thank Dr A. D. Friedman for C/EBPδ expression constructs; Dr J. W. DeWille for C/EBPδ promoter constructs, and Dr G. Behre for C/EBPα promoter construct.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal