Abstract
Recently, a novel mechanism introducing genetic instability, termed aberrant somatic hypermutation (ASHM), has been described in diffuse large B-cell lymphoma. To further investigate whether ASHM also occurs in mucosa-associated lymphoid tissue type (MALT) lymphoma, we studied the mutation profile of PIM1, PAX5, RhoH/TTF, and c-MYC in 17 MALT lymphomas and 17 extranodal diffuse large B-cell lymphomas (DLBCLs) still exhibiting a low-grade MALT lymphoma component (transformed MALT lymphoma). Mutations in one or more genes were detected in 13 (76.5%) of 17 cases of MALT lymphomas and in all of 17 (100%) cases of extranodal DLBCL. A total of 100 sequence variants were found in 30 of 34 cases, 28 in the MALT lymphomas and 72 in extranodal DLBCL. Further, in PIM1 and c-MYC some of the mutations were found to affect coding exons, leading to amino acid exchanges, thus potentially altering gene function. Expression levels of activation-induced cytidine deaminase (AID), an enzyme essential for somatic hypermutation (SHM), was associated with the mutational load. These data indicate that aberrant SHM is associated with extranodal DLBCL and MALT lymphoma, likewise. By mutating regulatory and coding sequences of the targeted genes, ASHM may represent a major contributor to their pathogenesis.
Introduction
Extranodal marginal zone B-cell lymphoma of the mucosa-associated lymphoid tissue type (MALT lymphoma) preferentially arises in the gastrointestinal tract, especially the stomach where it is closely linked to a Helicobacter pylori–induced chronic gastritis.1,2 In general, MALT lymphoma may affect virtually every organ in the body.3,4 Because of the interaction between lymphoma cells and mucosal adhesion molecules,5 MALT lymphomas display an indolent clinical behavior; however, transformation in diffuse large B-cell lymphoma (DLBCL) occurs but, according to the WHO criteria, is considered as a separate entity.6 Foci of extranodal DLBCL may be seen in MALT lymphoma, termed transformed MALT lymphoma, suggestive of transformation from one to the other entity.
MALT lymphomas demonstrate a number of distinct cytogenetic alterations,7 but a molecular mechanism inducing genetic instability has not been described. In DLBCL somatic hypermutation aberrantly targets the 5′ sequences of several proto-oncogenes relevant to lymphomagenesis, including PIM1, PAX5, RhoH/TTF, and cMYC. This phenomenon, termed aberrant somatic hypermutation (ASHM) occurs in over 50% of DLBCL but is rare in indolent lymphomas.8 Although the molecular mechanism of the SHM is still unknown, studies identified the activation-induced cytidine deaminase (AID) as an absolute requirement for the SHM.9
The present study was aimed at investigating the role of the aberrant somatic hypermutation in MALT lymphomas and in MALT lymphomas transformed to extranodal DLBCL and to elucidate the role of AID in this process.
Materials and methods
Materials, diagnoses, and DNA extraction
DNA extraction (DNA Mini Kit; Qiagen, Valencia, CA) was performed from formalin-fixed and paraffin-embedded macrodissected tissue containing at least 90% malignant cells. Seventeen MALT lymphoma specimens, 6 of gastric origin and 11 of extragastric origin (2 orbita, 7 parotid gland, 1 thyroid, and 1 soft tissue), and 17 samples of extranodal DLBCL, 10 of gastric origin and 7 of extragastric origin (2 thyroid and 5 parotid gland), and 6 normal controls, including 5 surrounding nonneoplastic tissues samples and one of peripheral blood mononuclear cells were included. All lymphomas were classified according to the WHO classification of lymphoid neoplasms.6 Attention was made that cases of extranodal DLBCL comprised at least one focus of a low-grade lymphoma component considering these lymphomas as “transformed MALT lymphomas.”10 Immunohistochemistry was performed using monoclonal antibodies to CD20, CD21, CD3, κ, λ, CD30, MUM1, BCL-6 (Dako, Glostrup, Denmark), CD10 (Novocastra, Newcastle upon Tyne, United Kingdom), MIB-1 (Dianova, Hamburg, Germany).1,11
Sequencing analysis of PAX5, RhoH/TTF, c-MYC, PIM1, and IgHV
Mutational profile by direct DNA sequencing of PAX-5, RhoH/TTF, c-MYC, and PIM1 was generated on selected regions previously described to contain greater than 90% of mutations found in DLBCL.8 Rearranged IgHV genes were amplified with a seminested polymerase chain reaction (PCR) approach using primers as previously described.12 The IMGT database13 was used for calculating the IgHV mutational status. PCR products were purified and sequenced from both sides using the BigDye terminator chemistry 3.1 (Applied Biosystems, Foster City, CA). Sequences were run on an ABI3130-xl automated sequencer (Applied Biosystems). All oligonucleotides used in the initial PCR were also used for sequencing and are available in Table S1 (available on the Blood website; see the Supplemental Table link at the top of the online article). Sequences were confirmed by 2 independent PCR reactions. Nucleotide changes corresponding to previously published polymorphism were excluded from analysis. Further, all changes occurring more than once in separate cases were considered as polymorphic variants and were disregarded. Subcloning for intraclonal heterogeneity of selected cases using Pyrococcus furiosus (Pfu) Turbo polymerase (Stratagene, La Jolla, CA) was carried out as described previously.8
Analysis of AID mRNA/protein expression
Total RNA was extracted using Trizol method (Invitrogen, Carlsbad, CA) as previously described14 and reverse transcribed using cDNA Archive kit (Applied Biosystems). Quantitative real-time PCR assay was performed with the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) in triplicates. For quantification the AID mRNA TaqMan based Assay-on-Demand Gene Expression assays (Applied Biosystems) was used and normalized with β-actin. cDNA of centroblasts (CBs) was used as positive control and nontemplate, and reverse transcriptase (RT) minus control-containing RNA only served as negative control. For expression analysis the ΔΔCT values of each sample in comparison to the CBs were determined. The values were calculated as mean values of 3 independent measurements. Immunohistochemical staining for AID was performed using antibody EK2-5G9 as previously described.15
Statistical analysis
Statistical analysis was performed using SPSS 11.5 (SPSS, Chicago, IL). To calculate differences in AID mRNA levels in the experimental groups, Mann-Whitney U test was used. Spearman correlation test was performed to examine a correlation of AID expression with overall frequency of mutations.
Results and discussion
Seventeen samples of MALT lymphoma and 17 extranodal DLBCL samples were subjected to DNA sequence analysis of PAX5, RhoH/TTF, c-MYC, and PIM1 (Table 1). Overall mutations in at least one of the 4 genes were detected in 13 (76.5%) of 17 MALT lymphoma cases and in 17 (100%) of 17 extranodal DLBLC cases. Each of the 4 genes investigated was altered in a significant fraction (Table 2): in MALT lymphomas PIM1 was mutated in 8 (47.1%) of 17, RhoH/TTF in 2 (11.8%) of 17, cMYC in 10 (58.8%) of 17, and PAX5 in 3 (17.6%) of 17 cases. In DLBCL PIM1 was mutated in 11 (64.7%) of 17, RhoH/TTF in 8 (47.1%) of 17, cMYC in 13 (76.5%) of 17, and PAX5 in 10 (58.8%) of 17 cases. No mutations were found in control tissues. To confirm the somatic nature of these mutations the corresponding germ line DNA was analyzed in selected cases and demonstrated the tumor-specific origin of mutations. Fluorescence in situ hybridization (FISH) analysis using the IGH/MALT1 probe showed one translocation in an extranodal MALT lymphoma case but no other indication of involvement of the immunoglobulin heavy chain locus in 12 of 17 evaluable cases of MALT lymphomas and in 17 DLBC lymphomas. A detailed characterization of immunophenotypic markers11 revealed 5 of 17 cases of extranodal DLBCL of germinal center B (GCB) cell-like phenotype and the remaining of non-GCB cell origin.
Mutation analysis of PAX5, RhoH/TTF, cMYC, and PIM1 in extranodal DLBCL and MALT lymphoma
| . | PAX5 . | RhoH/TTF . | cMYC . | PIM1 . | IgH gene . | IgH mutations . |
|---|---|---|---|---|---|---|
| DLBCL | ||||||
| GHM1 | 799 C>T | 707 A>G | 3341 G>A | 1147 T>C | NA | NA |
| 810 C>T | 3377 A>T | 1157 T>C | ||||
| 1831 C>T | ||||||
| GHM2 | — | — | 3384 G>A | — | 1-69 | 7.62 |
| 3539 C>A | ||||||
| 5045 C>T | ||||||
| GHM3 | 819 G>A | — | 5021 C>T | — | 3-23 | 0 |
| 827 T>A | ||||||
| GHM4 | 1362 G>A | — | 5009 C>T | — | NA | NA |
| 5026 G>T* | ||||||
| 5044 G>T* | ||||||
| GHM5 | — | 755 A>G | 4568 C>T | — | 3-30-3 | 6.45 |
| GHM6 | 789 C>T | 764 A>T | 3613 A>G | 1355 C>A | NA | NA |
| 841 G>A | 1409 C>T* | |||||
| GHM7 | 718 C>T | — | 4638 C>T | 2041 G>A* | 4-34 | 2.98 |
| 2079 C>T | ||||||
| GHM8 | 890 G>A | 452 A>C | — | — | 3-30 | 4.04 |
| GHM9 | — | — | — | 1285 G>A | NA | NA |
| 1292 G>A | ||||||
| GHM10 | 1264 G>A | 755 A>G | — | — | NA | NA |
| EHM11 | — | — | 3472 T>C | 1905 G>T* | NA | NA |
| 5012 C>T | ||||||
| 5041 C>T* | ||||||
| EHM12 | 733 C>T | — | — | 1465 C>A* | 6-1 | 2.97 |
| 735 G>A | 1849 G>C* | |||||
| EHM15 | 1142 C>T | 329 C>T | 3522 A>G | 1747 G>A* | 3-30-3 | 8.16 |
| 1149 G>C | 4969 C>T* | 1749 G>A | ||||
| 1271 G>A | 4972 C>T* | |||||
| EHM16 | — | 443 C>T | 2577 C>T* | 1610 C>T* | 3-30-3 | 2.4 |
| 850 C>G | 2859 T>C* | |||||
| 4747 C>A* | ||||||
| EHM17 | — | — | 2746 C>T* | 1165 G>A | NA | NA |
| 4512 C>T | 1364 C>A | |||||
| EHM18 | 904 G>A | — | 3561 C>T | 1162 G>T | 2-70 | 2.12 |
| 1755 G>A | ||||||
| 1769 G>A* | ||||||
| EHM19 | — | 640 G>A | 4606 A>G* | 1998 ins G | 1-69 | 4.21 |
| 700 G>A | ||||||
| MALT | ||||||
| ENM14 | 1148 G>A | — | 2601 G>A* | — | 3-30-3 | 6.06 |
| 1150 G>T | 2640 G>A* | |||||
| ENM20 | 813 G>A | 486 T>G | 2821 C>G* | — | NA | NA |
| 1324 A>G | ||||||
| ENM21 | — | — | 3207 C>A | — | 1-69 | 5.52 |
| ENM29 | — | — | — | 1637 T>C | 1-69 | 0 |
| 1987 G>T* | ||||||
| ENM30 | — | — | — | 1802 A>G* | 4-34 | 0 |
| ENM33 | — | — | 2792 C>T | — | 1-69 | 2.17 |
| ENM35 | — | 649 C>T | 5042 C>T | 1148 G>T | 4-34 | 2.4 |
| GNM36 | — | — | 2494 G>A* | 1863 T>A* | 5-51 | 0 |
| GNM37 | 1366 G>A | — | — | 1123 G>C | 4-34 | 0 |
| GNM38 | — | — | 3388 C>T | — | 6-1 | 0 |
| GNM39 | — | — | 4700 C>G | 1598 A>T* | 5-51 | 0 |
| 4961 G>C* | ||||||
| GNM40 | — | — | 4618 A>C* | 1441 C>A | NA | NA |
| GNM41 | — | — | 2402 G>C* | 1463 G>A* | 4-34 | 0 |
| . | PAX5 . | RhoH/TTF . | cMYC . | PIM1 . | IgH gene . | IgH mutations . |
|---|---|---|---|---|---|---|
| DLBCL | ||||||
| GHM1 | 799 C>T | 707 A>G | 3341 G>A | 1147 T>C | NA | NA |
| 810 C>T | 3377 A>T | 1157 T>C | ||||
| 1831 C>T | ||||||
| GHM2 | — | — | 3384 G>A | — | 1-69 | 7.62 |
| 3539 C>A | ||||||
| 5045 C>T | ||||||
| GHM3 | 819 G>A | — | 5021 C>T | — | 3-23 | 0 |
| 827 T>A | ||||||
| GHM4 | 1362 G>A | — | 5009 C>T | — | NA | NA |
| 5026 G>T* | ||||||
| 5044 G>T* | ||||||
| GHM5 | — | 755 A>G | 4568 C>T | — | 3-30-3 | 6.45 |
| GHM6 | 789 C>T | 764 A>T | 3613 A>G | 1355 C>A | NA | NA |
| 841 G>A | 1409 C>T* | |||||
| GHM7 | 718 C>T | — | 4638 C>T | 2041 G>A* | 4-34 | 2.98 |
| 2079 C>T | ||||||
| GHM8 | 890 G>A | 452 A>C | — | — | 3-30 | 4.04 |
| GHM9 | — | — | — | 1285 G>A | NA | NA |
| 1292 G>A | ||||||
| GHM10 | 1264 G>A | 755 A>G | — | — | NA | NA |
| EHM11 | — | — | 3472 T>C | 1905 G>T* | NA | NA |
| 5012 C>T | ||||||
| 5041 C>T* | ||||||
| EHM12 | 733 C>T | — | — | 1465 C>A* | 6-1 | 2.97 |
| 735 G>A | 1849 G>C* | |||||
| EHM15 | 1142 C>T | 329 C>T | 3522 A>G | 1747 G>A* | 3-30-3 | 8.16 |
| 1149 G>C | 4969 C>T* | 1749 G>A | ||||
| 1271 G>A | 4972 C>T* | |||||
| EHM16 | — | 443 C>T | 2577 C>T* | 1610 C>T* | 3-30-3 | 2.4 |
| 850 C>G | 2859 T>C* | |||||
| 4747 C>A* | ||||||
| EHM17 | — | — | 2746 C>T* | 1165 G>A | NA | NA |
| 4512 C>T | 1364 C>A | |||||
| EHM18 | 904 G>A | — | 3561 C>T | 1162 G>T | 2-70 | 2.12 |
| 1755 G>A | ||||||
| 1769 G>A* | ||||||
| EHM19 | — | 640 G>A | 4606 A>G* | 1998 ins G | 1-69 | 4.21 |
| 700 G>A | ||||||
| MALT | ||||||
| ENM14 | 1148 G>A | — | 2601 G>A* | — | 3-30-3 | 6.06 |
| 1150 G>T | 2640 G>A* | |||||
| ENM20 | 813 G>A | 486 T>G | 2821 C>G* | — | NA | NA |
| 1324 A>G | ||||||
| ENM21 | — | — | 3207 C>A | — | 1-69 | 5.52 |
| ENM29 | — | — | — | 1637 T>C | 1-69 | 0 |
| 1987 G>T* | ||||||
| ENM30 | — | — | — | 1802 A>G* | 4-34 | 0 |
| ENM33 | — | — | 2792 C>T | — | 1-69 | 2.17 |
| ENM35 | — | 649 C>T | 5042 C>T | 1148 G>T | 4-34 | 2.4 |
| GNM36 | — | — | 2494 G>A* | 1863 T>A* | 5-51 | 0 |
| GNM37 | 1366 G>A | — | — | 1123 G>C | 4-34 | 0 |
| GNM38 | — | — | 3388 C>T | — | 6-1 | 0 |
| GNM39 | — | — | 4700 C>G | 1598 A>T* | 5-51 | 0 |
| 4961 G>C* | ||||||
| GNM40 | — | — | 4618 A>C* | 1441 C>A | NA | NA |
| GNM41 | — | — | 2402 G>C* | 1463 G>A* | 4-34 | 0 |
NA indicates not applicable; and —, not mutated.
Missense mutations.
Distribution and features of PAX5, RhoH/TTF, cMYC, and PIM1 mutations in extranodal DLBCL and MALT lymphoma
| Locus Entity . | Distribution, no. mutated/no. tested (%) . | Mutation frequency per 100 bp (range) . | Single base pair substitution . | G + C/A + T . | Transitions over transversions . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| DLBCL . | MALT . | DLBCL . | MALT . | DLBCL . | MALT . | DLBCL . | MALT . | DLBCL . | MALT . | |
| PAX5 | 10/17 (58.8) | 3/17 (17.6) | 0.12* (0.06-0.33) | 0.12* (0.11-0.49) | 15 | 5 | 14:1 | 4:1 | 13:2 | 4:1 |
| RhoH/TTF | 8/17 (47.1) | 2/17 (11.8) | 0.16* (0.07-0.38) | 0.09* (0.08-0.10) | 11 | 2 | 6:5 | 1:1 | 7:4 | 1:1 |
| cMYC | 13/17 (76.5) | 10/17 (58.8) | 0.04* (0.02-0.07) | 0.03* (0.02-0.08) | 25 | 12 | 20:5 | 11:1 | 20:5 | 6:6 |
| PIM1 | 11/17 (64.7) | 8/17 (64.7) | 0.14* (0.05-0.38) | 0.07* (0.05-0.10) | 20 | 9 | 18:2 | 5:4 | 14:6 | 3:6 |
| All genes | 17/17 (100) | 13/17 (76.5) | 0.12* | 0.08* | 71 | 28 | 58:13 | 21:7 | 54:17 | 14:14 |
| IgH | — | — | 3.98† (2.12-8.16) | 1.22† (0.97-5.52) | 65 | 28 | 51:14 | 23:5 | 30:35 | 17:11 |
| Locus Entity . | Distribution, no. mutated/no. tested (%) . | Mutation frequency per 100 bp (range) . | Single base pair substitution . | G + C/A + T . | Transitions over transversions . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| DLBCL . | MALT . | DLBCL . | MALT . | DLBCL . | MALT . | DLBCL . | MALT . | DLBCL . | MALT . | |
| PAX5 | 10/17 (58.8) | 3/17 (17.6) | 0.12* (0.06-0.33) | 0.12* (0.11-0.49) | 15 | 5 | 14:1 | 4:1 | 13:2 | 4:1 |
| RhoH/TTF | 8/17 (47.1) | 2/17 (11.8) | 0.16* (0.07-0.38) | 0.09* (0.08-0.10) | 11 | 2 | 6:5 | 1:1 | 7:4 | 1:1 |
| cMYC | 13/17 (76.5) | 10/17 (58.8) | 0.04* (0.02-0.07) | 0.03* (0.02-0.08) | 25 | 12 | 20:5 | 11:1 | 20:5 | 6:6 |
| PIM1 | 11/17 (64.7) | 8/17 (64.7) | 0.14* (0.05-0.38) | 0.07* (0.05-0.10) | 20 | 9 | 18:2 | 5:4 | 14:6 | 3:6 |
| All genes | 17/17 (100) | 13/17 (76.5) | 0.12* | 0.08* | 71 | 28 | 58:13 | 21:7 | 54:17 | 14:14 |
| IgH | — | — | 3.98† (2.12-8.16) | 1.22† (0.97-5.52) | 65 | 28 | 51:14 | 23:5 | 30:35 | 17:11 |
— indicates not mutated.
Mutation frequencies were calculated on the entire region analyzed and on mutated cases only, considering 2 alleles/gene/case.
Mutation frequencies for IgH were calculated on the entire region analyzed and on mutated cases only
The detailed characterization of the mutational profile is shown in Table 1 and their features are summarized in Table 2. A total of 28 sequence variants were detected in 13 of 17 MALT lymphoma cases and 72 sequence variants in 17 DLBCL cases. Although the mutations were almost exclusively single base pair substitutions, one insertion was also present. The average frequency of mutation taking into account only mutated cases for all 4 genes together was 1.5 times higher for the extranodal DLBCLs compared with the MALT lymphomas (MALT lymphomas: 0.08 × 10−2/bp [base pair] versus 0.12 × 10−2 /bp in extranodal DLBCL [P < .01]) but approximately 15 and approximately 33 times lower when compared with their respective IgHV mutations. No difference in frequency of mutation could be observed between GCB and non-GCB phenotype.
Of the 28 single base pair substitutions observed in the MALT lymphoma cases, 14 were transitions and 14 were transversions, corresponding to a transition-to-transversion ratio of 1.0 (expected, 0.5). In the DLBCL group 71 single base pair substitutions were found, 54 transitions and 17 transversions, with a ratio of 3.2. In addition the ratio of G + C to A + T was shifted toward a clear predominance of G + C mutations for both entities (3.0 and 4.5 for MALT and DLBC lymphoma, respectively). In PIM1 and cMYC a number of mutations were located in the coding regions and led to amino acid substitutions with potential functional consequences (Table 1). In PIM1 13 mutations in 5 MALT and 7 extranodal DLBC lymphoma cases represented missense mutation, 8 of them affected exon 4, which predicts a change in the structure and, possibly, the function of the PIM1 protein.16 In cMyc 9 of 17 missense mutations found in 2 MALT lymphomas and 5 extranodal DLBCLs affected exon 2.
cMYC exon 2 encodes for the transactivation domain and represents a mutational hot spot in translocated cMYC in Burkitt lymphomas.16,17 Alterations introduced by the ASHM in this region are likely to have a serious effect on c-MYC function.18,19 To investigate whether the aberrant mutational activity is ongoing in PIM1, PAX5, and RhoH/TTF, we selected a subset of cases (n = 7) for sequencing of their cloned PCR products generated using the proofreading Pfu polymerase. In all tumors analyzed one or 2 predominant alleles recapitulated the mutations observed by direct sequencing, confirming their presence in the tumor clone and revealing their frequent biallelic distribution. In 5 of 7 cases a significant number of intraclonal variants were found, indicating ongoing hypermutation in at least a fraction of patients.
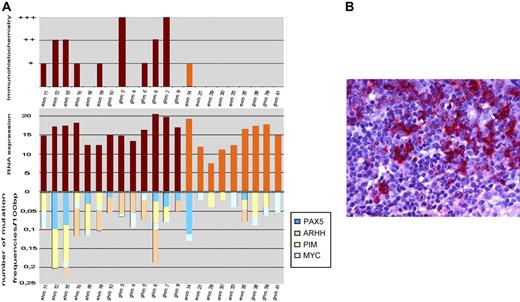
Because AID is an absolute requirement for SHM of Ig genes in GC B cells we analyzed AID mRNA and protein expression levels (Figure 1). Compared with centroblast RNA AID was expressed in all evaluable MALT lymphomas and DLCBLs, likewise. Among DLBCL no significant difference in expression levels between GCB cell and non-GCB cell lymphomas was visible. This is in line with previous reports in nodal DLBCL showing expression of AID mRNA or protein at various levels irrespective of GCB cell-like or activated B-cell–like subtype.20,21 There was a trend for increased AID expression levels in DLBCLs compared with MALT lymphomas. Comparing average frequency of mutations of non-Ig genes with AID RNA expression levels, a significant positive correlation could be observed (P = .016; correlation coefficient = 0.49 Spearmann Rho). However, this correlation could not be maintained when analyzing MALT lymphoma and extranodal DLBCL separately. This observation is in accordance with a publication reporting that AID protein expression does not correlate with ongoing SHM or ASHM in DLBCL,21 whereas most extranodal DLBCL expressed AID protein in only 2 of 13 evaluable MALT lymphomas stained positive for AID (Figure 1). Different sensitivity of test systems (immunohistochemistry versus RT-PCR) or post-transcriptional modifications may account for the lower incidence in MALT lymphomas. Comparing AID immunohistochemistry levels with IgHV mutation frequency, a significant correlation could be observed (P = .011; correlation coefficient = 0.67 Spearmann Rho).
Expression of AID in MALT lymphoma and extranodal DLBCL. (A) The comparison of activation-induced cytidine deaminase (AID) mRNA expression, immunohistochemistry, and frequency of mutations is shown. Compared with centroblast mRNA AID/β-actin was expressed in all mutated evaluable MALT lymphomas and DLCBL, likewise. (B) The positive staining for AID protein in neoplastic B cells of extranodal DLBCL is shown. Images were captured with a Leica DMLB microscope (Leica, Bensheim, Germany) using a Leica PL FLUOTAR objective lens (40×/0.70) and a Leica DC200 camera. Images were imported directly into PowerPoint (Microsoft, Redmond, WA) using the Leica DC200 camera software (version 2.51).
Expression of AID in MALT lymphoma and extranodal DLBCL. (A) The comparison of activation-induced cytidine deaminase (AID) mRNA expression, immunohistochemistry, and frequency of mutations is shown. Compared with centroblast mRNA AID/β-actin was expressed in all mutated evaluable MALT lymphomas and DLCBL, likewise. (B) The positive staining for AID protein in neoplastic B cells of extranodal DLBCL is shown. Images were captured with a Leica DMLB microscope (Leica, Bensheim, Germany) using a Leica PL FLUOTAR objective lens (40×/0.70) and a Leica DC200 camera. Images were imported directly into PowerPoint (Microsoft, Redmond, WA) using the Leica DC200 camera software (version 2.51).
The identification of an active aberrant somatic hypermutation mechanism in extranodal DLBCL, which, in our series, are exclusively transformed MALT lymphomas still displaying a low-grade component, expands the type of aggressive lymphoma associated with this molecular abnormality.22-24 Notably, MALT lymphomas representing putative precursor lesions of extranodal DLBCL are also targeted by the aberrant somatic hypermutation process albeit at lower frequencies. Because frequency of mutations and AID protein/RNA expression are both lower in MALT lymphomas, one might speculate an active involvement of ASHM in the high-grade transformation process of MALT lymphoma to overt DLBCL, resembling that described for Richter and prolymphocytic transformation in chronic lymphocytic leukemia.25 By mutating regulatory and coding sequences of the targeted genes, ASHM may represent a major contributor to their pathogenesis.
Authorship
Contribution: P.N. designed and supervised the project and wrote the manuscript; A.J.A.D. performed most of the experiments, analyzed data, and wrote the manuscript; A.A., A.B., and C.B.-S. rediagnosed and selected all specimen; P.B.S. performed the statistics; M.F. and W.E. performed and analyzed the cytogentics; C.G. performed the real-time PCR of the AID gene; M.B. made the immunohistochemistry; R.I.B. contributed to data analyses; W.L. provided the laboratory facility.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter Neumeister, Auenbruggerplatz 38, A-8036 Graz, Austria; e-mail: peter.neumeister@meduni-graz.at.
Presented in abstract form at the 47th annual meeting of the American Society of Hematology, Atlanta, GA, December 12, 2005.26
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Riccardo Dalla-Favera, Institute of Cancer Genetics, Columbia University, NY, for providing centroblast RNA.
This work was supported by grants from Leukämiehilfe Steiermark, Österreichische Krebshilfe, and Jubiläumsfond der ÖNB (N11181).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal