Abstract
B-cell chronic lymphocytic leukemia (CLL) is characterized by the accumulation of clonal B cells that are resistant to apoptosis as a result of bcl2 oncogene overexpression. Studies were done to determine the mechanism for the up-regulation of bcl-2 protein observed in CD19+ CLL cells compared with CD19+ B cells from healthy volunteers. The 11-fold higher level of bcl-2 protein in CLL cells was positively correlated with a 26-fold elevation in the cytosolic level of nucleolin, a bcl2 mRNA–stabilizing protein. Measurements of the bcl2 heterogeneous nuclear/bcl2 mRNA (hnRNA)/mRNA ratios and the rates of bcl2 mRNA decay in cell extracts indicated that the 3-fold higher steady-state level of bcl2 mRNA in CLL cells was the result of increased bcl2 mRNA stability. Nucleolin was present throughout the nucleus and cytoplasm of CLL cells, whereas in normal B cells nucleolin was only detected in the nucleus. The addition of recombinant human nucleolin to extracts of normal B cells markedly slowed the rate of bcl2 mRNA decay. SiRNA knockdown of nucleolin in MCF-7 cells resulted in decreased levels of bcl2 mRNA and protein but no change in β-actin. These results indicate that bcl-2 overexpression in CLL cells is related to stabilization of bcl2 mRNA by nucleolin.
Introduction
Overexpression of bcl-2 protein is thought to allow cells that are genetically unstable to avoid apoptosis and become tumorigenic. In addition to its importance in cancer development, high bcl2 expression in hematologic tumors is frequently an obstacle to cancer chemotherapy, since bcl2 overexpression has been shown to confer cellular resistance to a variety of anticancer drugs.1,2 The clinical relevance of bcl2 overexpression is clearly evident in the development of B-cell chronic lymphocytic leukemia (CLL). CLL is the most prevalent form of adult leukemia in the Western world and is considered an incurable disease. CLL is indolent during most of its clinical course and the clonal B cells accumulate during the indolent phase by avoiding apoptosis.3 High-level expression of bcl2 mRNA and protein is often seen in CLL despite the absence of evidence for gene rearrangements that are known to enhance bcl2 transcription.4-6 This raised the question whether overexpression of bcl-2 protein in CLL is a consequence of increased bcl2 mRNA stability.
The majority of the elements that regulate mRNA stability map to the 3′-untranslated region (3′-UTR) of mRNAs. The 3′-UTR has been described as “a molecular hotspot for pathology,”7 and modifications of specific elements within the 3′-UTR can profoundly affect the expression and metabolic fate of the mRNA.8 Prominent among these elements are the AU-rich elements (AREs). AREs generally contain multiple copies of the AUUUA pentamer and have a high content of U or A-U. AUUUA motifs are often associated with destabilization of short-lived cytokine and protooncogene mRNAs.9,10 The 3′-UTR of bcl2 mRNA contains 4 potential AREs. ARE-1 has the highest concentration of AUUUA pentamers of the 4 AREs and has potent bcl2 mRNA–destabilizing activity.11,12 Examination of different mRNAs suggests that the destabilizing effects of an ARE and AUUUA motifs can be increased or decreased by interactions with ARE-binding proteins.13,14 The manner in which ARE-binding proteins modulate mRNA decay is not completely clear. With some mRNAs, binding of specific proteins to the ARE stabilizes the mRNA in a circular form and impedes deadenylation of the poly(A) tail by poly(A) ribonuclease (PARN).15,16 In contrast, following shortening of the poly(A) tail, the binding of destabilizing proteins to the ARE can result in recruitment of an exosome to the ARE mRNAs, leading to rapid degradation of the mRNA.17,18
Nucleolin is a multifunctional protein that is a member of the RNP-containing family of RNA-binding proteins. This protein binds to the 3′-UTR of amyloid precursor protein (APP) mRNA and enhances APP mRNA stability.19,20 Nucleolin is also required for the stabilization of IL-2 mRNA that occurs during T-cell activation.21 Recent studies have identified nucleolin as a bcl2 mRNA–stabilizing protein in HL-60 leukemia cells.22,23 It binds specifically to the ARE-1 instability element in the 3′-UTR of bcl2 mRNA and protects bcl2 mRNA from ribonuclease degradation. However, it is not known whether the increased levels of bcl2 mRNA and protein in CLL cells are related to stabilization of bcl2 mRNA by nucleolin. To address this question, the studies described herein examined the stability of bcl2 mRNA in CLL cells isolated from patients compared with normal B cells from healthy volunteers. This novel, posttranscriptional mechanism of bcl2 overexpression in CLL proposed here may provide an answer to the long-standing question regarding the mechanism by which bcl2 mRNA and protein are overexpressed in CLL in the absence of enhanced bcl2 transcription. Stabilization of bcl2 mRNA by nucleolin would also be consistent with the indolent nature of this disease in which CLL cells accumulate by avoiding apoptosis.
Patients, materials, and methods
Isolation of CD19+ CLL cells and CD19+ normal B cells
Peripheral-blood samples were obtained from CLL patients and healthy volunteers after informed consent according to our human research protocol approved by the institutional review board (IRB) of the Medical University of South Carolina (Human Research [HR] no. 10967). Mononuclear cells were isolated from the blood samples by Ficoll-Isopaque centrifugation and the B lymphocytes were purified from this fraction by immunomagnetic separation using CD19 microbeads (Miltenyi Biotec, Auburn, CA). Flow-cytometric analysis revealed that at least 90% of either the normal or the CLL cells in the purified B-cell fractions were CD19+ but negative for the T-cell antigen CD3.
Immunoblot analysis
Immunoblotting was done as previously described.23 For determination of cytosolic nucleolin and total cellular bcl-2 proteins, freshly isolated CD19+ B cells were lysed for 15 minutes on ice in lysis buffer,23 followed by centrifugation at 10 000g for 15 minutes at 4°C to yield S10 extracts. Protein concentrations were determined by the BCA assay (Pierce, Rockford, IL). Aliquots of the S10 extracts containing various amounts of protein (5 μg to 40 μg) were electrophoresed on an 8% to 16% polyacrylamide SDS gel and transblotted. All antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The amounts of each protein were determined by counting the total numbers of pixels in each band (integrated density value) with a ChemiImager digital imaging system (Alpha Innnotech, San Leandro, CA) and/or a Typhoon PhosphorImager (GE Healthcare, Piscataway, NJ). Values that were within the linear range of the assay were normalized to known amounts of external standards of nucleolin or bcl-2 proteins from HL-60 cell extracts that were run on every gel. This allowed for a more accurate comparison of the results among different patient samples.
Confocal microscopy
CLL cells and normal B cells were placed on poly-d-lysine–coated microwell dishes, fixed in 4% paraformaldehyde in PBS for 15 minutes at room temperature, and then permeabilized with 0.2% Triton X-100 in PBS for 10 minutes. Nonspecific binding of antibody was blocked with 1% bovine serum albumin and 5% goat serum in PBS for 1 hour at room temperature. The dishes were incubated overnight at 4°C with primary antinucleolin antibody (1:100 dilution in blocking buffer), washed 3 times in PBS, and incubated with secondary FITC-conjugated goat anti–mouse IgG (diluted 1:500 in blocking buffer) for 1 hour at room temperature. RNA was digested with RNase A (100 μg/mL for 15 minutes at room temperature), and propidium iodide (1 μg/mL) was used to stain DNA. The cells were washed 3 times in PBS and then observed under a Carl Zeiss LSM Pascal confocal microscope (Thornwood, NY). Confocal images (1024 × 768 pixels) were obtained using a 63× objective lens, and the images were overlaid using Carl Zeiss LSM Pascal image browser 4.0 software.
Bcl2 mRNA and hnRNA determination by reverse transcriptase–polymerase chain reaction (RT-PCR)
Total RNA was isolated from freshly purified B lymphocytes (0.5 × 107 to 1 × 107 cells) using an RNeasy mini kit (Qiagen, Valencia, CA), according to the manufacturer's protocol. The samples were subjected to on-column digestion of DNA with RNase-free DNase (Qiagen) during RNA purification. No genomic DNA contamination was detected in the RNA samples after PCR amplification without prior reverse transcription (data not shown). RNA concentrations were determined spectrophotometrically at 260 nm. Equal amounts of total RNA (2 μg) from each sample were reverse transcribed as previously described.23 To ensure that PCR product formation was linear with respect to the amount of cDNA, typically 6 PCR reactions were carried out containing various amounts of cDNA from each CLL and normal B-cell sample. The primer pairs for bcl2 mRNA cDNAs (5′ GAGGATTGTGGCCTTCTTTG 3′ and 5′AGCCTGCAGCTTTGTTCCAT 3′) were used to amplify a 424-bp sequence overlapping the first and second exons. Primer pairs for bcl2 heterogeneous nuclear bcl2 mRNA (hnRNA) cDNAs (5′ TGATGTGAGTCTGGGCTGAG 3′ and 5′ GAACGCTTTGTCCAGAGGAG 3′) were used to amplify a 152-bp sequence found in the first intron of bcl2 hnRNA, which was specific for a primary unprocessed transcript. All PCR primers were obtained from Integrated DNA Technologies (Coralville, IA). PCR amplifications for both bcl2 mRNA and hnRNA were performed in a single reaction under the following conditions: HotStar Taq polymerase activation for 15 minutes at 95°C, 28 cycles of template denaturation for 1 minute at 94°C, primer-template annealing for 1 minute at 55°C, and primer extension for 1 minute at 72°C, and a final extension reaction for an additional 7 minutes at 72°C. PCR amplification of a 232-bp sequence of β-actin gene was performed similarly using specific primer pairs (5′GCGGGAAATCGTGCGTGACAT 3′ and 5′ GATGGAGTTGAAGGTAGTTC 3′) with the exceptions that 23 cycles of PCR amplification and an annealing temperature at 57°C were used. The PCR products were resolved on a 2% agarose gel, stained with ethidium bromide, and visualized with a Typhoon PhosphorImager. Product formation was quantitated by determining the integrated density value of each band using ImageQuant TL software (GE Healthcare) and normalized to the amount of β-actin gene product.
Binding of Bcl2 ARE-1 to nucleolin in extracts of CLL and normal B cells
[32P]ARE-1 RNA transcripts were synthesized in in vitro transcription reactions using [32P]uridine triphosphate (GE Healthcare) as previously described.22 CD19+ B cells were obtained from either 2 CLL patients or 2 healthy human volunteers. Cytoplasmic S100 extracts were prepared from the freshly isolated cells by incubating the cells in lysis buffer for 20 minutes on ice, followed by successive centrifugation of the supernatants at 10 000g for 2 minutes at 4°C and then at 100 000g for 1 hour at 4°C. Aliquots of the S100 fractions containing 100 μg of protein were precleared with protein G agarose beads (Santa Cruz Biotechnology) and mouse IgG (1 μg) in lysis buffer containing 150 mM KCl. The S100 fractions were then incubated with 0.25 nM (25 fmol) [32P]ARE-1 RNA (final specific radioactivity of 90 mCi (3.33 ×109 Bq)/mmol) and 2 μg of antinucleolin monoclonal antibody or mouse IgG (control antibody) for 3.5 hours at 4°C. The immunocomplexes were precipitated with protein G agarose beads, washed twice with lysis buffer, and then analyzed by liquid scintillation counting. Results were from 2 healthy volunteers and 2 CLL patients with 3 determinations per individual.
In vitro mRNA decay assays
Spe-I linearized pCR4-bcl-CR ARE plasmid22 was used as a template for synthesis of a transcript containing a portion of the bcl2 coding region (nucleotides 600-750) and the ARE (nucleotides 751-1057). The 5′-capped, 32 P-labeled transcripts were synthesized by T7 RNA polymerase, using a mMessage mMachine T7 kit (Ambion, Austin, TX), following the manufacturer's instructions. Poly(A) tails of approximately 150 nucleotides were added to the 3′ ends of the transcript using a poly(A) tailing kit (Ambion), and unincorporated ribonucleoside triphosphates (NTPs) were removed by G-25 spin column chromatography. Approximately 150 000 counts per minute (cpm) of capped and polyadenylated CR-ARE RNAs were used per decay reaction, which was performed as described by Ford and Wilusz.24 Typically, a 70-μL reaction mixture contained 16 μL of 10% polyvinyl alcohol, 5 μL of a 12.5 mM ATP/250 mM phosphocreatine mixture, 5 μL of 500 ng/μL poly(A) (GE Healthcare), 5 μL 32P-labeled transcript (∼ 175 nM), and 8 to 10 μg of protein from either CLL or normal B-cell S100 extracts. In some of the reactions, purified recombinant nucleolin was added. Recombinant nucleolin was generated using a bacterial expression vector (pET21a) containing cDNA sequences that code for residues 284-707 of human nucleolin [Δ1-283 Nuc-(His)6].25 The histidine-tagged nucleolin fragment was expressed in Escherichia coli and purified on a Ni++-NTA column as previously described.22 Samples were incubated at 30°C and the reaction was stopped at various times by transferring 15-μL aliquots to 100 μL of stop buffer (400 mM NaCl; 25 mM Tris-HCl, pH 7.5; 0.1% SDS) and immediately extracting with 100 μL of phenol-chloroform. RNA was ethanol precipitated and then electrophoresed on 7 M urea–6% polyacrylamide gels. After electrophoresis, gels were fixed, dried, and analyzed by phosphorimaging.
Generation of nucleolin SiRNA transfectants
The Ambion web-based target sequence converter was used to convert siRNA target sites into double-stranded DNA fragments with BamHI and HindIII sticky ends. A negative control vector that expresses a hairpin siRNA with limited homology to any known sequences in the human genome was provided with the vector kit. Briefly, a double-stranded oligonucleotide targeting the nucleotides of the human nucleolin sequence 5′ AAGACAGTGATGAAGAGGAGG 3′ (GenBank accession no. NM_005381) was cloned into the BamHI and HindIII sites of the pSilencer 2.0-U6 vector (Ambion). The cells were transfected with 10 μg of either Hnuc (nucleolin siRNA) or scrambled siRNA plasmids using Lipofectamine2000 (Invitrogen, Carlsbad, CA), and after 48 hours the cells were selected in medium containing 500 μg/mL of G418. The medium was replaced daily with fresh G418 medium. After 8 days, resistant clones were picked and the cells were propagated for 20 days prior to analysis of nucleolin and bcl-2 protein levels. The nucleolin and scrambled siRNA sequences in the G418-resistant clones were confirmed by sequencing.
Total RNA was extracted from the transfectants using Trizol reagent (Invitrogen) according to the manufacturer's protocol. Expression of nucleolin and bcl2 mRNA levels was analyzed by quantitative polymerase chain reaction (qPCR) in the stable clones. cDNA synthesis was performed using 1 μg total RNA as described.23 The primers for nucleolin and bcl2 were 5′ CCA GCCATCCAAAACTCTGT 3′ and 5′ TAACTATCCTTGCCCGAACG 3′ and 5′ ATGTGTGTGGAGAGCGTCAA 3′ and 5′ ACAGTTCCACAAAGGCATCC 3′, respectively. The primers for β-actin were 5′ AAATCTGGCACCACACCTTC3′ and 5′ GGGGTGTTGAAGGTCTCAAA 3′. All primers were from Integrated DNA Technologies. cDNA (1 μg) was amplified using a Brilliant SYBR Green QPCR Master Mix from Stratagene (La Jolla, CA). The reaction was carried out at 95°C for 10 minutes, followed by 40 cycles of 95°C for 30 seconds, 53°C for 90 seconds, and 72°C for 60 seconds. Nucleolin and bcl2 mRNAs were quantified and normalized relative to β-actin mRNA. Each reaction was performed in duplicate and the comparative cycle threshold (Ct) method was used for relative quantification of gene expression.
Results
Patient characteristics
Seventeen patients with CLL were studied. Although selected only on the basis of willingness to donate blood cells, the subjects were predominantly early stage and untreated (Table 1). Median age, sex, and absolute lymphocyte counts were typical of CLL patients in general. Interphase cytogenetic analysis by fluorescent in situ hybridization, using a limited panel of probes, was available for 8 subjects: 2 had no abnormalities, and 5 had chromosome 13 deletions involving band q14, either alone (3 patients) or with mutation of ATM (1 patient), or duplicated chromosome 12 (1 patient). A single patient had an isolated p53 mutation.
Subject characteristics
| Characteristics . | Data . |
|---|---|
| Total, no. | 17 |
| No. men/no. women | 9/8 |
| Median age, y (range) | 68 (49-88) |
| Rai stage, no. | |
| 0 | 5 |
| 1 | 5 |
| 2 | 3 |
| 3 | 3 |
| 4 | 1 |
| Prior therapy, no. | |
| None | 11 |
| Chlorambucil | 2 |
| Chlorambucil, rituximab | 3 |
| Cyclophosphamide, prednisone, rituximab | 1 |
| Median absolute lymphocyte count × 109/L (range) | 32.5 (1.8-224.4) |
| CD38−, no. | 16 |
| CD38+, no. | 1 |
| Interphase cytogenetics, FISH, no. | |
| del 13q14 | 3 |
| del 13q14, +12 | 1 |
| del 13q14, ATM | 1 |
| p53 | 1 |
| No abnormalities | 2 |
| Not available | 9 |
| Characteristics . | Data . |
|---|---|
| Total, no. | 17 |
| No. men/no. women | 9/8 |
| Median age, y (range) | 68 (49-88) |
| Rai stage, no. | |
| 0 | 5 |
| 1 | 5 |
| 2 | 3 |
| 3 | 3 |
| 4 | 1 |
| Prior therapy, no. | |
| None | 11 |
| Chlorambucil | 2 |
| Chlorambucil, rituximab | 3 |
| Cyclophosphamide, prednisone, rituximab | 1 |
| Median absolute lymphocyte count × 109/L (range) | 32.5 (1.8-224.4) |
| CD38−, no. | 16 |
| CD38+, no. | 1 |
| Interphase cytogenetics, FISH, no. | |
| del 13q14 | 3 |
| del 13q14, +12 | 1 |
| del 13q14, ATM | 1 |
| p53 | 1 |
| No abnormalities | 2 |
| Not available | 9 |
Overexpression of nucleolin and Bcl2 proteins in CLL cells compared with normal B cells
Nucleolin has recently been identified as a bcl2 mRNA–stabilizing protein in HL-60 leukemia cells.22,23 Thus, the initial studies compared the relative levels of nucleolin and bcl-2 proteins in purified CD19+ CLL cells and normal CD19+ B cells by immunoblotting S10 extracts of these cells. We chose to analyze S10 extracts in order to permit measurement of nonnuclear nucleolin and mitochondrial bcl-2 protein in the same cell extract. In particular, nucleolin in the cytoplasm would be directly involved in bcl2 mRNA stabilization.
To accurately compare the immunoblot results from different patients, the integrated density values (IDVs) of the nucleolin and the bcl-2 protein bands in the immunoblots were normalized to the IDVs obtained from known amounts of nucleolin and bcl-2 protein external standards. This analysis revealed that bcl-2 and nucleolin proteins were 11-fold elevated (P < .001) and 26-fold elevated (P < .001), respectively, in CLL cells from 17 patients compared with B cells from 9 healthy volunteers (Figure 1). In addition, the enhanced bcl-2 protein levels were positively correlated with the increased nucleolin levels (Pearson correlation = 0.83, P < .001). Total cellular nucleolin levels were higher in CLL cells than normal B cells, primarily as a result of the much higher levels of cytoplasmic nucleolin in the CLL cells. No statistically significant differences in the nuclear levels of nucleolin were observed between CLL and normal B cells (P > .05). The nuclear protein histone 2B was not detected in Western blots of the S10 extracts from either CLL or normal B cells (data not shown). Thus, the presence of high levels of nucleolin in the S10 cytoplasmic extracts of CLL cells was not consistent with contamination of the S10 extracts with nuclear nucleolin.
Overexpression of nucleolin and bcl-2 proteins in CLL cells compared with normal B cells. Peripheral-blood lymphocytes were isolated from CLL patients and healthy volunteers by density gradient centrifugation and the B cells were purified by positive selection with magnetic-activated cell separation (MACS) CD19+ immunomagnetic beads. Nucleolin and bcl-2 protein levels were measured in S10 extracts of the cells by Western blotting. The results were normalized to the values obtained from known amounts of nucleolin and bcl-2 protein external standards. The labels N and C along the X-axis refer to normal B cells and CLL cells, respectively, from individual subjects.
Overexpression of nucleolin and bcl-2 proteins in CLL cells compared with normal B cells. Peripheral-blood lymphocytes were isolated from CLL patients and healthy volunteers by density gradient centrifugation and the B cells were purified by positive selection with magnetic-activated cell separation (MACS) CD19+ immunomagnetic beads. Nucleolin and bcl-2 protein levels were measured in S10 extracts of the cells by Western blotting. The results were normalized to the values obtained from known amounts of nucleolin and bcl-2 protein external standards. The labels N and C along the X-axis refer to normal B cells and CLL cells, respectively, from individual subjects.
Results from confocal microscopy studies (Figure 2) were consistent with the immunoblotting data. Figure 2 shows representative images from 30 images of CLL cells from each of 3 patients and 30 images of B cells from each of 2 healthy human volunteers. The intracellular localization of nucleolin was determined by indirect immunofluorescence using primary antibody against nucleolin and an FITC-conjugated anti–mouse IgG secondary antibody (green fluorescence). The DNA was stained with propidium iodide (red fluorescence). The overlay images in Figure 2 (yellow fluorescence) indicate that nucleolin was present throughout the nucleus and cytoplasm of CLL cells, whereas in normal B cells nucleolin was concentrated in nucleoli and also located in the nucleoplasm. It is interesting that in CLL cells, extensive staining of nucleolin was observed along the periphery of the cells. This observation seems consistent with reports that nucleolin is present in the plasma membranes of certain cells,26,27 although further studies will be required to verify the localization of nucleolin in the plasma membrane of CLL cells.
Confocal microscopy images of nucleolin localization in normal B cells and CLL cells. The localization of nucleolin was determined by indirect immunofluorescence using a primary antibody against nucleolin and FITC-conjugated antimouse secondary antibody. DNA was stained with propidium iodide.
Confocal microscopy images of nucleolin localization in normal B cells and CLL cells. The localization of nucleolin was determined by indirect immunofluorescence using a primary antibody against nucleolin and FITC-conjugated antimouse secondary antibody. DNA was stained with propidium iodide.
Bcl2 mRNA stability is increased in CLL cells relative to normal B cells
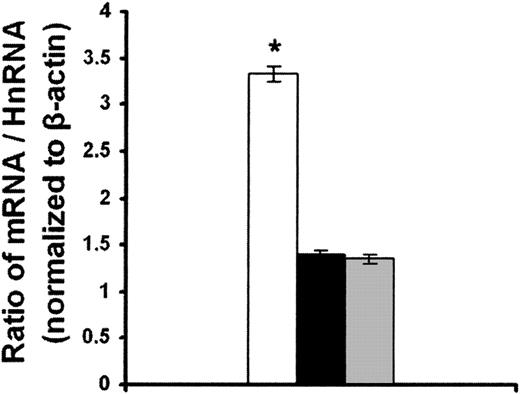
Overexpression of bcl-2 protein in CLL cells compared with normal B cells could result from enhanced bcl2 mRNA transcription, increased bcl2 mRNA stability, or increased efficiency of bcl2 mRNA translation. It is difficult to measure mRNA stability in primary CLL cells with the standard method using actinomycin D to block transcription. bcl2 mRNA is very stable in CLL cells requiring long incubation times with actinomycin D, which is toxic to the cells. To circumvent this problem, we measured the levels of nascent, unspliced hnRNA and mature bcl2 mRNA in CLL cells and normal B cells from healthy volunteers. This method has been used successfully to determine the relative rate of mRNA transcription and mRNA decay in a variety of cells.28,29 Equal amounts of total RNA from each sample were reverse transcribed, and PCR was performed with 2 sets of primers. One reaction contained primers that anneal to the first intron (to selectively amplify hnRNA) and one contained primers that anneal to sequences in 2 adjacent exons (to selectively amplify spliced, mature RNA). We found that the ratio of bcl2 mRNA to bcl2 hnRNA was about 3-fold higher for CLL cells compared with normal B cells (P < .001; Figure 3). The 3-fold higher bcl2 mRNA/bcl2 hnRNA ratio for CLL cells was entirely the result of an increase in the bcl2 mRNA level in CLL cells (3.3 ± 0.4 SEM relative to β-actin mRNA) compared with the bcl2 mRNA level in normal B cells (1.1 ± 0.2 SEM relative to β-actin mRNA). No significant difference was observed in the level of bcl2 hnRNA in CLL cells (6.5 ± 1.4 SEM relative to β-actin mRNA) versus normal B cells (5.5 ± 1.4 SEM relative to β-actin mRNA). These results indicate that bcl2 mRNA is relatively more stable in CLL cells compared with normal B cells. In contrast, if the rate of bcl2 mRNA transcription was relatively higher in CLL cells compared with normal B cells, then the bcl2 mRNA/hnRNA ratio would have been lower in CLL versus normal B cells.
Relative levels of expression of bcl2 hnRNA and bcl2 mRNA in CLL and normal B cells. The levels of bcl2 hnRNA and bcl2 mRNA in CLL and normal B cells from 4 CLL patients and 4 healthy volunteers were determined by RT-PCR. Results are expressed as the means of 4 determinations per group ± SEM. *P < .001 compared with normal B cells.
Relative levels of expression of bcl2 hnRNA and bcl2 mRNA in CLL and normal B cells. The levels of bcl2 hnRNA and bcl2 mRNA in CLL and normal B cells from 4 CLL patients and 4 healthy volunteers were determined by RT-PCR. Results are expressed as the means of 4 determinations per group ± SEM. *P < .001 compared with normal B cells.
To further investigate whether bcl2 mRNA is stabilized in CLL cells, the stability of bcl2 RNA transcripts was examined in extracts prepared from purified CLL cells and normal B cells using an in vitro RNA decay system.24 Capped and polyadenylated mRNAs were used in these assays to mimic in vivo decay, which involves cap-stimulated deadenylation by poly (A) ribonuclease (PARN) followed by rapid decay of the mRNA body by the exosome.8 32P-labeled bcl-2–CR RNA or bcl-2–CR-ARE RNA transcripts were incubated with cytoplasmic S100 extracts from CLL and normal B cells in the presence of poly(A) to activate deadenylation. As shown in Figure 4, bcl2 transcripts decayed more rapidly in extracts of normal B cells than in extracts of CLL cells. The average half-life of bcl2 RNA in cytoplasmic extracts of CLL cells from 4 patients was estimated to be 72 minutes by extrapolation of the data from Figure 4, whereas the average half-life of the transcript in extracts of normal B cells from 3 healthy volunteers was 12 minutes. The rapid decay of the bcl2–CR-ARE RNA transcripts in normal B-cell extracts was highly ARE dependent, since the rates of decay of bcl2 mRNA coding region transcripts lacking the ARE (bcl2–CR RNA) were similar in normal B-cell and CLL cell extracts. It is important to note that addition of 280 nM purified recombinant nucleolin [Δ1-283 Nuc-(His)6] to extracts of normal B cells greatly slowed the decay rate of bcl2-ARE (extrapolated half-life of 62 minutes; Figure 4). Thus, in vitro decay assays also support the conclusion that bcl-2 mRNA is stabilized in CLL cells relative to normal B cells and suggest a role for nucleolin in modulating bcl2 mRNA stability.
Decay of bcl2 mRNA in extracts of CLL and normal B cells. The 5′-capped and polyadenylated [32P]bcl-2–CR RNA and [32P]bcl-2–CR-ARE RNA were incubated with S100 extracts prepared from either CLL cells from 4 patients or normal B cells from 3 human volunteers. At the indicated times, aliquots of the reaction mixtures were removed and analyzed by polyacrylamide gel electrophoresis and filmless phosphorimaging. The results are expressed as the mean percentage of full-length RNA remaining ± SEM as a function of time. ○ indicates CLL cell extract + [32P]bcl-2–CR RNA; •, CLL cell extract + [32P]bcl-2–CR-ARE RNA; □, normal B-cell extract + [32P]bcl-2–CR RNA; ▪, normal B-cell extract + [32P]bcl-2–CR-ARE RNA; ▴, normal B-cell extract + [32P]bcl-2–CR-ARE RNA + 280 nM purified recombinant nucleolin.
Decay of bcl2 mRNA in extracts of CLL and normal B cells. The 5′-capped and polyadenylated [32P]bcl-2–CR RNA and [32P]bcl-2–CR-ARE RNA were incubated with S100 extracts prepared from either CLL cells from 4 patients or normal B cells from 3 human volunteers. At the indicated times, aliquots of the reaction mixtures were removed and analyzed by polyacrylamide gel electrophoresis and filmless phosphorimaging. The results are expressed as the mean percentage of full-length RNA remaining ± SEM as a function of time. ○ indicates CLL cell extract + [32P]bcl-2–CR RNA; •, CLL cell extract + [32P]bcl-2–CR-ARE RNA; □, normal B-cell extract + [32P]bcl-2–CR RNA; ▪, normal B-cell extract + [32P]bcl-2–CR-ARE RNA; ▴, normal B-cell extract + [32P]bcl-2–CR-ARE RNA + 280 nM purified recombinant nucleolin.
Binding of exogenous Bcl2 ARE-1 mRNA to nucleolin is greater in extracts of CLL cells compared with extracts of normal B cells
Further experiments were done to address the question whether the up-regulation of cytoplasmic nucleolin in CLL cells (Figures 1–2) results in increased interaction of nucleolin with bcl2 ARE-1 RNA. S100 fractions from CLL and normal B cells were incubated with [32P]ARE-1 RNA, and the nucleolin–ARE-1 complexes were coimmunoprecipitated with antinucleolin monoclonal antibody. The immunoprecipitates were analyzed by liquid scintillation counting. Precipitation of [32P]ARE-1 RNA was about 5-fold greater in CLL extracts incubated with antinucleolin antibody (7.8 ± 2.5 SE fmol) compared with CLL extracts incubated with control IgG antibody (1.6 ± 1 SE fmol). Importantly, the amount of [32P]ARE-1 RNA precipitated in extracts of CLL cells was 6.5-fold greater than that recovered from normal B-cell extracts (7.8 ± 2.5 SE fmol vs 1.2 ± 0.7 SE fmol).
Knockdown of nucleolin decreases Bcl2 mRNA stability and Bcl-2 protein levels in stable clones of MCF-7 cells
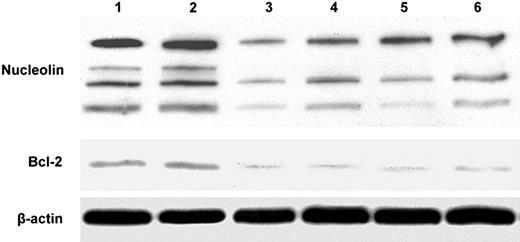
If nucleolin is an important transacting factor required for the stabilization of bcl2 mRNA, then knockdown of nucleolin with a siRNA should lead to destabilization of bcl2 mRNA. Because it is very difficult to generate stable transfectants of CLL cells obtained directly from patients, nucleolin was knocked down in MCF-7 breast cancer cells by transfecting the cells with a plasmid containing a nucleolin siRNA. Control MCF-7 cells were transfected with a plasmid expressing a scrambled siRNA with limited homology to any known human genomic sequence. Confocal microscopy using an FITC-conjugated antinucleolin monoclonal antibody revealed high-level expression of nucleolin in the cytoplasm of MCF-7 cells. Nucleolin and bcl2 mRNA levels were determined by real-time PCR analysis in 2 MCF-7 clones transfected with a scrambled siRNA and 5 clones transfected with the nucleolin siRNA. The levels of nucleolin mRNA and bcl2 mRNA in the stable nucleolin siRNA transfected clones were reduced to 24% ± 3% SEM and 17% ± 5% SEM, respectively, of the corresponding levels measured in the 2 MCF-7 clones transfected with the scrambled siRNA. To compare the relative degrees of bcl2 mRNA stability in the nucleolin siRNA transfectants versus the scrambled siRNA transfectants, we measured the levels of heterogeneous nuclear bcl2 RNA and mature bcl2 mRNA in these transfectants as described in Figure 3. The ratio of the bcl2 mRNA level to the bcl2 hnRNA level was about 2.5-fold higher for the scrambled siRNA transfectant compared with the 2 nucleolin siRNA transfectants (Figure 5). This suggests that bcl2 mRNA is relatively less stable in the nucleolin siRNA-transfected MCF-7 clones than in a scrambled siRNA transfectant. Western-blot analysis of the transfectants revealed that the levels of full-length nucleolin (106 kDa) and its proteolysis products30 were down-regulated in all 5 clones transfected with the nucleolin siRNA compared with the 2 clones transfected with the scrambled siRNA (Figure 6). Equally important, nucleolin knockdown was accompanied by down-regulation of bcl-2 protein, but not β-actin, in the 5 clones transfected with the nucleolin siRNA. These results indicate that down-regulation of the bcl2 mRNA–binding protein nucleolin leads to bcl2 mRNA instability and decreased levels of bcl-2 protein.
Relative levels of expression of bcl2 hnRNA and bcl2 mRNA in stable clones of MCF-7 cells. The levels of bcl2 hnRNA and bcl2 mRNA in MCF-7 cells transfected with either a scrambled siRNA (□) or a nucleolin siRNA (▪) and (▒) were determined by real-time quantitative PCR. Results are expressed as the means of 4 determinations per group ± SEM. *P < .001 compared with either of the nucleolin siRNA transfected clones.
Relative levels of expression of bcl2 hnRNA and bcl2 mRNA in stable clones of MCF-7 cells. The levels of bcl2 hnRNA and bcl2 mRNA in MCF-7 cells transfected with either a scrambled siRNA (□) or a nucleolin siRNA (▪) and (▒) were determined by real-time quantitative PCR. Results are expressed as the means of 4 determinations per group ± SEM. *P < .001 compared with either of the nucleolin siRNA transfected clones.
Knockdown of nucleolin decreases bcl-2 protein levels in stable clones of MCF-7 cells. The cells were transfected with either a scrambled siRNA (lanes 1 and 2) or a nucleolin siRNA (lanes 3-6). S10 cell extracts were prepared and the amounts of immunoreactive nucleolin, bcl2, and β-actin proteins were determined by Western blotting.
Knockdown of nucleolin decreases bcl-2 protein levels in stable clones of MCF-7 cells. The cells were transfected with either a scrambled siRNA (lanes 1 and 2) or a nucleolin siRNA (lanes 3-6). S10 cell extracts were prepared and the amounts of immunoreactive nucleolin, bcl2, and β-actin proteins were determined by Western blotting.
Discussion
B-cell CLL is the most prevalent form of adult leukemia in the Western Hemisphere, partly because the indolent nature of this disease allows for prolonged survival of the patient. Nevertheless, the clonal B cells accumulate by circumventing the normal B-cell apoptotic pathways.3 The ability of CLL cells to avoid apoptosis complicates the clinical management of the disease with conventional anticancer drugs.
It is known that CLL cells from the majority of patients overexpress bcl-2 protein.4-6 However, the molecular basis for the overexpression of bcl-2 protein is less clear, since no consistent gene mutations or rearrangements have been discovered that lead to increased transcription of the bcl2 gene. The results reported herein show for the first time that bcl2 mRNA is highly stabilized in CD19+ CLL cells compared with CD19+ B cells from healthy volunteers. In addition, we show that the enhanced stability of bcl2 mRNA in CLL cells is related, at least in part, to the overexpression of the protein nucleolin in the cytoplasm of CLL cells. Nucleolin was overexpressed in the cytoplasm of CLL cells from all of the 17 patients examined, with the average expression level being 26-fold higher in the CLL cells than normal B cells. This finding is remarkable in that nucleolin is predominately a nuclear protein in most cell types, although a few studies have shown that nucleolin is also present in the cytoplasm and plasma membrane of some tumor cells.31-33 The increase in S10 nucleolin levels in the CLL cells was not the result of an increased proliferation rate of the CLL cells compared with the normal B cells, since the CLL cells were clinically indolent and showed no significant thymidine incorporation into DNA (data not shown). The fact that nucleolin was uniformly overexpressed in all our CLL patients, including those in early stages without prior therapy for CLL, suggests that nucleolin stabilization of bcl2 mRNA is an early event in CLL pathogenesis rather than a feature of disease evolution or an epiphenomenon caused by cytotoxic chemotherapy.
We have previously demonstrated with mRNA decay assays using extracts of HL-60 leukemia cells that exogenous nucleolin binds to an ARE instability element in the 3′-UTR of bcl2 mRNA and protects this mRNA from degradation.22 It is thought that stabilizing ARE-binding proteins, such as nucleolin, may enhance binding of poly (A) binding protein to translation initiation factors elF4E and elF4G, which circularizes the mRNA.8 This promotes mRNA stability by inhibiting deadenylation and/or blocking exosome-mediated decay.8 Our results are consistent with the idea that nucleolin is a bcl2 mRNA–stabilizing protein in CLL cells. Of particular importance is that the overexpressed nucleolin occurs in the cytoplasmic compartment of CLL cells, where it is potentially available to bind to bcl2 mRNA. Nucleolin has also been shown to bind to the 3′-UTRs of amyloid precursor protein mRNA34,35 and human preprorenin mRNA,36 thereby promoting stabilization of these messages. This protein is likewise involved in the stabilization of IL-2 mRNA that occurs during T-cell activation.21
It was recently reported that about 65% of B-cell CLL patients showed either down-regulation or deletion of microRNAs miR-15a and miR-16-1.37 These microRNAs target bcl2 mRNA and probably interfere with its translation, since they do not appear to affect bcl2 mRNA levels when transfected into the human megakaryocytic cell line MEG-O1. However, Cimmino et al37 did not address the possibility of increased bcl2 mRNA levels or stability in CLL cells from patients. Thus, it is possible that bcl-2 protein is up-regulated in CLL cells as a result of both increased bcl2 mRNA stability (nucleolin up-regulation) and increased bcl2 mRNA translation (miR-15a, miR-16-1 down-regulation). Nevertheless, the results reported herein strongly suggest that increased bcl2 mRNA stability is an important mechanism involved in the altered expression of this gene in CLL cells. We observed up-regulation of the bcl2 mRNA–stabilizing protein nucleolin in 17 of 17 CLL patients. In addition, our analysis of bcl2 hnRNA and bcl2 mRNA levels, as well as the kinetics of bcl2 mRNA decay, is consistent with enhanced bcl2 mRNA stability in CLL cells.
CLL cells are frequently resistant to chemotherapeutic drugs because of their enhanced bcl-2 protein levels and low growth fraction. Clinical trials have examined the effectiveness of bcl-2 antisense oligonucleotides (Genasense; Genta, Berkeley Heights, NJ) in inducing bcl2 mRNA down-regulation and antitumor responses in various hematologic and solid tumors.38,39 While the bcl-2 antisense approach in general is conceptually straightforward, it has not yet been validated in these clinical trials. In particular, it is not clear how antitumor selectivity will be achieved with bcl-2 antisense compounds, since some normal tissues also depend on bcl-2 protein for survival. However, nucleolin that is overexpressed in the cytoplasm of CLL cells may represent a specific target for the development of drugs active in CLL. One strategy for exploiting this target would be to identify small molecules that interfere with the binding of cytoplasmic nucleolin to bcl2 mRNA in CLL cells. In this regard, certain G-rich oligodeoxynucleotides, which apparently target nucleolin,32 may be useful for validating this approach for inducing apoptosis in CLL cells.
Authorship
Contribution: Y.O. performed most of the experiments and data reduction and assisted in writing the manuscript. S.S. carried out the confocal microscopy and nucleolin knockdown studies. E.A.K., J.C.S., M.P.-R., and R.K.S. recruited patients and healthy volunteers to the study. E.A.K. and M.P.-R. purified CLL cells from some of the blood samples. T.K.S. performed the RNA decay assays. E.K.S. and R.K.S. contributed to experimental design and data analysis. D.J.F. conceived and designed the research plan and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel J. Fernandes, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC 29425; e-mail: fernand@musc.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grant 6006-06 from the Leukemia and Lymphoma Society and by grants CA109254 and CA83925 from the National Cancer Institute.




![Figure 4. Decay of bcl2 mRNA in extracts of CLL and normal B cells. The 5′-capped and polyadenylated [32P]bcl-2–CR RNA and [32P]bcl-2–CR-ARE RNA were incubated with S100 extracts prepared from either CLL cells from 4 patients or normal B cells from 3 human volunteers. At the indicated times, aliquots of the reaction mixtures were removed and analyzed by polyacrylamide gel electrophoresis and filmless phosphorimaging. The results are expressed as the mean percentage of full-length RNA remaining ± SEM as a function of time. ○ indicates CLL cell extract + [32P]bcl-2–CR RNA; •, CLL cell extract + [32P]bcl-2–CR-ARE RNA; □, normal B-cell extract + [32P]bcl-2–CR RNA; ▪, normal B-cell extract + [32P]bcl-2–CR-ARE RNA; ▴, normal B-cell extract + [32P]bcl-2–CR-ARE RNA + 280 nM purified recombinant nucleolin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/7/10.1182_blood-2006-08-043257/4/m_zh80070710530004.jpeg?Expires=1764998669&Signature=YDkWeiAa7P1tnK05Vgok~PDc34t5B9-0RU2KpmeUo0y5IUGsiSEyx3uAOImTC7pBCXIhXSHrEHDsm~D4zf3PzKiGk9fjqR7cIGNDre7e4kZ3t7qbaTO-jvBo3lYpldai5pMMa7i3Cis7OU-mPRusA959noj0MCKFH9tSYzmouyFiJYVdkEBT2b4VbJYOrS2LCayF1NCFlJPYfYkQ9tjn4L8czpG2h5VSKZ6CeRkKBBK2hlUaZCaUao4X844PEEQKFuocrXv9TpU2x~BqJUx508zYSwaL8J1f~xNYMP8lFfnhPw6lc2VcNoYj5cQ07VSgoFXs4hjhOua1ZFoTnElICw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal