Abstract
We previously reported a lower risk of non-Hodgkin lymphoma (NHL) associated with high consumption of vitamin B6 and methionine, dietary determinants of one-carbon metabolism. Evidence has linked genetic variants involved in one-carbon metabolism to NHL. We investigated 30 polymorphisms in 18 genes for their main effect on NHL among 1141 incident cases and 949 population-based controls and examined gene-nutrient interactions in a subgroup of 386 cases and 319 controls who provided detailed food-frequency information. Odds ratios (ORs) and 95% confidence intervals (CIs) were adjusted for age, sex, and race. We observed a decreased risk of NHL overall with BHMT Ex8+453A>T and increased risk with CBS Ex13+41C>T, FPGS Ex15-263T>C, and SHMT1 Ex12+138C>T and Ex12+236C>T. Furthermore, significant gene-nutrient interactions limited the protective association comparing high versus low vitamin B6 to FPGS Ex15-263T>C CC (OR = 0.22; 95% CI = 0.10-0.52), MTHFS IVS2-1411T>G TT/TG (OR = 0.54; 95% CI = 0.36-0.81), and MTR Ex26-20A>G AA (OR = 0.55; 95% CI = 0.35-0.86) genotypes, and the protective association of methionine to FTHFD Ex10-40G>T GG (OR = 0.63; 95% CI = 0.44-0.91), MTHFR Ex8-62A>C CC (OR = 0.13; 95% CI = 0.04-0.39), and MTRR Ex5+136T>C TT (OR = 0.67; 95% CI = 0.47-0.97) genotypes. Warranting replication, our finding of gene-nutrient interactions in one-carbon metabolism supports their etiologic involvement in lymphomagenesis.
Introduction
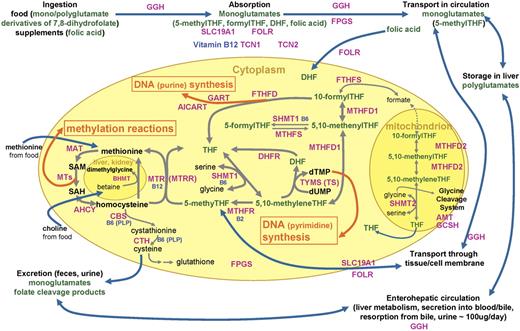
One-carbon metabolism in eukaryotic cells involves reactions to transfer single carbon units for DNA synthesis and for methylation of biologic compounds, including DNA (Figure 1). 1 One-carbon transfer reactions are mediated by numerous enzymes that require nutritional coenzymes, most notably folate—a B vitamin that serves as a one-carbon carrier/donor, and also vitamins B12, B6, and B2 and methionine. Disruptions in one-carbon metabolism due to deficiency of the nutrients or genetic polymorphisms of the enzymes involved have been linked to cancer etiology through insufficient DNA synthesis/repair and aberrant gene expression.2
Diagram of folate and one-carbon metabolism in mammalian organisms. Intracellular one-carbon transfer reactions are essential for nucleotide (thymidylate and purine) synthesis and methylation of numerous compounds, including DNA, RNA, proteins, and phospholipids. These one-carbon transfer reactions are mainly supported by folate, a B vitamin that serves as a one-carbon carrier/donor. This diagram depicts absorption, transport, and metabolism of folate around the intracellular one-carbon metabolism as well as enzymes/proteins and other nutritional factors involved.1-5 Folates in food, mostly polyglutamates, are hydrolyzed to monoglutamates by GGH in the gut and are absorbed across the intestinal mucosa with folic acid from fortified foods and supplements mostly by a saturable pH-dependent process, via reduced folate carrier (encoded by SLC19A1), and by passive diffusion at high concentrations. Once absorbed into the portal circulation, folates are taken up by the liver, where they are metabolized to polyglutamates by FPGS and retained or released into blood or bile as 5-methylTHF. Folate released in bile is reabsorbed in the small intestine. About two thirds of 5-methylTHF, the predominant form of folate in circulation, is bound to low-affinity proteins, mostly albumin: low levels of high-affinity folate binders are also found in blood. Blood 5-methylTHF is transported into the cell by carrier-mediated or receptor-mediated mechanisms. Reduced folate carrier has a higher affinity for reduced folate than oxidized folic acid and accounts for the transport of most folate and methotrexate. Membrane-bound folate receptors, including folate receptor 1 encoded by FOLR1, with high affinity for folic acid are expressed in epithelial tissues, and its expression is elevated in malignant epithelial tumors.6 The predominant cytoplasmic folate, 5-methylTHF, donates its one-carbon moiety to methylate homocysteine to methionine, yielding THF. THF is a much preferred substrate to FPGS that lengthens the glutamate chain of the monoglutamate folate so folates can be retained in the cell. This polyglutamylation also enables folates to be used by one-carbon metabolizing enzymes that have much higher affinities for polyglutamates than monoglutamates. In deficiency of vitamin B12, which is a coenzyme for methionine synthase (MTR) that converts 5-methylTHF to THF, or with insufficient transcobalamins (TCN1, TCN2) for vitamin B12 absorption, deficiency of functional folate (THF) occurs despite sufficient folate in circulation (“methyl-trap”). MTR loses its activity when its vitamin B12–derived coenzyme, cobalamin, gets oxidized: MTRR reactivates MTR using the methyl supply from SAM. Homocysteine can be remethylated via an alternative mechanism of BHMT using betaine, supplied from dietary choline, in kidney and liver. Methionine, from homocysteine and also supplied from diet, is converted to SAM, a universal donor of one-carbon unit to numerous methylation reactions via MTs in part for DNA methylation. Resulting SAH is hydrolyzed to homocysteine, which then gets remethylated or catabolyzed via the transsulfuration pathway initiated by CBS. The active coenzyme THF obtains one-carbon moiety from amino acid serine via SHMT1 catalysis, yielding 5,10-methyleneTHF, which is an important common substrate to methylation pathway described (remethylation of homocysteine to methionine) via MTHFR or to nucleic acid synthesis pathways via TYMS (uridylate to thymidylate conversion; pyrimidine synthesis) or MTHFD1/FTHFD (purine synthesis). DHF, the remnant of TYMS reaction on THF, is also supplied from folic acid that is reported to be found in blood in higher proportion than usual when a large dose is consumed from fortified foods or supplements. 5,10-MethenylTHF can be interconverted with 5-formylTHF (also known as folinic acid or leucovorin; thought to be the storage form of folate) via SHMT1/MTHFS. Although less understood, mitochondrial one-carbon metabolism is proposed to be in equilibrium with cytoplasmic metabolism and contains glycine cleavage system. AHCY indicates S-adenosylhomocysteine hydrolase; AICART, phosphoribosylaminoimidazolecarboxamide formyltransferase; AMT, aminomethyltransferase; B2, vitamin B2; B6, vitamin B6; B12, vitamin B12; BHMT, betaine-homocysteine methyltransferase; CBS, cystathionine-beta-synthase; CTH, cystathionase; DHF, dihydrofolate; DHFR, dihydrofolate reductase; FPGS, folylpolyglutamate synthase; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; FOLR, folate receptor; FTHFD, 10-formyltetrahydrofolate dehydrogenase; FTHFS, 10-formyltetrahydrofolate synthase; FTHFSDC1, 10-formyltetrahydrofolate synthetase domain containing 1; GART, glycinamide ribonucleotide formyltransferase; GCPII, glutamate carboxypeptidase II; GCSH, glycine cleavage system protein H; GGH, gamma-glutamylhydrolase; MAT, methionine S-adenosyltransferase; MTs, a group of methyltransferases; MTHFD1, cytoplasmic 5,10-methylenetetrahydrofolate dehydrogenase; MTHFD2, mitochondrial 5,10-methylenetetrahydrofolate dehydrogenase; MTHFR, 5,10-methylenetetrahydrofolate reductase; MTHFS, 5,10-methenyltetrahydrofolate synthetase; MTR, methionine synthase; MTRR, methionine synthase reductase; PLP, pyridoxal 5′-phosphate; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SHMT1, cytoplasmic serine hydroxymethyltransferase; SHMT2, mitochondrial serine hydroxymethyltransferase; SLC19A1, reduced folate carrier; TCN1, transcobalamin 1; TCN2, transcobalamin 2; THF, tetrahydrofolate; and TYMS, thymidylate synthetase.
Diagram of folate and one-carbon metabolism in mammalian organisms. Intracellular one-carbon transfer reactions are essential for nucleotide (thymidylate and purine) synthesis and methylation of numerous compounds, including DNA, RNA, proteins, and phospholipids. These one-carbon transfer reactions are mainly supported by folate, a B vitamin that serves as a one-carbon carrier/donor. This diagram depicts absorption, transport, and metabolism of folate around the intracellular one-carbon metabolism as well as enzymes/proteins and other nutritional factors involved.1-5 Folates in food, mostly polyglutamates, are hydrolyzed to monoglutamates by GGH in the gut and are absorbed across the intestinal mucosa with folic acid from fortified foods and supplements mostly by a saturable pH-dependent process, via reduced folate carrier (encoded by SLC19A1), and by passive diffusion at high concentrations. Once absorbed into the portal circulation, folates are taken up by the liver, where they are metabolized to polyglutamates by FPGS and retained or released into blood or bile as 5-methylTHF. Folate released in bile is reabsorbed in the small intestine. About two thirds of 5-methylTHF, the predominant form of folate in circulation, is bound to low-affinity proteins, mostly albumin: low levels of high-affinity folate binders are also found in blood. Blood 5-methylTHF is transported into the cell by carrier-mediated or receptor-mediated mechanisms. Reduced folate carrier has a higher affinity for reduced folate than oxidized folic acid and accounts for the transport of most folate and methotrexate. Membrane-bound folate receptors, including folate receptor 1 encoded by FOLR1, with high affinity for folic acid are expressed in epithelial tissues, and its expression is elevated in malignant epithelial tumors.6 The predominant cytoplasmic folate, 5-methylTHF, donates its one-carbon moiety to methylate homocysteine to methionine, yielding THF. THF is a much preferred substrate to FPGS that lengthens the glutamate chain of the monoglutamate folate so folates can be retained in the cell. This polyglutamylation also enables folates to be used by one-carbon metabolizing enzymes that have much higher affinities for polyglutamates than monoglutamates. In deficiency of vitamin B12, which is a coenzyme for methionine synthase (MTR) that converts 5-methylTHF to THF, or with insufficient transcobalamins (TCN1, TCN2) for vitamin B12 absorption, deficiency of functional folate (THF) occurs despite sufficient folate in circulation (“methyl-trap”). MTR loses its activity when its vitamin B12–derived coenzyme, cobalamin, gets oxidized: MTRR reactivates MTR using the methyl supply from SAM. Homocysteine can be remethylated via an alternative mechanism of BHMT using betaine, supplied from dietary choline, in kidney and liver. Methionine, from homocysteine and also supplied from diet, is converted to SAM, a universal donor of one-carbon unit to numerous methylation reactions via MTs in part for DNA methylation. Resulting SAH is hydrolyzed to homocysteine, which then gets remethylated or catabolyzed via the transsulfuration pathway initiated by CBS. The active coenzyme THF obtains one-carbon moiety from amino acid serine via SHMT1 catalysis, yielding 5,10-methyleneTHF, which is an important common substrate to methylation pathway described (remethylation of homocysteine to methionine) via MTHFR or to nucleic acid synthesis pathways via TYMS (uridylate to thymidylate conversion; pyrimidine synthesis) or MTHFD1/FTHFD (purine synthesis). DHF, the remnant of TYMS reaction on THF, is also supplied from folic acid that is reported to be found in blood in higher proportion than usual when a large dose is consumed from fortified foods or supplements. 5,10-MethenylTHF can be interconverted with 5-formylTHF (also known as folinic acid or leucovorin; thought to be the storage form of folate) via SHMT1/MTHFS. Although less understood, mitochondrial one-carbon metabolism is proposed to be in equilibrium with cytoplasmic metabolism and contains glycine cleavage system. AHCY indicates S-adenosylhomocysteine hydrolase; AICART, phosphoribosylaminoimidazolecarboxamide formyltransferase; AMT, aminomethyltransferase; B2, vitamin B2; B6, vitamin B6; B12, vitamin B12; BHMT, betaine-homocysteine methyltransferase; CBS, cystathionine-beta-synthase; CTH, cystathionase; DHF, dihydrofolate; DHFR, dihydrofolate reductase; FPGS, folylpolyglutamate synthase; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; FOLR, folate receptor; FTHFD, 10-formyltetrahydrofolate dehydrogenase; FTHFS, 10-formyltetrahydrofolate synthase; FTHFSDC1, 10-formyltetrahydrofolate synthetase domain containing 1; GART, glycinamide ribonucleotide formyltransferase; GCPII, glutamate carboxypeptidase II; GCSH, glycine cleavage system protein H; GGH, gamma-glutamylhydrolase; MAT, methionine S-adenosyltransferase; MTs, a group of methyltransferases; MTHFD1, cytoplasmic 5,10-methylenetetrahydrofolate dehydrogenase; MTHFD2, mitochondrial 5,10-methylenetetrahydrofolate dehydrogenase; MTHFR, 5,10-methylenetetrahydrofolate reductase; MTHFS, 5,10-methenyltetrahydrofolate synthetase; MTR, methionine synthase; MTRR, methionine synthase reductase; PLP, pyridoxal 5′-phosphate; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SHMT1, cytoplasmic serine hydroxymethyltransferase; SHMT2, mitochondrial serine hydroxymethyltransferase; SLC19A1, reduced folate carrier; TCN1, transcobalamin 1; TCN2, transcobalamin 2; THF, tetrahydrofolate; and TYMS, thymidylate synthetase.
Epidemiologic evidence linking genetic susceptibility in one-carbon metabolism to cancer risk is most extensive for colorectum7 and growing for cancers of breast,8 stomach,9 and other sites. Non-Hodgkin lymphoma (NHL), like other cancers, exhibits genetic instability10 and aberrant DNA methylation patterns,11 suggesting a key role of one-carbon metabolism in lymphoid tissues with high turnover rates and supporting its involvement in lymphomagenesis. In fact, genetic polymorphisms of some one-carbon metabolism enzymes have been associated with an altered risk of adult lymphomas12-20 and leukemias15,21-23 : lower risk was associated with MTHFR Ex5+79C>T (commonly known as 677C>T)15,17,21 and Ex8-62A>C (1298A>C)21 (or the combination15,17,21 ), MTR Ex26-20A>G (2756A>G),14,16,20 MTRR Ex2-64A>G (66A>G),15 SHMT1 Ex12+138C>T (1420C>T),13,22 and TYMS variants of variable number tandem repeats (VNTRs) Ex1+52-28base (3R versus 2R),13,19,22 494 6-bp deletion,18 IVS6-68 C>T,18 1053C>T,18 and their haplotypes.18 Also, potential gene-gene interactions have been suggested.13,15,20,22
However, studies to date have considered only a few of these enzymes out of many involved in the metabolic pathway (Figure 1). Also, findings on these genetic variants have not been consistent in all studies: some studies showed conflicting findings for MTHFR Ex8-62A>C or the combination of its 2 single nucleotide polymorphisms (SNPs),20 MTR Ex26-20A>G12,17 and TYMS VNTR Ex1+52-28base,20 and some showed null results for CBS,16 MTHFR,12,14-16,18,19 MTR,15,18,22 and SHMT1.18,19 This inconsistency in past studies may be in part due to the small size of most studies or to unavailability of dietary information to examine potential gene-nutrient interactions that have been reported for other cancers.24-26
In a US population–based case-control study, we previously reported that the highest quartiles of vitamin B6 and methionine intake were associated with about 50% lower risk of NHL overall and that folate intake was inversely associated with diffuse large B-cell lymphoma (DLBCL).27 In this report, we investigated genetic susceptibility of selected one-carbon metabolism enzymes and their interaction with diet using a comprehensive assessment of the metabolic pathways (Figure 1).
Materials and methods
Study population
As described in detail previously,28 newly-diagnosed and human immunodeficiency virus (HIV)–negative NHL cases were identified from 4 Surveillance, Epidemiology, and End Results (SEER) registries among women and men aged 20 to 74 years during the period of July 1998 through June 2000. On all NHL cases from local SEER registries, we obtained pathology and subtype information that was based on abstracted reports of the diagnosing pathologists. We then classified the histologically confirmed cases into subtypes of DLBCL, follicular lymphoma, and small lymphocytic lymphoma (SLL) according to the International Classification of Diseases-Oncology (ICD-O-2; codes for NHL: 9590-9595, 9670-9717).29 Although pathology samples were not reviewed centrally, we consider pathological diagnosis of these main NHL subtypes to be reliable and comparable across SEER study centers.30,31 Population controls who were aged 20 to 74 years, HIV negative, and with no history of NHL were identified among the study area residents via random-digit-dialing and Health Care Financing Administration (Medicare) files. The study was approved by the human subjects review boards at all participating institutions (NCI and SEER centers of Detroit, Iowa, Los Angeles, and Seattle), and we obtained written informed consent from all participants, in accordance with the Declaration of Helsinki.
We identified 2248 potentially eligible cases and interviewed 1321 cases (participation rate, 76%; response rate, 59%)28 : we did not contact 520 cases (death, inability to locate, physician refusal, or relocation outside of the study area) and could not acquire participation of 407 cases. Of 2409 potentially eligible controls frequency matched to cases on sex, age, race, and SEER centers, 2046 were contacted, and 1057 were interviewed (participation rate, 52%; response rate, 44%). Among the interviewed participants, 1172 cases and 982 controls provided biologic samples for genotyping32 : 773 cases and 668 controls provided blood samples, and 399 cases and 314 controls provided mouthwash buccal cell samples. Demographic characteristics (age, sex, education) were comparable among individuals who provided blood, buccal samples, or neither.32 Among the 1141 cases and 949 controls who were genotyped successfully, 517 cases and 434 controls (about 50% of non–African American participants) were queried for diet/lifestyle history using a split-sample design described earlier,27 of which 386 cases (75%) and 319 controls (74%) returned the questionnaire.
Genotyping
We considered enzymes and proteins involved in folate absorption and transport or intracellular one-carbon metabolism (Figure 1) and chose 14 genes that have been previously studied or are otherwise believed to play an important role in one-carbon metabolism.3 In addition, we included 4 DNA repair genes (MBD2, MGMT, MLH1, and MSH2) that in part depend on one-carbon supplies, and therefore, may have a synergistic effect with nutritional or genetic factors of one-carbon metabolism on carcinogenesis. Twenty-nine SNPs and one insertion polymorphism in these genes were selected (Table 1), based on prior functional data from previous reports18,22,34-37 or expected functional consequences in that the polymorphisms result in amino acid change or they are located within the 3′ untranslated region (UTR), which contains regulatory sequences and binding sites for other molecules that could alter the stability of the mRNA transcript of the gene.
Genes involved in one-carbon metabolism and their polymorphic variants investigated in relation to non-Hodgkin lymphoma, the NCI-SEER Case-Control Study, 1998-2000
| Gene . | Enzyme/protein . | Location . | dbSNP ID [amino acid change] . |
|---|---|---|---|
| BHMT | Betaine-homocysteine methyltransferase | 5q13.1-q15 | rs585800 (Ex8+453A>T)† |
| CBS | Cystathionine-beta-synthase | 21q22.3 | rs234706 (Ex9+33C>T)‡§ [Y233Y]; rs12613 (Ex18-39IG>A)‡; rs1801181 (Ex13+41C>T)‡ [A360A] |
| FPGS | Folylpolyglutamate synthase | 9q34.1 | rs10106 (Ex15-263T>C)‡ |
| FTHFD | 10-Formyltetrahydrofolate dehydrogenase | 3q21.3 | rs2305230 (Ex10-40G>T)‡ [L395L]; rs1127717 (Ex21+31A>G)† [D793G] |
| GGH | Gamma-glutamyl hydrolase | 8q12.1 | rs719236 (−423G>T)‡; rs1031552 (IVS7-3001C>T)‡ |
| MBD2 | Methyl-CpG binding domain protein 2 | 18q21 | rs7614 (Ex8+438A>G)†; rs603097 (−2176T>C)‡ |
| MGMT* | O-6-Methylguanine-DNA methyltransferase | 10q26 | rs2308321 (Ex4+13A>G)‡ [I143V]; rs2308327 (Ex4+119A>G)† [K178R]; rs12917 (Ex2-25C>T)† [L84F] |
| MLH1 | MutLhomolog 1 | 3p21.3 | rs1799977 (Ex8-23A>G)† [I219V]; rs2286940 (IVS12-169C>T)† |
| MSH2 | MutShomolog 2 | 2p22-p21 | rs4987188 (Ex6+23G>A)ठ[G322D] |
| MTHFD2 | 5,10-Methylenetetrahydrofolate dehydrogenase 2 | 2p13.1 | rs1667627 (IVS1+3323T>C)† |
| MTHFR | 5,10-Methylenetetrahydrofolate reductase | 1p36.3 | rs1801131 (Ex8-62A>C or 1298A>C)‡ [E429A]; rs1801133 (Ex5+79C>T or 677C>T)‡ [A222V] |
| MTHFS | 5,10-Methenyltetrahydrofolate synthetase (or 5-formyltetrahydrofolate cyclo-ligase) | 15q24.3 | rs622506 (IVS2-1411T>G)† |
| MTR | 5-Methyltetrahydrofolate-homocysteine methyltransferase (or methionine synthase) | 1q43 | rs1805087 (Ex26-20A>G or 2756A>G)†§ [D919G] |
| MTRR | 5-Methyltetrahydrofolate-homocysteine methyltransferase reductase (or methionine synthase reductase) | 5p15.3-15.2 | rs161870 (Ex5+136T>C)‡ [L206L] |
| SHMT1 | Cytoplasmic serine hydroxymethyltransferase 1 | 17p11.2 | rs1979277 (Ex12+138C>T or 1420C>T)† [L435F]; rs1979276 (Ex12+236C>T)† |
| SLC19A1 | Solute carrier family 19 (folate transporter), member 1 (or reduced folate carrier 1) | 21q22.3 | rs12659 (Ex4-254C>T)† [P232P]; rs1051296 (Ex8-233T>G)† |
| TCN1 | Transcobalamin I | 11q11-q12 | rs526934 (IVS1+372T>C)† |
| TYMS | Thymidylate synthetase | 18p11.32 | rs699517 (Ex8+157C>T)‡; VNTR (Ex1+52-28base 3R>2R)‡ |
| Gene . | Enzyme/protein . | Location . | dbSNP ID [amino acid change] . |
|---|---|---|---|
| BHMT | Betaine-homocysteine methyltransferase | 5q13.1-q15 | rs585800 (Ex8+453A>T)† |
| CBS | Cystathionine-beta-synthase | 21q22.3 | rs234706 (Ex9+33C>T)‡§ [Y233Y]; rs12613 (Ex18-39IG>A)‡; rs1801181 (Ex13+41C>T)‡ [A360A] |
| FPGS | Folylpolyglutamate synthase | 9q34.1 | rs10106 (Ex15-263T>C)‡ |
| FTHFD | 10-Formyltetrahydrofolate dehydrogenase | 3q21.3 | rs2305230 (Ex10-40G>T)‡ [L395L]; rs1127717 (Ex21+31A>G)† [D793G] |
| GGH | Gamma-glutamyl hydrolase | 8q12.1 | rs719236 (−423G>T)‡; rs1031552 (IVS7-3001C>T)‡ |
| MBD2 | Methyl-CpG binding domain protein 2 | 18q21 | rs7614 (Ex8+438A>G)†; rs603097 (−2176T>C)‡ |
| MGMT* | O-6-Methylguanine-DNA methyltransferase | 10q26 | rs2308321 (Ex4+13A>G)‡ [I143V]; rs2308327 (Ex4+119A>G)† [K178R]; rs12917 (Ex2-25C>T)† [L84F] |
| MLH1 | MutLhomolog 1 | 3p21.3 | rs1799977 (Ex8-23A>G)† [I219V]; rs2286940 (IVS12-169C>T)† |
| MSH2 | MutShomolog 2 | 2p22-p21 | rs4987188 (Ex6+23G>A)ठ[G322D] |
| MTHFD2 | 5,10-Methylenetetrahydrofolate dehydrogenase 2 | 2p13.1 | rs1667627 (IVS1+3323T>C)† |
| MTHFR | 5,10-Methylenetetrahydrofolate reductase | 1p36.3 | rs1801131 (Ex8-62A>C or 1298A>C)‡ [E429A]; rs1801133 (Ex5+79C>T or 677C>T)‡ [A222V] |
| MTHFS | 5,10-Methenyltetrahydrofolate synthetase (or 5-formyltetrahydrofolate cyclo-ligase) | 15q24.3 | rs622506 (IVS2-1411T>G)† |
| MTR | 5-Methyltetrahydrofolate-homocysteine methyltransferase (or methionine synthase) | 1q43 | rs1805087 (Ex26-20A>G or 2756A>G)†§ [D919G] |
| MTRR | 5-Methyltetrahydrofolate-homocysteine methyltransferase reductase (or methionine synthase reductase) | 5p15.3-15.2 | rs161870 (Ex5+136T>C)‡ [L206L] |
| SHMT1 | Cytoplasmic serine hydroxymethyltransferase 1 | 17p11.2 | rs1979277 (Ex12+138C>T or 1420C>T)† [L435F]; rs1979276 (Ex12+236C>T)† |
| SLC19A1 | Solute carrier family 19 (folate transporter), member 1 (or reduced folate carrier 1) | 21q22.3 | rs12659 (Ex4-254C>T)† [P232P]; rs1051296 (Ex8-233T>G)† |
| TCN1 | Transcobalamin I | 11q11-q12 | rs526934 (IVS1+372T>C)† |
| TYMS | Thymidylate synthetase | 18p11.32 | rs699517 (Ex8+157C>T)‡; VNTR (Ex1+52-28base 3R>2R)‡ |
The main effect of MGMT has been reported previously.33
Genotyping was done in subjects with DNA from blood samples only.
Genotyping was done in all of the subjects with DNA from blood or buccal tissue samples.
P < .05 for deviation from Hardy-Weinberg equilibrium among controls.
Genotyping details are available elsewhere.32 We extracted DNA using Puregene Autopure DNA extraction kits (Gentra Systems, Minneapolis, MN) from buffy coats and using phenol chloroform extraction methods for buccal cell samples.38 All genotyping was conducted at the NCI Core Genotyping Facility (CGF, Advanced Technology Corporation, Gaithersburg, MD), using the Taqman platform (http://snp500cancer.nci.nih.gov; National Cancer Institute).39 In order to conserve DNA, we genotyped the blood-based DNA samples first (∼ 75%) and expanded to buccal cell samples if the blood-based DNA data yielded significant (P < .05) or suggestive (ie, borderline significance or significant linear trend) associations with NHL. As a result, 16 polymorphisms from 11 genes were not genotyped in buccal cell–based DNA due to null results in blood-based DNA (noted in Table 1). Genotype frequencies in general were similar for individuals who provided blood versus buccal cells and also similar by participation status.40
Genotyping was successful in 96% to 100% of DNA samples, similarly in blood- and buccal cell–based samples. Only 3 SNPs were not in Hardy-Weinberg equilibrium in white controls (CBS Ex9+33C>T, MSH2 Ex6+23G>A, and MTR Ex26-20A>G; Table 1). To assure quality control, 40 replicate samples from 2 blood donors and duplicate samples from 100 study subjects were interspersed blindly among study samples for all genotyping, which yielded an agreement of 99% or more.32
Dietary assessment
Dietary intake was assessed using a modified version of the self-administered Block food frequency questionnaire.27 Participants were queried on 107 food and beverage items for their “usual eating habits (as an adult and before one year ago, not including any recent dietary changes)” by giving responses for 9 frequencies and 3 portion sizes. The instrument was validated against multiple diet records (correlations were 0.5-0.6 for most nutrients).41,42
Statistical analyses
We used the Wilcoxon nonparametric test for continuous variables and chi-squared tests for categoric variables to compare the descriptive characteristics of (1) cases and controls with genetic information, separately by the availability of diet information (Table 2); and (2) controls with and without diet information.
Descriptive characteristics of non-Hodgkin lymphoma (NHL) cases and controls with genotype and diet information, the NCI-SEER Case-Control Study, 1998-2000
| Characteristic . | Among subjects with genotype data, no. (%)† . | Among subjects with both genotype and diet data, no. (%)† . | ||
|---|---|---|---|---|
| NHL cases . | Controls . | NHL cases . | Controls . | |
| No. of cases | 1141 | 949 | 386 | 319 |
| Men | 629 (55) | 506 (53) | 215 (56) | 166 (52) |
| Women | 512 (45) | 443 (47) | 171 (44) | 153 (48) |
| Age at enrollment | ||||
| Younger than 45 y | 203 (18) | 150 (16) | 67 (17) | 40 (13) |
| 45 y to younger than 55 y | 252 (22) | 178 (19) | 90 (23) | 54 (17) |
| 55 y to younger than 65 y | 311 (27) | 228 (24) | 105 (27) | 79 (25) |
| 65 y to 74 y | 375 (33)* | 393 (41) | 124 (32)* | 146 (46) |
| Race‡ | ||||
| White | 981 (86)* | 760 (80) | 366 (95) | 305 (96) |
| African American | 80 (7) | 127 (13) | 0 (0) | NA |
| Others | 80 (7) | 62 (7) | 20 (5) | 14 (4) |
| Study center | ||||
| Detroit | 236 (21) | 170 (18) | 38 (10) | 26 (8) |
| Iowa | 333 (29) | 272 (29) | 151 (39) | 118 (37) |
| Los Angeles | 283 (25) | 238 (25) | 87 (23) | 69 (22) |
| Seattle | 289 (25) | 269 (28) | 110 (29) | 106 (33) |
| Education | ||||
| Less than 12 y | 109 (10) | 89 (10) | 26 (7) | 28 (9) |
| 12 to less than 16 y | 693 (61) | 543 (58) | 237 (61) | 178 (56) |
| 16 y or more | 334 (29) | 297 (32) | 123 (32) | 113 (35) |
| Family history of NHL | 48 (4) | 30 (3) | 20 (5) | 16 (5) |
| Body mass index | ||||
| Normal, less than 25 kg/m2 | 459 (40) | 389 (41) | 142 (37) | 112 (35) |
| Overweight, 25 kg/m2 or greater to less than 30 kg/m2 | 408 (36) | 336 (35) | 160 (41) | 132 (41) |
| Obese, 30 kg/m2 or greater | 274 (24) | 224 (24) | 84 (22) | 75 (24) |
| DNA source | ||||
| Blood | 735 (64) | 631 (66) | 271 (70) | 242 (76) |
| Buccal tissue | 395 (35) | 313 (33) | 112 (29) | 75 (24) |
| Both | 11 (1) | 5 (1) | 3 (1) | 2 (1) |
| Patient pathology | ||||
| All B-cell | 927 (81) | NA | 327 (85) | NA |
| Diffuse large B-cell | 360 (32) | NA | 142 (37) | NA |
| Follicular | 271 (24) | NA | 98 (25) | NA |
| SLL | 147 (13) | NA | 48 (12) | NA |
| Marginal zone | 92 (8) | NA | 26 (7) | NA |
| Other | 57 (5) | NA | 13 (3) | NA |
| All T-cell | 73 (6) | NA | 21 (5) | NA |
| NOS | 141 (12) | NA | 38 (10) | NA |
| Characteristic . | Among subjects with genotype data, no. (%)† . | Among subjects with both genotype and diet data, no. (%)† . | ||
|---|---|---|---|---|
| NHL cases . | Controls . | NHL cases . | Controls . | |
| No. of cases | 1141 | 949 | 386 | 319 |
| Men | 629 (55) | 506 (53) | 215 (56) | 166 (52) |
| Women | 512 (45) | 443 (47) | 171 (44) | 153 (48) |
| Age at enrollment | ||||
| Younger than 45 y | 203 (18) | 150 (16) | 67 (17) | 40 (13) |
| 45 y to younger than 55 y | 252 (22) | 178 (19) | 90 (23) | 54 (17) |
| 55 y to younger than 65 y | 311 (27) | 228 (24) | 105 (27) | 79 (25) |
| 65 y to 74 y | 375 (33)* | 393 (41) | 124 (32)* | 146 (46) |
| Race‡ | ||||
| White | 981 (86)* | 760 (80) | 366 (95) | 305 (96) |
| African American | 80 (7) | 127 (13) | 0 (0) | NA |
| Others | 80 (7) | 62 (7) | 20 (5) | 14 (4) |
| Study center | ||||
| Detroit | 236 (21) | 170 (18) | 38 (10) | 26 (8) |
| Iowa | 333 (29) | 272 (29) | 151 (39) | 118 (37) |
| Los Angeles | 283 (25) | 238 (25) | 87 (23) | 69 (22) |
| Seattle | 289 (25) | 269 (28) | 110 (29) | 106 (33) |
| Education | ||||
| Less than 12 y | 109 (10) | 89 (10) | 26 (7) | 28 (9) |
| 12 to less than 16 y | 693 (61) | 543 (58) | 237 (61) | 178 (56) |
| 16 y or more | 334 (29) | 297 (32) | 123 (32) | 113 (35) |
| Family history of NHL | 48 (4) | 30 (3) | 20 (5) | 16 (5) |
| Body mass index | ||||
| Normal, less than 25 kg/m2 | 459 (40) | 389 (41) | 142 (37) | 112 (35) |
| Overweight, 25 kg/m2 or greater to less than 30 kg/m2 | 408 (36) | 336 (35) | 160 (41) | 132 (41) |
| Obese, 30 kg/m2 or greater | 274 (24) | 224 (24) | 84 (22) | 75 (24) |
| DNA source | ||||
| Blood | 735 (64) | 631 (66) | 271 (70) | 242 (76) |
| Buccal tissue | 395 (35) | 313 (33) | 112 (29) | 75 (24) |
| Both | 11 (1) | 5 (1) | 3 (1) | 2 (1) |
| Patient pathology | ||||
| All B-cell | 927 (81) | NA | 327 (85) | NA |
| Diffuse large B-cell | 360 (32) | NA | 142 (37) | NA |
| Follicular | 271 (24) | NA | 98 (25) | NA |
| SLL | 147 (13) | NA | 48 (12) | NA |
| Marginal zone | 92 (8) | NA | 26 (7) | NA |
| Other | 57 (5) | NA | 13 (3) | NA |
| All T-cell | 73 (6) | NA | 21 (5) | NA |
| NOS | 141 (12) | NA | 38 (10) | NA |
NA indicates not applicable.
P < .05 for comparison of cases versus controls, within the group of genetic and genetic + diet information availability, using Wilcoxon nonparametric test for continuous variables and using Pearson chi-squared test for categoric variables.
Number and percent for each category of descriptive characteristic variables.
By study design, study participants were divided into 2 groups for detailed questionnaires, and all African Americans and a random half of the rest were assigned to one group for information on self/family medical history, while the other participants were assigned to fill out diet information.
For the main effect of the genotype on NHL, we estimated odds ratios (ORs) and 95% confidence intervals (CIs) comparing heterozygote and homozygous variant (or less prevalent genotype) to homozygous wild-type (or more prevalent genotype) in unconditional logistic regression models (Table 3). Based on the risk estimates, heterozygotes were combined with either homozygous variants or homozygous wild types to explore a dominant or recessive model, respectively. We assigned ordinal scores (0, 1, 2) to homozygous wild-type, heterozygote, and homozygous variant, respectively, to obtain the P value for linear trend in regression. Heterogeneity among NHL subtypes for the main effect of each polymorphism was assessed by contrasting one subtype as “cases” and another as “controls” in the logistic regression model (eg, SLL versus DLBCL or SLL versus follicular lymphoma for the main effect of CBS Ex13+41C>T or FPGS Ex15-263T>C). We adjusted for the matching factors of age (continuous), sex, and race, but not study centers, to have the most parsimonious estimates.
Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the association between selected genetic polymorphisms involved in one-carbon metabolism and overall and subtype non-Hodgkin lymphoma (NHL) among participants with diet information, the NCI-SEER Case-Control Study, 1998-2000
| Gene, nucleotide change . | No. of controls*; n = 319 . | NHL cases . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Overall . | Diffuse large B-cell lymphoma . | Follicular lymphoma . | Small lymphocytic lymphoma . | ||||||
| No. of cases*; n = 386 . | OR (95% CI) . | No. of cases*; n = 142 . | OR (95% CI) . | No. of cases*; n = 98 . | OR (95% CI) . | No. of cases*; n = 48 . | OR (95% CI) . | ||
| BHMT Ex8+453A>T | |||||||||
| AA | 122 | 160 | 1.0 (reference) | 46 | 1.0 (reference) | 46 | 1.0 (reference) | 23 | 1.0 (reference) |
| AT | 99 | 98 | 0.76 (0.53-1.10) | 30 | 0.83 (0.49-1.43) | 28 | 0.76 (0.44-1.33) | 13 | 0.65 (0.31-1.35) |
| TT | 19 | 13 | 0.54 (0.25-1.14) | 5 | 0.74 (0.26-2.13) | 2 | — | 3 | — |
| AT/TT | 118 | 111 | 0.73 (0.51-1.04) | 35 | 0.82 (0.49-1.37) | 30 | 0.68 (0.40-1.17) | 16 | 0.67 (0.34-1.33) |
| P trend | .05 | .43 | .08 | .34 | |||||
| CBS Ex13+41C>T | |||||||||
| CC | 142 | 149 | 1.0 (reference) | 56 | 1.0 (reference) | 36 | 1.0 (reference) | 16 | 1.0 (reference) |
| CT | 136 | 165 | 1.15 (0.83-1.59) | 58 | 1.08 (0.70-1.68) | 51 | 1.57 (0.96-2.58) | 17 | 1.14 (0.55-2.35) |
| TT | 33 | 59 | 1.68 (1.03-2.73) | 22 | 1.59 (0.85-2.98) | 8 | 1.01 (0.42-2.40) | 12 | 3.32 (1.43-7.71) |
| CT/TT | 169 | 224 | 1.25 (0.92-1.70) | 80 | 1.18 (0.78-1.79) | 59 | 1.46 (0.91-2.36) | 29 | 1.56 (0.81-3.01) |
| P trend | .05 | .21 | .35 | .02 | |||||
| FPGS Ex15-263T>C | |||||||||
| TT | 108 | 126 | 1.0 (reference) | 47 | 1.0 (reference) | 30 | 1.0 (reference) | 12 | 1.0 (reference) |
| CT | 160 | 171 | 0.91 (0.65-1.28) | 57 | 0.81 (0.51-1.28) | 45 | 1.04 (0.61-1.77) | 24 | 1.35 (0.65-2.83) |
| CC | 44 | 79 | 1.58 (1.00-2.49) | 31 | 1.64 (0.91-2.94) | 21 | 1.93 (0.98-3.80) | 11 | 2.47 (1.00-6.15) |
| CT/CC | 204 | 250 | 1.06 (0.77-1.45) | 88 | 0.98 (0.64-1.50) | 66 | 1.22 (0.74-2.01) | 35 | 1.57 (0.78-3.16) |
| P trend | .12 | .23 | .10 | .06 | |||||
| FTHFD Ex10-40T>G | |||||||||
| GG | 224 | 284 | 1.0 (reference) | 101 | 1.0 (reference) | 72 | 1.0 (reference) | 34 | 1.0 (reference) |
| GT | 76 | 85 | 0.90 (0.63-1.29) | 31 | 0.92 (0.57-1.50) | 23 | 0.93 (0.54-1.60) | 11 | 0.95 (0.46-1.97) |
| TT | 10 | 9 | 0.67 (0.26-1.68) | 5 | 1.08 (0.36-3.26) | 2 | — | 2 | — |
| GT/TT | 86 | 94 | 0.87 (0.62-1.23) | 36 | 0.94 (0.59-1.49) | 25 | 0.90 (0.53-1.52) | 13 | 1.00 (0.50-1.99) |
| P trend | .35 | .87 | .60 | .88 | |||||
| MTHFR Ex8-62A>C (1298A>C) | |||||||||
| AA | 135 | 183 | 1.0 (reference) | 66 | 1.0 (reference) | 48 | 1.0 (reference) | 24 | 1.0 (reference) |
| AC | 141 | 165 | 0.86 (0.63-1.19) | 61 | 0.90 (0.59-1.39) | 40 | 0.79 (0.48-1.29) | 20 | 0.79 (0.42-1.51) |
| CC | 38 | 35 | 0.67 (0.40-1.13) | 12 | 0.63 (0.31-1.30) | 10 | 0.72 (0.33-1.58) | 4 | — |
| AC/CC | 179 | 200 | 0.82 (0.61-1.11) | 73 | 0.85 (0.56-1.27) | 50 | 0.78 (0.49-1.23) | 24 | 0.75 (0.41-1.39) |
| P trend | .12 | .25 | .29 | .31 | |||||
| MTHFS IVS2-1411T>G | |||||||||
| TT | 80 | 99 | 1.0 (reference) | 35 | 1.0 (reference) | 21 | 1.0 (reference) | 16 | 1.0 (reference) |
| GT | 108 | 135 | 0.99 (0.67-1.46) | 37 | 0.80 (0.46-1.40) | 43 | 1.55 (0.84-2.85) | 17 | 0.79 (0.37-1.66) |
| GG | 42 | 32 | 0.58 (0.34-1.02) | 11 | 0.57 (0.26-1.24) | 9 | 0.78 (0.33-1.88) | 5 | 0.58 (0.20-1.71) |
| GT/GG | 150 | 167 | 0.87 (0.60-1.27) | 48 | 0.73 (0.44-1.24) | 52 | 1.33 (0.74-2.38) | 22 | 0.73 (0.36-1.47) |
| P trend | .11 | .15 | .95 | .30 | |||||
| MTR Ex26-20A>G | |||||||||
| AA | 169 | 186 | 1.0 (reference) | 58 | 1.0 (reference) | 53 | 1.0 (reference) | 26 | 1.0 (reference) |
| AG | 62 | 79 | 1.15 (0.77-1.71) | 23 | 1.09 (0.61-1.93) | 21 | 1.01 (0.56-1.84) | 12 | 1.18 (0.56-2.48) |
| GG | 10 | 7 | 0.65 (0.24-1.76) | 2 | — | 2 | — | 1 | — |
| AG/GG | 72 | 86 | 1.08 (0.74-1.58) | 25 | 1.02 (0.59-1.77) | 23 | 0.96 (0.54-1.71) | 13 | 1.10 (0.53-2.27) |
| P trend | .98 | .85 | .76 | .97 | |||||
| MTRR Ex5+136C>T | |||||||||
| TT | 230 | 278 | 1.0 (reference) | 101 | 1.0 (reference) | 69 | 1.0 (reference) | 36 | 1.0 (reference) |
| CT | 70 | 91 | 1.03 (0.72-1.48) | 35 | 1.05 (0.65-1.69) | 26 | 1.19 (0.69-2.03) | 10 | 0.90 (0.42-1.93) |
| CC | 8 | 11 | 1.19 (0.47-3.02) | 2 | — | 2 | — | 2 | — |
| CT/CC | 78 | 102 | 1.04 (0.74-1.48) | 37 | 0.99 (0.63-1.58) | 28 | 1.15 (0.69-1.94) | 12 | 0.99 (0.48-2.01) |
| P trend | .75 | .79 | .69 | .83 | |||||
| SHMT1 Ex12+138C>T | |||||||||
| CC | 127 | 125 | 1.0 (reference) | 37 | 1.0 (reference) | 30 | 1.0 (reference) | 21 | 1.0 (reference) |
| CT | 87 | 128 | 1.53 (1.05-2.22) | 40 | 1.59 (0.94-2.71) | 40 | 1.86 (1.07-3.24) | 16 | 1.11 (0.54-2.26) |
| TT | 26 | 17 | 0.66 (0.34-1.29) | 5 | 0.70 (0.25-1.96) | 3 | — | 3 | — |
| CT/TT | 113 | 145 | 1.33 (0.93-1.89) | 45 | 1.39 (0.83-2.32) | 43 | 1.54 (0.90-2.65) | 19 | 0.99 (0.51-1.95) |
| P trend | .68 | .65 | .66 | .71 | |||||
| SHMT1 Ex12+236C>T | |||||||||
| CC | 121 | 122 | 1.0 (reference) | 37 | 1.0 (reference) | 31 | 1.0 (reference) | 20 | 1.0 (reference) |
| CT | 93 | 134 | 1.46 (1.01-2.12) | 41 | 1.46 (0.86-2.47) | 42 | 1.68 (0.97-2.90) | 17 | 1.10 (0.54-2.25) |
| TT | 26 | 17 | 0.64 (0.33-1.25) | 5 | 0.66 (0.23-1.86) | 3 | — | 3 | — |
| CT/TT | 119 | 151 | 1.28 (0.90-1.82) | 46 | 1.28 (0.77-2.13) | 45 | 1.40 (0.82-2.39) | 20 | 0.99 (0.51-1.95) |
| P trend | .81 | .83 | .89 | .70 | |||||
| Gene, nucleotide change . | No. of controls*; n = 319 . | NHL cases . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Overall . | Diffuse large B-cell lymphoma . | Follicular lymphoma . | Small lymphocytic lymphoma . | ||||||
| No. of cases*; n = 386 . | OR (95% CI) . | No. of cases*; n = 142 . | OR (95% CI) . | No. of cases*; n = 98 . | OR (95% CI) . | No. of cases*; n = 48 . | OR (95% CI) . | ||
| BHMT Ex8+453A>T | |||||||||
| AA | 122 | 160 | 1.0 (reference) | 46 | 1.0 (reference) | 46 | 1.0 (reference) | 23 | 1.0 (reference) |
| AT | 99 | 98 | 0.76 (0.53-1.10) | 30 | 0.83 (0.49-1.43) | 28 | 0.76 (0.44-1.33) | 13 | 0.65 (0.31-1.35) |
| TT | 19 | 13 | 0.54 (0.25-1.14) | 5 | 0.74 (0.26-2.13) | 2 | — | 3 | — |
| AT/TT | 118 | 111 | 0.73 (0.51-1.04) | 35 | 0.82 (0.49-1.37) | 30 | 0.68 (0.40-1.17) | 16 | 0.67 (0.34-1.33) |
| P trend | .05 | .43 | .08 | .34 | |||||
| CBS Ex13+41C>T | |||||||||
| CC | 142 | 149 | 1.0 (reference) | 56 | 1.0 (reference) | 36 | 1.0 (reference) | 16 | 1.0 (reference) |
| CT | 136 | 165 | 1.15 (0.83-1.59) | 58 | 1.08 (0.70-1.68) | 51 | 1.57 (0.96-2.58) | 17 | 1.14 (0.55-2.35) |
| TT | 33 | 59 | 1.68 (1.03-2.73) | 22 | 1.59 (0.85-2.98) | 8 | 1.01 (0.42-2.40) | 12 | 3.32 (1.43-7.71) |
| CT/TT | 169 | 224 | 1.25 (0.92-1.70) | 80 | 1.18 (0.78-1.79) | 59 | 1.46 (0.91-2.36) | 29 | 1.56 (0.81-3.01) |
| P trend | .05 | .21 | .35 | .02 | |||||
| FPGS Ex15-263T>C | |||||||||
| TT | 108 | 126 | 1.0 (reference) | 47 | 1.0 (reference) | 30 | 1.0 (reference) | 12 | 1.0 (reference) |
| CT | 160 | 171 | 0.91 (0.65-1.28) | 57 | 0.81 (0.51-1.28) | 45 | 1.04 (0.61-1.77) | 24 | 1.35 (0.65-2.83) |
| CC | 44 | 79 | 1.58 (1.00-2.49) | 31 | 1.64 (0.91-2.94) | 21 | 1.93 (0.98-3.80) | 11 | 2.47 (1.00-6.15) |
| CT/CC | 204 | 250 | 1.06 (0.77-1.45) | 88 | 0.98 (0.64-1.50) | 66 | 1.22 (0.74-2.01) | 35 | 1.57 (0.78-3.16) |
| P trend | .12 | .23 | .10 | .06 | |||||
| FTHFD Ex10-40T>G | |||||||||
| GG | 224 | 284 | 1.0 (reference) | 101 | 1.0 (reference) | 72 | 1.0 (reference) | 34 | 1.0 (reference) |
| GT | 76 | 85 | 0.90 (0.63-1.29) | 31 | 0.92 (0.57-1.50) | 23 | 0.93 (0.54-1.60) | 11 | 0.95 (0.46-1.97) |
| TT | 10 | 9 | 0.67 (0.26-1.68) | 5 | 1.08 (0.36-3.26) | 2 | — | 2 | — |
| GT/TT | 86 | 94 | 0.87 (0.62-1.23) | 36 | 0.94 (0.59-1.49) | 25 | 0.90 (0.53-1.52) | 13 | 1.00 (0.50-1.99) |
| P trend | .35 | .87 | .60 | .88 | |||||
| MTHFR Ex8-62A>C (1298A>C) | |||||||||
| AA | 135 | 183 | 1.0 (reference) | 66 | 1.0 (reference) | 48 | 1.0 (reference) | 24 | 1.0 (reference) |
| AC | 141 | 165 | 0.86 (0.63-1.19) | 61 | 0.90 (0.59-1.39) | 40 | 0.79 (0.48-1.29) | 20 | 0.79 (0.42-1.51) |
| CC | 38 | 35 | 0.67 (0.40-1.13) | 12 | 0.63 (0.31-1.30) | 10 | 0.72 (0.33-1.58) | 4 | — |
| AC/CC | 179 | 200 | 0.82 (0.61-1.11) | 73 | 0.85 (0.56-1.27) | 50 | 0.78 (0.49-1.23) | 24 | 0.75 (0.41-1.39) |
| P trend | .12 | .25 | .29 | .31 | |||||
| MTHFS IVS2-1411T>G | |||||||||
| TT | 80 | 99 | 1.0 (reference) | 35 | 1.0 (reference) | 21 | 1.0 (reference) | 16 | 1.0 (reference) |
| GT | 108 | 135 | 0.99 (0.67-1.46) | 37 | 0.80 (0.46-1.40) | 43 | 1.55 (0.84-2.85) | 17 | 0.79 (0.37-1.66) |
| GG | 42 | 32 | 0.58 (0.34-1.02) | 11 | 0.57 (0.26-1.24) | 9 | 0.78 (0.33-1.88) | 5 | 0.58 (0.20-1.71) |
| GT/GG | 150 | 167 | 0.87 (0.60-1.27) | 48 | 0.73 (0.44-1.24) | 52 | 1.33 (0.74-2.38) | 22 | 0.73 (0.36-1.47) |
| P trend | .11 | .15 | .95 | .30 | |||||
| MTR Ex26-20A>G | |||||||||
| AA | 169 | 186 | 1.0 (reference) | 58 | 1.0 (reference) | 53 | 1.0 (reference) | 26 | 1.0 (reference) |
| AG | 62 | 79 | 1.15 (0.77-1.71) | 23 | 1.09 (0.61-1.93) | 21 | 1.01 (0.56-1.84) | 12 | 1.18 (0.56-2.48) |
| GG | 10 | 7 | 0.65 (0.24-1.76) | 2 | — | 2 | — | 1 | — |
| AG/GG | 72 | 86 | 1.08 (0.74-1.58) | 25 | 1.02 (0.59-1.77) | 23 | 0.96 (0.54-1.71) | 13 | 1.10 (0.53-2.27) |
| P trend | .98 | .85 | .76 | .97 | |||||
| MTRR Ex5+136C>T | |||||||||
| TT | 230 | 278 | 1.0 (reference) | 101 | 1.0 (reference) | 69 | 1.0 (reference) | 36 | 1.0 (reference) |
| CT | 70 | 91 | 1.03 (0.72-1.48) | 35 | 1.05 (0.65-1.69) | 26 | 1.19 (0.69-2.03) | 10 | 0.90 (0.42-1.93) |
| CC | 8 | 11 | 1.19 (0.47-3.02) | 2 | — | 2 | — | 2 | — |
| CT/CC | 78 | 102 | 1.04 (0.74-1.48) | 37 | 0.99 (0.63-1.58) | 28 | 1.15 (0.69-1.94) | 12 | 0.99 (0.48-2.01) |
| P trend | .75 | .79 | .69 | .83 | |||||
| SHMT1 Ex12+138C>T | |||||||||
| CC | 127 | 125 | 1.0 (reference) | 37 | 1.0 (reference) | 30 | 1.0 (reference) | 21 | 1.0 (reference) |
| CT | 87 | 128 | 1.53 (1.05-2.22) | 40 | 1.59 (0.94-2.71) | 40 | 1.86 (1.07-3.24) | 16 | 1.11 (0.54-2.26) |
| TT | 26 | 17 | 0.66 (0.34-1.29) | 5 | 0.70 (0.25-1.96) | 3 | — | 3 | — |
| CT/TT | 113 | 145 | 1.33 (0.93-1.89) | 45 | 1.39 (0.83-2.32) | 43 | 1.54 (0.90-2.65) | 19 | 0.99 (0.51-1.95) |
| P trend | .68 | .65 | .66 | .71 | |||||
| SHMT1 Ex12+236C>T | |||||||||
| CC | 121 | 122 | 1.0 (reference) | 37 | 1.0 (reference) | 31 | 1.0 (reference) | 20 | 1.0 (reference) |
| CT | 93 | 134 | 1.46 (1.01-2.12) | 41 | 1.46 (0.86-2.47) | 42 | 1.68 (0.97-2.90) | 17 | 1.10 (0.54-2.25) |
| TT | 26 | 17 | 0.64 (0.33-1.25) | 5 | 0.66 (0.23-1.86) | 3 | — | 3 | — |
| CT/TT | 119 | 151 | 1.28 (0.90-1.82) | 46 | 1.28 (0.77-2.13) | 45 | 1.40 (0.82-2.39) | 20 | 0.99 (0.51-1.95) |
| P trend | .81 | .83 | .89 | .70 | |||||
We omitted estimates based on case counts less than 5 (indicated by —).
Unconditional logistic regression model adjusted for age at study entry (continuous), sex, and race (white, non–African American others).
Counts of controls and overall/subtype cases vary across polymorphisms due to (1) 2-step genotyping strategy (genotyping was not completed for certain polymorphisms [Table 1] in participants who provided only buccal cell samples [∼25%] if genotype information from blood-based DNA samples did not yield significant or suggestive associations with overall NHL) and (2) error in genotyping or insufficient DNA (2%-4% in general).
We examined gene-nutrient and gene-gene interactions regardless of significance of the main effects of genetic polymorphisms. Gene-nutrient interactions were assessed by including ordinal score variables of each genotype and nutrient (dichotomized at the median based on the nutrient distribution among control individuals) along with their product term in the regression model and were concluded significant by the P value less than .10 of the cross-product term. For significant gene-nutrient interactions, the nutrient-NHL association comparing above versus below median of the nutrient is presented stratified by genotypes (Table 4). All nutrients except alcohol were adjusted for total energy intake by the nutrient-density method.43 Regression models of folate, vitamin B6, and methionine were adjusted for each other for potential confounding. Additional adjustments for energy intake and other nutrients did not materially change the risk estimates (ie, change < 10%).
Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the association between nutritional factors of one-carbon metabolism and non-Hodgkin lymphoma, stratified by selected genetic polymorphisms, the NCI-SEER Study, 1998-2000
| Nutritional factor, genotype . | Dietary intake levels of nutrients . | P interaction . | ||
|---|---|---|---|---|
| Low, less than median . | High, median or higher . | |||
| No. of cases/controls* . | No. of cases/controls* . | OR (95% CI) . | ||
| Vitamin B6 | ||||
| Combined genotypes | 237/162 | 149/157 | 0.70 (0.51-0.95) | N/A |
| FPGS Ex15-263T>C | ||||
| TT | 69/59 | 57/49 | 1.03 (0.61-1.74) | |
| CT | 104/81 | 67/79 | 0.71 (0.46-1.12) | |
| CC | 59/17 | 20/27 | 0.22 (0.10-0.52) | .002 |
| MTHFS IVS2-1411T>G | ||||
| TT/TG | 149/89 | 85/99 | 0.54 (0.36-0.81) | |
| GG | 18/26 | 14/16 | 1.68 (0.61-4.65) | .06 |
| MTR Ex26-20A>G | ||||
| AA | 117/78 | 69/91 | 0.55 (0.35-0.86) | |
| AG/GG | 50/44 | 36/28 | 1.20 (0.62-2.33) | .05 |
| Methionine | ||||
| Combined genotypes | 225/164 | 161/155 | 0.82 (0.60-1.11) | N/A |
| FTHFD Ex10-40G>T | ||||
| GG | 179/112 | 105/112 | 0.63 (0.44-0.91) | |
| GT/TT | 43/44 | 51/42 | 1.34 (0.74-2.46) | .04 |
| MTHFR Ex8-62A>C (1298A>C) | ||||
| AA | 104/77 | 79/58 | 1.04 (0.66-1.63) | |
| AC | 92/72 | 73/69 | 0.93 (0.58-1.49) | |
| CC | 27/11 | 8/27 | 0.13 (0.04-0.39) | .007 |
| MTRR Ex5+136T>C | ||||
| TT | 165/108 | 113/122 | 0.67 (0.47-0.97) | |
| CT/CC | 57/48 | 45/30 | 1.28 (0.69-2.35) | .05 |
| Nutritional factor, genotype . | Dietary intake levels of nutrients . | P interaction . | ||
|---|---|---|---|---|
| Low, less than median . | High, median or higher . | |||
| No. of cases/controls* . | No. of cases/controls* . | OR (95% CI) . | ||
| Vitamin B6 | ||||
| Combined genotypes | 237/162 | 149/157 | 0.70 (0.51-0.95) | N/A |
| FPGS Ex15-263T>C | ||||
| TT | 69/59 | 57/49 | 1.03 (0.61-1.74) | |
| CT | 104/81 | 67/79 | 0.71 (0.46-1.12) | |
| CC | 59/17 | 20/27 | 0.22 (0.10-0.52) | .002 |
| MTHFS IVS2-1411T>G | ||||
| TT/TG | 149/89 | 85/99 | 0.54 (0.36-0.81) | |
| GG | 18/26 | 14/16 | 1.68 (0.61-4.65) | .06 |
| MTR Ex26-20A>G | ||||
| AA | 117/78 | 69/91 | 0.55 (0.35-0.86) | |
| AG/GG | 50/44 | 36/28 | 1.20 (0.62-2.33) | .05 |
| Methionine | ||||
| Combined genotypes | 225/164 | 161/155 | 0.82 (0.60-1.11) | N/A |
| FTHFD Ex10-40G>T | ||||
| GG | 179/112 | 105/112 | 0.63 (0.44-0.91) | |
| GT/TT | 43/44 | 51/42 | 1.34 (0.74-2.46) | .04 |
| MTHFR Ex8-62A>C (1298A>C) | ||||
| AA | 104/77 | 79/58 | 1.04 (0.66-1.63) | |
| AC | 92/72 | 73/69 | 0.93 (0.58-1.49) | |
| CC | 27/11 | 8/27 | 0.13 (0.04-0.39) | .007 |
| MTRR Ex5+136T>C | ||||
| TT | 165/108 | 113/122 | 0.67 (0.47-0.97) | |
| CT/CC | 57/48 | 45/30 | 1.28 (0.69-2.35) | .05 |
Unconditional logistic regression model adjusted for age at study entry (continuous), sex, race (white, non-African American others), and mutually for dietary intake of methionine and vitamin B6. Median dietary intake is defined as 0.97 mg/1000 kcal for vitamin B6 and 0.97 g/1000 kcal for methionine.
For low dietary intakes of both vitamin B6 and methionine, all ORs were 1.0 (reference).
Counts of cases and controls vary across polymorphisms due to (1) two-step genotyping strategy (genotyping was not completed for certain polymorphisms [Table 1] in participants who provided only buccal cell samples [∼25%] if genotype information from blood-based DNA samples did not yield significant or suggestive associations with overall NHL) and (2) error in genotyping or insufficient DNA (2%-4% in general).
We used SAS/Genetics (version 9.1.3; SAS Institute, Cary, NC) to assess Hardy-Weinberg equilibrium and linkage disequilibrium among SNPs from the same gene. We explored haplotype analyses for genes with 2 or more polymorphisms but did not detect any stronger associations, and therefore, present the findings from individual polymorphism analyses. To evaluate the probability of false-positive associations, we computed the false discovery rate (FDR),44 which controls the proportion of false positives out of all significant findings using the P values from the regression of score variables (ie, the trend test, also referred to as the additive model) using SAS software: we considered FDR less than 0.2 as noteworthy. We also computed the false-positive report probabilities (FPRPs)45 using prior probabilities ranging from 0.1 to 0.001 based on gene selection criteria described in “Genotyping” and considered values below a criterion of 0.2 noteworthy as recommended in the initial description of the method.45
Results
Descriptive characteristics and histologic subgroup distributions were comparable between all participants who were genotyped and the subgroup who had both genotype and diet information (Table 2): controls with and without diet information were similar in the characteristics compared, except the racial distribution due to study design. In the subgroup with diet data, cases tended to be younger, but were otherwise similar to controls with respect to sex, race, study center, education, family history of NHL, body mass index (weight in kilograms/height in meters squared), and the availability of biospecimens.
Table 3 shows the genotype-NHL association of polymorphisms that exhibited a significant main effect, linear trend, or interaction with a nutrient among people with both genotype and diet data. Associations for all polymorphisms are available on the Blood website; see the Supplemental Materials link at the top of the online article (Table S1, for participants with diet data; Table S2, for all participants with genotype data). The rarer homozygous genotype for BHMT Ex8+453A>T (TT versus AA) was associated with a lower risk of NHL overall, whereas the rarer homozygotes for CBS Ex13+41C>T (TT versus CC) and FPGS Ex15-263T>C (CC versus TT) were associated with an elevated risk. Heterozygotes for SHMT1 Ex12+138C>T and Ex12+236C>T (CT versus CC for both) were associated with a higher risk of NHL overall. Findings were similar in pattern for main subtypes: although the results reached statistical significance for the positive association of CBS Ex13+41C>T and FPGS Ex15-263T>C with the risk of SLL and of SHMT1 Ex12+138C>T with follicular lymphoma (Table 3), the difference in risk estimates among subtypes was not significant in case-patient comparisons. Main effects among all genotyped data (Table S2) showed similar association patterns and identified additional variants in FTHFD, MTHFS, and MTR for significant associations or trends with NHL overall (ie, P value < .05). FDR and FPRP estimates for all our findings were above the cut-point value .2.
Based on similar main effects across main NHL subtypes, we assessed gene-nutrient interactions on combined NHL. We examined interactions between all genetic variants and nutritional determinants, specifically vitamin B6 and methionine that were previously found protective against NHL.27 We detected a number of significant interactions (by P value < .1). The main nutrient-NHL association in these data was attenuated from the previous diet analyses27 due to loss of participants without genotype information and also less detailed categories applied. The inverse association between vitamin B6 intake and NHL was apparent only in the FPGS Ex15-263T>C CC (P interaction = .002), MTHFS IVS2-1411T>G TT/TG (P interaction = .06), and MTR Ex26-20A>G AA (P interaction = .05) genotypes. Also, the inverse association between dietary methionine and overall NHL appeared to be limited to FTHFD Ex10-40G>T GG (P interaction = .04), MTHFR Ex8-62A>C CC (P interaction = .007), and MTRR Ex5+136T>C TT genotypes (P interaction = .05). There were no meaningful interactions for other nutrients, including folate. The main associations and gene-nutrient interactions were not confounded by other risk factors in combined and individual subtype analyses.
Among all participants with genotype information, we detected an interaction between CBS and FPGS (P interaction = .007), but without synergistic effects (all other genotypes compared with CBS Ex13+41C>T CC/FPGS Ex15-263T>C TT showed similarly elevated risk for NHL by about 50%) and without a further interaction with vitamin B6 (data not shown).
Discussion
We observed that the protective association of high dietary intake of one-carbon nutrients with NHL varied by genetic polymorphisms of the one-carbon metabolizing enzymes: especially, vitamin B6 by polymorphisms in FPGS, MTHFS, and MTR and methionine by polymorphisms in FTHFD, MTHFR, and MTRR. To our knowledge, this is the first investigation of NHL that examined potential gene-nutrient interactions involving one-carbon metabolism.
Our findings were especially strong for polymorphisms in FPGS and MTHFR: their interactions with vitamin B6 and methionine intake, respectively, determined by the logistic regression methods were also evident using the case-only analysis (data not shown), but other borderline findings, which are more likely due to chance variation in controls, went away in the case-only analysis. The enzyme FPGS catalyzes an essential and rate-limiting polyglutamylation step in the intracellular one-carbon metabolism46,47 : it adds several glutamyl residues to all reduced folates supplied from the circulation. The polyglutamylation by FPGS improves both retention of folates within the cell and their affinity to one-carbon metabolizing enzymes. Expression of FPGS was found to be low in acute lymphoblastic leukemias,48 and the low expression in normal-appearing mucosa of colorectal carcinoma cases has been associated with poor survival.49 Common genetic variants of FPGS, although not studied to date, may convey functional attributes: our data suggest a detrimental effect associated with its Ex15-263T>C variant located in the 3′ UTR, which may reflect the functional significance of the variant (eg, stability of mRNA50 ) or its linkage with a true functional variant.51 The risk associated with the variant was moderate in the group consuming high vitamin B6, yielding the protective association of vitamin B6 only among the homozygous variant genotype. We currently do not have explanations for the mechanism.
MTHFR is the most studied enzyme in one-carbon metabolism that irreversibly “commits” 5,10-methylenetetrahydrofolate, a common substrate to both methylation pathways and nucleotide synthesis, toward the methylation cycle and away from nucleotide/DNA synthesis.52 Its 2 common variants, Ex8-62A>C (also known as 1298A>C) and Ex5+79C>T (or 677C>T), are in linkage disequilibrium,53 have shown diminished enzyme activity compared with the wild types,34,35 and have been associated with a lower risk of leukemia.21 The protective association may be due to preferential DNA synthesis with these MTHFR polymorphisms than with wild types, especially when one-carbon nutrients are sufficient, which was clearly demonstrated in a recent in vivo experiment.52 Such promotion of DNA synthesis over methylation with adequate one-carbon supply may be critical to prevent DNA aberrations and carcinogenesis in lymphoid cells with high rate of turnover. Our study is the first to show an interaction between a variant in MTHFR and one-carbon nutrients, specifically methionine, in relation to NHL. This finding emulates the previous epidemiologic reports of such interactions in colorectal cancer.24,54-56
Markedly different incidence patterns57 as well as pathologic and clinical heterogeneity among NHL subtypes58 support subtype-specific investigations of etiologic factors. At the same time, some risk factors, including family history and occupational/environmental factors, have shown fairly consistent positive or inverse associations across histologic subtypes.58 For example, in large pooled analyses from the InterLymph, a genetic polymorphism in tumor necrosis factor was associated with DLBCL, but not follicular lymphoma.30 Further, we previously reported a protective association of dietary folate with DLBCL, but not follicular lymphoma.27 However, dietary methionine was inversely associated with main subtypes similarly.27 In the current study, we explored the associations with all NHL as well as by main subtypes and found the main effect of one-carbon–related genetic polymorphisms to be mostly consistent across subtypes.
We did not detect a significant main effect of the variants in MTHFR, MTRR, or TYMS12,14-16,18,22 and found approximately 50% lower risk associated with MTR Ex26-20A>G (Table S2),14,16,20 which is consistent with previous findings. The moderate inverse association of the variants in MTHFR or MTR with NHL appeared contingent on methionine or vitamin B6 intake, respectively, in our data. Our investigation of additional enzymes that had not been explored led to a discovery of moderate associations regarding BHMT, CBS, and FPGS (and FTHFD and MTHFS in Table S2), of which FPGS, FTHFD, and MTHFS showed an interaction with nutritional status. These findings for novel main effects and interactions are intriguing and warrant replications, especially in pooled analyses in order to reduce the possibility of chance findings based on small numbers.
For a number of cancers that one-carbon metabolism has been linked to, gene-nutrient interactions are considered to be an integral part of the epidemiologic investigation. Folate has been reported to compensate the reduced enzymatic activity of MTHFR with the Ex5+79C>T (or 677C>T) variant by changing the conformation and structure of the enzyme.59 However, we did not observe any independent interaction of folate when simultaneously adjusted for vitamin B6. The lack of an independent interaction may be due to the correlation between dietary folate and vitamin B6 from common food sources (r = 0.65 in our data) or due to improved folate status since fortification above the range where one-carbon metabolism relies on folate dose-dependently: the manifestation of genetic susceptibility may rather vary by the overall availability of one-carbon moieties/nutrients.60 For example, the protective association between MTHFR variants and childhood acute lymphoblastic leukemia was apparent only among children who were born before the prenatal folate supplementation was widely put into practice in Canada.61
This study had a number of strengths. The population-based design of this case-control study might have reduced selection bias compared with hospital-based recruitment. The median consumption of vitamin B6 (1.9 mg/2000 kcal) and methionine (1.9 g/2000 kcal) among our control individuals was largely comparable with the Recommended Dietary Allowances and the median intake levels of the US population.4,62 We used histologically confirmed cases, reducing misclassification bias. We incorporated a detailed paradigm of one-carbon metabolism compared with previous studies, which is an important consideration for studying a composite effect of a number of genes involved in complex metabolic pathways.
On the other hand, our findings of gene-nutrient interactions have similar limitations that were discussed in our previous study of dietary associations.27 We had assessed whether the low response rates of our study could have driven the inverse associations between one-carbon nutrients and NHL and did not find a strong indication for such selection bias, based on similar associations across sex and study centers with different response rates or by education levels (a correlate of participation among controls), and also based on no confounding by these factors. Our findings based on postdiagnostic assessment of diet need to be replicated in prospective data with less influence of potential bias associated with recalled diet and selective participation of healthier controls. Although this is one of the largest polymorphism studies to date regarding one-carbon metabolism, it was still limited to examine gene-nutrient interactions.
Our strategy of targeting few SNPs per gene based on literature evidence and limited variation data available at the time of genotyping might have missed important variants for NHL risk that are not in linkage disequilibrium with the ones we studied. This strategy also limited our ability to construct and perform a meaningful haplotype analysis. Similarly, future work could expand to nutritional hypotheses in the context of other genes involved in one-carbon metabolism, including DHFR,63 FOLR2,6 and MTHFD164 (Figure 1).
In conclusion, this study provides further evidence that the one-carbon pathway could contribute to lymphomagenesis, specifically through interactions between common genetic variation and dietary intake. However, these results require replication in further large studies as well as pooled analyses. As the understanding of gene-nutrient interactions in one-carbon metabolism advances in relation to lymphomagenesis, our findings could be incorporated into evaluation of nutritional intervention trials and development of suitable biomarkers for detection of high-risk individuals (eg, CpG island methylation phenotype65 ). Lastly, this line of investigation could also have implications for prognosis and survival of NHL.
Authorship
Contribution: The principal investigator of the NCI-SEER NHL Study is P.H.; questionnaire and cancer outcome data from the 4 NCI-SEER registries were obtained and provided by W.C., L.E.K., S.D., and M.S.; the one-carbon metabolism polymorphism project was conceived and led by Q.L., N.R., and U.L.; bioinformatics support and genotyping were supervised by S.C. at the NCI Core Genotyping Facility; the statistical analysis was performed by U.L. and Q.L. with input from S.S.W., P.H., and N.R.; the paper was drafted and revised by U.L., S.S.W., P.H., L.E.K., A.B., N.R., and Q.L.; and all authors reviewed and approved the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Unhee Lim, Nutritional Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd, EPS 320, Rockville, MD 20852; e-mail: limu@mail.nih.gov.
Appendix
Abbreviations of enzymes or proteins (genes) are as follows: BHMT (BHMT) indicates betaine-homocysteine methyltransferase; CBS (CBS), cystathionine-beta-synthase; DHFR (DHFR), dihydrofolate reductase; FOLR2 (FOLR2), folate receptor 2 (or folate receptor beta); FPGS (FPGS), folylpolyglutamate synthase; FTHFD (FTHFD), 10-formyltetrahydrofolate dehydrogenase (or aldehyde dehydrogenase 1 family, member L1; ALDH1L1); GGH (GGH), gamma-glutamyl hydrolase; MBD2 (MBD2), methyl-CpG binding domain protein 2; MGMT (MGMT), O-6-methylguanine-DNA methyltransferase; MLH1 (MLH1), mutL homolog 1; MSH2 (MSH2), mutS homolog 2; MTHFD1 (MTHFD1), (cytoplasmic) 5,10-methylenetetrahydrofolate dehydrogenase 1; MTHFD2 (MTHFD2), (mitochondrial) 5,10-methylenetetrahydrofolate dehydrogenase 2; MTHFR (MTHFR), 5,10-methylenetetrahydrofolate reductase; MTHFS (MTHFS), 5,10-methenyltetrahydrofolate synthetase (or 5-formyltetrahydrofolate cyclo-ligase); MTR (MTR), 5-methyltetrahydrofolate-homocysteine methyltransferase (or methionine synthase); MTRR (MTRR), 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (or methionine synthase reductase); SHMT1 (SHMT1), (cytoplasmic) serine hydroxymethyltransferase 1; SLC19A1 (SLC19A1), solute carrier family 19 (folate transporter), member 1 (or reduced folate carrier 1); TCN1 (TCN1), transcobalamin 1; and TYMS (TYMS), thymidylate synthetase.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by the Intramural Research Program of the NIH at the NCI and by contracts with the NCI (N01-PC-67010, N01-PC-67008, N02-PC-71105, N01-PC-67009, and N01-PC-65064).
We thank our study participants for their involvement. We gratefully acknowledge the contributions of the SEER centers of Los Angeles, Iowa, Seattle, and Detroit for rapid identification of cases; the Centers for Medicare & Medicaid Services for selection of older controls; Drs Meredith Yeager and Robert Welch at the Core Genotyping Facility at the Advanced Technology Center for carrying out genotyping and providing bioinformatics support; Dr Nilanjan Chatterjee for input on statistical analyses; Drs Min Shen and Sonja Berndt for assistance with genotype and haplotype analyses; Carol Haines (Westat) for development of study materials and procedures, for selection of younger controls, and for study coordination; Steve Palladino and Peter Hui (IMS) for computer support; Carla Chorley (BBI Biotech Research Laboratories) for specimen handling; and Geoffrey Tobias for research assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal