Abstract
α-defensins are antibiotic peptides that act as natural inhibitors of HIV-1 infection. However, the mechanisms of such inhibition are still unclear. Here we demonstrate that α-defensins block the earliest steps in the viral infectious cycle, as documented using an HIV-1 envelope-mediated cell-fusion assay. A broad-spectrum inhibitory activity was observed on primary and laboratory-adapted HIV-1 isolates irrespective of their coreceptor specificity and genetic subtype. A primary mechanism of such inhibition was identified as the ability of α-defensins to bind specifically both to the primary HIV-1 cellular receptor, CD4, and to the viral envelope glycoprotein, gp120. Moreover, treatment of CD4+ T cells with α-defensins caused a dramatic downmodulation of CD4 expression. By monoclonal antibody competition, the regions of interaction with α-defensins were mapped to the D1 domain of CD4 and to a surface contiguous to the CD4- and coreceptor-binding sites of gp120. Consistent with these findings, α-defensins inhibited the binding of gp120 to CD4. These data demonstrate that α-defensins specifically block the initial phase of the HIV infectious cycle and modulate the expression of CD4, a critical receptor in the physiology of T-cell activation.
Introduction
Evidence suggests that a concerted action of innate and adaptive immune responses is essential for the effective containment and, ultimately, the clearance of invading microorganisms.1 The innate immune system is phylogenetically more archaic and constitutes the first line of antimicrobial defense, characterized by a rapid response time; however, it is limited in its target specificity. By contrast, the adaptive immune system has evolved refined molecular devices for the recognition of a wide variety of specific epitopes, but a lag time is required for its functional activation. Once activated, most of the cells involved in both innate and cognate immune responses secrete soluble factors, including cytokines, chemokines, antibiotic peptides, and others, which can directly antagonize infectious microorganisms and/or contribute to the recruitment and activation of other immune cells, thereby amplifying the cascade of defense mechanisms and bolstering their effectiveness.2,3
In HIV infection, various host-derived soluble factors with antiviral activity have been described, which act by both specific and nonspecific mechanisms. These include chemokines such as RANTES, MIP-1α, and MIP-1β 4 ; cytokines such as the interferons, IL-10, IL-13, IL-16, transforming growth factor-β, and others5 ; as well as, more recently, defensins.6-9 Defensins are natural antibiotic peptides that play an important role in innate immune responses by acting as broad-spectrum antibacterial, antifungal, and antiviral effector molecules10 as well as by enhancing certain adaptive immune responses.11 α-defensins 1 to 3 are 3- to 4-kDa cationic peptides (29 to 30 amino acids [aa]) characterized by a conserved 6-cysteine motif, with 3 disulfide bonds that impose a characteristic β-sheet structure.12 α-defensin mRNA is expressed by natural killer (NK), B cells, and γδ T lymphocytes,13,14 while its expression by monocytes/macrophages is still debated.14,15 Neutrophilic granulocytes are by and large the major producers of α-defensins, which are stored at very high concentration in their azurophilic granules, accounting for up to 5% of the entire cellular protein content.2 Neutrophils, the most abundant leukocytes in the circulation, are the first cells to infiltrate inflammatory sites and prominent components of inflammatory responses induced by viral infections.16-18 The anti-HIV activity of α-defensins was first described in 1993,19 but it received little attention until 2002 when Zhang et al claimed that α-defensins 1, 2, and 3 constitute major components of the soluble HIV-suppressive activity produced by CD8+ T cells.20 Subsequently, however, it was shown that CD8+ T cells do not express α-defensin mRNA,13,15 suggesting that the α-defensins detected in CD8+ T-cell culture supernatants were derived from contaminating neutrophils.21,22 What remains undisputed, nevertheless, is that α-defensins are active against HIV-1, as recently confirmed by several authors.6,15,23,24 α-defensins have been identified as in vivo correlates of protection in mother-to-child transmission, with a significant association between high α-defensin concentrations and a decreased risk of intrapartum and postnatal HIV transmission.25
The mechanism of α-defensin–mediated HIV-1 inhibition is still uncertain. Based on the results obtained in different experimental models, it appears that α-defensins act at 2 different levels: inhibition of the viral replication cycle and blockade before HIV-1 entry into target cells. The first mechanism is supported by evidence that α-defensins inhibit the HIV infectious cycle after virus entry into target cells.23,24 This effect appears to be mediated by inhibition of the PKC signaling pathway and results in the blockade of proviral DNA nuclear import.24 In addition, α-defensins may act indirectly through the stimulation of HIV-inhibiting chemokine release by primary macrophages.26 On the other hand, several lines of evidence are compatible with the hypothesis that α-defensins also inhibit HIV-1 before its ingress into the target cell: (1) α-defensins are lectinlike molecules that interact with membrane glycoproteins6 ; consistent with this concept, both α- and θ-defensins were shown to bind to carbohydrate moieties in gp120 and CD4 as shown by surface plasmon resonance6,9 ; (2) retrocyclin, a cyclic antimicrobial peptide that shares high homology with α-defensins not only binds to glycoproteins but also crosslinks them, resulting in inhibition of fusion and entry of viruses as diverse as influenza virus, Sindbis virus, and baculovirus27 ; (3) α-defensins directly inactivate HIV-1 virion particles15,28 with a mode of action that is not yet fully understood and could involve disruption of virion lipids and/or interference with receptor binding. Despite these observations, no data on the effect of α-defensins on the early steps of HIV-1 infection have hitherto been reported.
To investigate the mechanism(s) of α-defensin–mediated HIV-1 inhibition, we evaluated the ability of α-defensin-1 and -2 to interfere with the earliest steps in the HIV life cycle using an envelope-mediated cell-fusion assay. We found that α-defensins directly interact with specific regions of the CD4 receptor and the gp120 envelope glycoprotein. Such binding results in the blockade of the reciprocal interaction between CD4 and gp120. Moreover, we demonstrated that α-defensins markedly downmodulate the expression of CD4, a receptor molecule that plays a critical role in the physiology of T-cell activation.
Materials and methods
Proteins and antibodies
Different human α-defensin preparations were used in this study, including synthetic α-defensin-1 (Sigma-Aldrich, St Louis, MO; lot no. 063K13021, more than 91% purity by high-performance liquid chromatography [HPLC]), synthetic α-defensin-2 (Sigma-Aldrich; lot no. 053K13771, 97% purity by HPLC), and recombinant human α-defensin-1 produced in Escherichia coli (Chemicon, Temecula, CA; lot no. 22101327, more than 98% purity by HPLC). In all preparations, bacterial lipopolysaccharide (LPS) contamination was determined to be less than 0.1 ng/μg protein. Recombinant human 4-domain soluble CD4 (sCD4); recombinant gp120s from HIV-1BaL, HIV-1SF162, and HIV-1MN; human anti–HIV-1 gp120 monoclonal antibodies (mAbs) 2G12, IgG1b12, 447-52D, 17b, 48d, and F105; and mouse anti-CXCR4 mAb 44717.111 all were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (ARRRP), Rockville, MD. Additional mouse mAbs used were anti-CD4 mAbs Leu3a (Becton Dickinson, San Jose, CA), 13B8 and Vit4 (Immunotech, Marseilles, France), OKT4 (Ortho Diagnostics, Raritan, NJ), and DB81 (S. Burastero and P.L., manuscript in preparation), anti-CCR5 2D7 (PharMingen, La Jolla, CA), anti-CD2, -CD26, -CD45RO, -CD46, and -HLA-DR (ImmunoTools, Friesoythe, Germany), anti-gp120 D19.29
Viruses and cells
Selected primary (91US714, 93BR020, 92US077) and laboratory-passaged (IIIB, BaL, JR-FL, MN) HIV-1 isolates were obtained through the NIH ARRRP. Primary isolate B117 was provided by Eva M. Fenyo, Lund University, Sweden. Two pediatric primary isolates (IT224.18 and IT193.21) were provided Gabriella Scarlatti, Department of Biological and Technological Research (DIBIT), San Raffaele Scientific Institute, Milan. All primary isolates were minimally passaged in vitro in primary human peripheral-blood mononuclear cells (PBMCs). For isolates CM235, BK132, and 89.6, recombinant vaccinia vectors expressing the cloned envelope genes (provided by Dr Edward A. Berger, Laboratory of Viral Diseases [LVD], National Institute of Allergy and Infectious Diseases [NIAID], Bethesda, MD) were used.
HeLa cells were obtained from the American Type Culture Collection, Rockville, MD. NIH-3T3 mouse fibroblast cells stably expressing human CD4 as well as either CCR5 or CXCR4 were provided by the NIH ARRRP. Chronically infected PM1 and SupT1 were derived as described.29
HIV-1 envelope-mediated cell-fusion assay
The fusion assay was performed as described29 using a modification of the test originally developed by Berger and coworkers.30 In the modified assay, the effector cells were chronically infected PM1 or SupT1 cells, whereas the target cells were NIH-3T3 CD4+CCR5+ or NIH-3T3 CD4+CXCR4+. For the cloned envelopes CM235, BK132, and 89.6, the classic assay was employed using as effectors HeLa cells infected with the appropriate vaccinia vectors (kindly provided by Dr Edward A. Berger).
Flow cytometry
PM1 cells were incubated with α-defensins in RPMI without FBS and then stained with the mAbs for 20 minutes at room temperature. The cells were then washed with phosphate-buffered saline (PBS) supplemented with 1% FBS and labeled with PE-conjugated polyclonal goat antimouse antibody (Sigma-Aldrich). Cells treated with an irrelevant, isotype-matched mAb were used as negative controls. Samples were analyzed on a FACScan cytometer (Becton Dickinson). In some experiments, the cells were fixed with 2% paraformaldehyde before treatment with α-defensins and mAb staining.
ELISA
Flat-bottom, 96-well enzyme-linked immunosorbent assay (ELISA) plates were coated with recombinant sCD4 (5 μg per well) or gp120 (2 μg per well) in PBS for 18 hours at 4°C. Bovine serum albumin (0.5%) was used for blocking the plates. Antibodies were added in 100 μL PBS and incubated for 1 hour at room temperature, after which the plate was washed and incubated for an additional hour with the secondary antibody. Peroxidase-conjugated, affinity-purified goat anti–mouse IgG or goat anti–human IgG (Sigma-Aldrich) was used as secondary antibodies. The reaction was revealed by using an appropriate substrate. The specific signal was calculated by subtracting background values obtained from replicate wells containing all the reagents except the specific ligand for each assay. Binding of mAbs 17b and 48d, directed to CD4-induced epitopes, was performed after preincubation of immobilized gp120 with sCD4 (2 μg per well) in PBS for 20 minutes at 37°C.29
Results
Inhibition of HIV-1 envelope-mediated cell-fusion by α-defensins
To investigate the mechanism of anti-HIV activity of human α-defensins, we used a cell-fusion assay based on vaccinia virus technology. Because α-defensins 1 and 2 are released simultaneously by neutrophils upon degranulation and have been reported to exert stronger anti-HIV effects in combination,20 a first set of experiments was performed using an equimolar mixture of synthetic human α-defensins 1 and 2. Figure 1A shows that α-defensins inhibited, in a dose-dependent fashion, cell fusion mediated by 3 HIV-1 envelopes with different coreceptor specificity (IIIB and MN[X4]; BaL[R5]). The potency of the effect was variable on the different envelopes, with half-maximal inhibitory concentrations (IC50) of 12.5 μM for IIIB, 26.5 μM for BaL, and 55.3 μM for MN. Although the level of bacterial LPS in the protein preparations was below 0.1 ng/μg, to exclude any possible effects of minimal endotoxin concentrations we tested LPS in the HIV-1 envelope-mediated cell fusion and observed no inhibitory effects at doses up to 1 μg/mL (data not shown). Because envelope-mediated cell fusion closely mimics the molecular events leading to viral entry, these results demonstrated that α-defensins block HIV-1 infection at its earliest stages, before the viral entry step.
α-defensins inhibit HIV-1 envelope-mediated cell fusion. (A) Dose-dependent inhibition of HIV-1IIIB (squares), HIV-1BaL (circles), and HIV-1MN (triangles) by α-defensins 1 and 2. The fusion assays were performed using as effectors human T cells persistently infected with HIV-1 (PM1 for BaL and MN, SupT1 for IIIB) and infected 18 hours earlier with a recombinant vaccinia vector expressing the lacZ reporter gene under control of the bacteriophage T7 promoter (vCB-21R); the target cells were NIH-3T3 mouse fibroblasts stably expressing human CD4 and either CXCR4 or CCR5 and infected 18 hours earlier with a recombinant vaccinia vector expressing the T7 RNA polymerase (vTF7-3). The fusion reaction was initiated by incubating effector and target cells together for 2 hours at 37°C in the presence or absence of different concentrations of an equimolar mixture of synthetic α-defensins 1 and 2 at the indicated concentrations (total dose). The fusion reaction was conducted in DMEM supplemented with 2.5% FBS (solid symbols) or no FBS (open symbols). (B) Dose-dependent inhibition of HIV-1MN envelope-mediated fusion by synthetic α-defensin-1 (○), synthetic α-defensin-2 (•), and recombinant α-defensin-1 (□) used individually. The assays were performed as in panel A in the absence of FBS.
α-defensins inhibit HIV-1 envelope-mediated cell fusion. (A) Dose-dependent inhibition of HIV-1IIIB (squares), HIV-1BaL (circles), and HIV-1MN (triangles) by α-defensins 1 and 2. The fusion assays were performed using as effectors human T cells persistently infected with HIV-1 (PM1 for BaL and MN, SupT1 for IIIB) and infected 18 hours earlier with a recombinant vaccinia vector expressing the lacZ reporter gene under control of the bacteriophage T7 promoter (vCB-21R); the target cells were NIH-3T3 mouse fibroblasts stably expressing human CD4 and either CXCR4 or CCR5 and infected 18 hours earlier with a recombinant vaccinia vector expressing the T7 RNA polymerase (vTF7-3). The fusion reaction was initiated by incubating effector and target cells together for 2 hours at 37°C in the presence or absence of different concentrations of an equimolar mixture of synthetic α-defensins 1 and 2 at the indicated concentrations (total dose). The fusion reaction was conducted in DMEM supplemented with 2.5% FBS (solid symbols) or no FBS (open symbols). (B) Dose-dependent inhibition of HIV-1MN envelope-mediated fusion by synthetic α-defensin-1 (○), synthetic α-defensin-2 (•), and recombinant α-defensin-1 (□) used individually. The assays were performed as in panel A in the absence of FBS.
Because various proteins present in bovine serum (eg, fetuin, α2-macroglobulin) are known to bind and sequester α-defensins,6,31,32 we tested the influence of serum on fusion inhibition by α-defensins by performing parallel tests in the presence and absence of bovine serum. Figure 1A shows that the antiviral activity of α-defensins was dramatically increased in the absence of bovine serum (IC50, 2.0 μM for IIIB, 3.1 μM for BaL, and 3.5 μM for MN), demonstrating that serum proteins impair the capacity of α-defensins to inhibit the early events of the HIV-1 infectious cycle.
To rule out that α-defensin–mediated inhibition of HIV-1–induced fusion was due to cytotoxicity or other nonspecific effects, we tested the ability of α-defensins to inhibit fusion between effector cells infected with a different virus (HHV-6) and target cells expressing the appropriate cellular receptor, CD46.33 HHV-6–mediated membrane fusion was not significantly affected by treatment with α-defensins at concentrations up to 15 μM in the absence of serum (data not shown). As a further proof of the lack of cytotoxic effects, no changes in the physical parameters (forward scatter [FSC] and side scatter [SSC]) were observed by flow cytometry in both continuous cell lines and primary PBMCs incubated for up to 24 hours at 37°C with 15 μM α-defensins. Likewise, no reduction of viability was seen in PBMCs cultured for up to 6 days in the presence of 15 μM α-defensins (data not shown).
Next, we compared the fusion-inhibitory activities of synthetic α-defensin-1 and -2 tested individually. As shown in Figure 1B, the potency of the 2 peptides was similar even though α-defensin-2 was slightly more effective than α-defensin-1 (IC50, 3.4 versus 3.9 μM on the HIV-1MN envelope), most likely reflecting modest differences in the level of purity between the 2 peptide preparations (97% versus 91% as assessed by HPLC). Of note, no cooperative effect between the 2 α-defensins was observed. To evaluate if the source of α-defensins may affect their fusion-inhibitory activity, synthetic α-defensin-1 was tested in parallel with recombinant α-defensin-1. As shown in Figure 1B, the synthetic molecule was slightly less effective than the recombinant one (IC50, 3.9 versus 3.1 μM) in blocking fusion mediated by the HIV-1MN envelope, again presumably reflecting the different purity levels of the 2 peptide preparations (91% versus more than 98% by HPLC). Altogether these results demonstrated that α-defensin-1 and -2 exert similar inhibitory effects on HIV-1 envelope-mediated fusion.
Broad-spectrum effect of α-defensins on primary HIV-1 envelopes of different coreceptor specificities and genetic subtypes
To evaluate the breadth of anti-HIV activity of α-defensins, we tested the fusion-inhibitory activity of synthetic α-defensins 1 and 2 on a panel of effector cells expressing HIV-1 envelopes of different coreceptor specificities (R5, R5X4, X4) and genetic subtypes (B, E, F). Seven of the 13 envelopes tested were derived from clinical HIV-1 isolates minimally passaged in vitro. As shown in Table 1, all the HIV-1 envelopes tested, irrespective of viral coreceptor usage phenotype and genetic subtype, were sensitive to the inhibitory effect of α-defensins, with IC50 ranging from 11.2 to 55.3 μM (mean ± SD, 24.0 ± 14.6 μM). No significant difference was observed in the inhibitory activity of α-defensins against HIV-1 isolates of different coreceptor specificity. When parallel assays were performed in the absence of bovine serum, the inhibitory activity of α-defensins was consistently and significantly higher (IC50 range, 1.4 to 11.5 μM; mean IC50 ± SD, 3.8 ± 2.9 μM; P < .001).
Inhibition of HIV-1 envelope-mediated cell fusion by α-defensins
| HIV-1 strain . | Subtype . | Origin . | Coreceptor usage . | IC50, μM* . | |
|---|---|---|---|---|---|
| FCS 2.5% . | No FCS . | ||||
| IT224.18 | B | Italy | R5 | 11.2 | nt |
| 91US714 | B | US | R5 | 33.2 | 2.4 |
| BaL | B | US | R5 | 26.5 | 3.1 |
| JR-FL | B | US | R5 | nt | 5.0 |
| CM235† | E | Thailand | R5 | 31.7 | nt |
| 92US077‡ | B | US | R5X4 | nt | 1.4 |
| B117 | B | Sweden | R5X4 | nt | 2.2 |
| IT193.21 | B | Italy | R5X4 | 17.0 | nt |
| 89.6† | B | US | R5X4 | nt | 11.5 |
| 93BR020 | F | Brazil | R5X4 | 14.0 | 3.8 |
| MN | B | US | X4 | 55.3 | 3.5 |
| IIIB | B | US | X4 | 12.5 | 2.0 |
| BK132† | B | Thailand | X4 | 12.6 | 2.3 |
| Mean ± SD | 24.0 ± 14.6 | 3.8 ± 2.9§ | |||
| HIV-1 strain . | Subtype . | Origin . | Coreceptor usage . | IC50, μM* . | |
|---|---|---|---|---|---|
| FCS 2.5% . | No FCS . | ||||
| IT224.18 | B | Italy | R5 | 11.2 | nt |
| 91US714 | B | US | R5 | 33.2 | 2.4 |
| BaL | B | US | R5 | 26.5 | 3.1 |
| JR-FL | B | US | R5 | nt | 5.0 |
| CM235† | E | Thailand | R5 | 31.7 | nt |
| 92US077‡ | B | US | R5X4 | nt | 1.4 |
| B117 | B | Sweden | R5X4 | nt | 2.2 |
| IT193.21 | B | Italy | R5X4 | 17.0 | nt |
| 89.6† | B | US | R5X4 | nt | 11.5 |
| 93BR020 | F | Brazil | R5X4 | 14.0 | 3.8 |
| MN | B | US | X4 | 55.3 | 3.5 |
| IIIB | B | US | X4 | 12.5 | 2.0 |
| BK132† | B | Thailand | X4 | 12.6 | 2.3 |
| Mean ± SD | 24.0 ± 14.6 | 3.8 ± 2.9§ | |||
nt indicates not tested.
IC50 values were calculated for each viral isolate from a dose-response curve obtained using an equimolar mixture of α-defensin-1 and -2.
Effector cells were infected with recombinant vaccinia viruses expressing the indicated HIV-1 envelope.
Effector cells expressing dual-tropic (R5X4) envelope were tested on CXCR4-expressing target cells.
P < .001 by the Mann-Whitney unpaired t test.
Dual interaction of α-defensins with cells expressing the HIV-1 envelope and the viral receptors
Next, we attempted to elucidate whether α-defensins block cell fusion through a direct interaction with the effector cells, which express the HIV-1 envelope, or with the target cells, which express the CD4 receptor and the coreceptor. For this purpose, α-defensins were preincubated with either effector or target cells and then removed before initiation of the fusion reaction. As a control, α-defensins were regularly added at the time of effector-target cell mixing and maintained in the culture medium throughout the fusion reaction period. As illustrated in Figure 2, α-defensins inhibited fusion induced by the HIV-1IIIB envelope irrespective of preincubation with effector or target cells. These data imply a direct interaction of α-defensins both with the viral envelope and with receptor molecules expressed on the target-cell surface.
Dual interaction of α-defensins with the HIV-1 envelope and the cellular receptors. α-defensins 1 and 2 were preincubated for 20 minutes either with effector cells (expressing the HIV-1 envelope) or with the target cells (expressing the CD4 and CXCR4 receptors) in the absence of serum. An equimolar mixture of α-defensins 1 and 2 was used (total concentration, 11.7 μM). The cells were washed twice prior to the initiation of the fusion reaction, and residual levels of α-defensins were determined by ELISA to be below the limit of detection of the assay. As a control, α-defensins were regularly added at the time of cell mixing and kept in the wells for the entire duration of the test. The fusion assay was performed using as effectors SupT1 cells chronically infected with HIV-1IIIB and as targets NIH-3T3 cells stably expressing human CD4 and CXCR4. Error bars indicate SD of mean values obtained from 3 replicate experiments.
Dual interaction of α-defensins with the HIV-1 envelope and the cellular receptors. α-defensins 1 and 2 were preincubated for 20 minutes either with effector cells (expressing the HIV-1 envelope) or with the target cells (expressing the CD4 and CXCR4 receptors) in the absence of serum. An equimolar mixture of α-defensins 1 and 2 was used (total concentration, 11.7 μM). The cells were washed twice prior to the initiation of the fusion reaction, and residual levels of α-defensins were determined by ELISA to be below the limit of detection of the assay. As a control, α-defensins were regularly added at the time of cell mixing and kept in the wells for the entire duration of the test. The fusion assay was performed using as effectors SupT1 cells chronically infected with HIV-1IIIB and as targets NIH-3T3 cells stably expressing human CD4 and CXCR4. Error bars indicate SD of mean values obtained from 3 replicate experiments.
α-defensins directly interact with human CD4: mapping of the binding regions
Because the CD4 glycoprotein is the primary receptor molecule for HIV-1 expressed on the cellular surface membrane,34,35 we investigated whether the mechanism of inhibition of HIV-1 envelope-mediated fusion was related to a direct interaction of α-defensins with CD4. The ability of α-defensins 1 and 2 to compete with mAb Leu3a, which recognizes a region of CD4 that is directly involved in the interaction with the external HIV-1 envelope glycoprotein, gp120, was assessed using an immunosorbent assay with a recombinant form of sCD4 immobilized on plastic. Increasing concentrations of synthetic α-defensin-1 or α-defensin-2 were added to sCD4 prior to Leu3a and maintained in the wells during the entire reaction period. As shown in Figure 3A, a dose-dependent inhibition of Leu3a binding was observed with both α-defensin-1 (IC50, 2.2 μM) and α-defensin-2 (IC50, 1.96 μM), demonstrating that both types of α-defensins directly interact with the CD4 molecule.
Binding of α-defensins to human CD4. (A) Competition of synthetic α-defensin-1 (○) and α-defensin-2 (•) with the anti-CD4 mAb Leu3a for binding to plastic-immobilized recombinant sCD4 in ELISA. The plates were coated with sCD4 at 5 μg/mL; α-defensins were added prior to Leu3a and kept in the wells throughout the reaction period. (B) Effect of synthetic α-defensin-2 on binding of different anti-CD4 mAbs to sCD4 with various epitope specificity (indicated on the x-axis) in ELISA. The assays were performed as in panel A. The epitope specificities of the anti-CD4 mAbs are indicated below. D indicates domain; CDR, complementarity-determining region. All antibodies were used at 1 μg/mL; α-defensin-2 was used at 7.3 μM. Error bars indicate SD of mean values obtained from 3 repeated assays.
Binding of α-defensins to human CD4. (A) Competition of synthetic α-defensin-1 (○) and α-defensin-2 (•) with the anti-CD4 mAb Leu3a for binding to plastic-immobilized recombinant sCD4 in ELISA. The plates were coated with sCD4 at 5 μg/mL; α-defensins were added prior to Leu3a and kept in the wells throughout the reaction period. (B) Effect of synthetic α-defensin-2 on binding of different anti-CD4 mAbs to sCD4 with various epitope specificity (indicated on the x-axis) in ELISA. The assays were performed as in panel A. The epitope specificities of the anti-CD4 mAbs are indicated below. D indicates domain; CDR, complementarity-determining region. All antibodies were used at 1 μg/mL; α-defensin-2 was used at 7.3 μM. Error bars indicate SD of mean values obtained from 3 repeated assays.
To define the α-defensin–binding region within the CD4 molecule, we tested the ability of synthetic α-defensin-2 to compete with a panel of CD4-specific mAbs in ELISA. Figure 3B shows that pretreatment with α-defensin-2 reduced the binding of various mAbs directed against different CDR-like regions of the first immunoglobulin-like domain of CD4, D1, which is directly implicated in the interaction with gp120: Vit-4 (CDR1 and CDR3), Leu3a and Sim4 (CDR2), and 13B8 (CDR3). By contrast, no inhibitory effect was seen with mAb OKT4, which recognizes an epitope within the D3 and D4 domains, which are dispensable for gp120 binding. These results demonstrated that α-defensins interact with the D1 domain of CD4, hindering the accessibility of various epitopes in the CDR-like regions 1 to 3 either by direct steric hindrance or by inducing conformational changes of the domain.
α-defensins directly interact with HIV-1 gp120: mapping of the binding region(s)
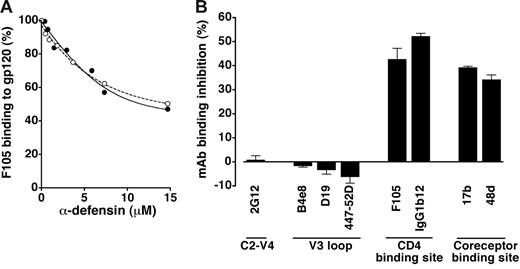
Because the results of fusion-inhibition assays demonstrated an interaction of α-defensins with effector cells (Figure 2), we investigated whether α-defensins can directly bind the external HIV-1 envelope glycoprotein, gp120. Figure 4A shows that both α-defensin-1 (IC50, 13.4 μM) and α-defensin-2 (IC50, 12.1 μM) competed in a dose-dependent fashion with a human mAb, F105, for binding to plastic-immobilized gp120. However, antibody displacement was only partial, suggesting an incomplete overlap between the antibody epitope and the α-defensin–binding site.
Binding of α-defensins to HIV-1 gp120. (A) Competition of synthetic α-defensin-1 (○) and α-defensin-2 (•) with the anti-gp120 mAb F105 (used at 5 μg/mL) for binding to plastic-immobilized recombinant gp120BaL in ELISA. (B) Effect of synthetic α-defensin-2 on binding of different anti-gp120 mAbs with various epitope specificities (indicated on the x-axis) in ELISA. All the mAbs were used at 5 μg/mL; α-defensin-2 was used at 7.3 μM. Error bars indicate SD of mean values obtained from 3 repeated assays.
Binding of α-defensins to HIV-1 gp120. (A) Competition of synthetic α-defensin-1 (○) and α-defensin-2 (•) with the anti-gp120 mAb F105 (used at 5 μg/mL) for binding to plastic-immobilized recombinant gp120BaL in ELISA. (B) Effect of synthetic α-defensin-2 on binding of different anti-gp120 mAbs with various epitope specificities (indicated on the x-axis) in ELISA. All the mAbs were used at 5 μg/mL; α-defensin-2 was used at 7.3 μM. Error bars indicate SD of mean values obtained from 3 repeated assays.
Next, we attempted to define the putative α-defensin–binding region(s) in gp120. For this purpose, a panel of anti-gp120 mAbs of human or murine origin was employed. Immunosorbent assays were performed using recombinant monomeric gp120 derived from the R5 isolate BaL (gp120BaL) immobilized on plastic. As shown in Figure 4B, treatment with synthetic α-defensin-2 failed to interfere with the binding of mAb 2G12, which recognizes a complex and discontinuous carbohydrate-dependent neutralization epitope mapping to the C2-C4 and V4 regions; likewise, no effect was observed on the binding of mAbs B4e8, D19, and 447-52D, all directed against conserved neutralization epitopes within the V3 domain. By contrast, a decreased binding activity was seen with mAbs directed against the CD4-binding region of gp120 (F105, IgG1b12) as well as against CD4-induced epitopes that overlap the coreceptor-binding site (17b, 48d). Of note, IgG1b12 is a broadly cross-reactive neutralizing antibody that potently inhibits the binding of CD4 to a wide variety of gp120 molecules.36 Similar data (not shown) were obtained with gp120SF162 (R5) and gp120MN (X4). These data confirmed that α-defensins directly interact with HIV-1 gp120, binding to a region that overlaps, is contiguous to, or influences the conformation of the CD4- and the coreceptor-binding regions.
α-defensin-2 inhibits the interaction between gp120 and CD4
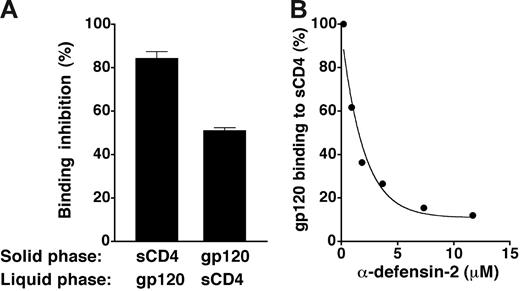
To evaluate the ability of α-defensins to interfere with gp120/CD4 binding, we performed reciprocal binding-inhibition experiments using either sCD4 or gp120BaL immobilized in the solid phase; synthetic α-defensin-2 was preincubated with the immobilized molecules and then removed by washing prior to the addition of the soluble ligands (either gp120BaL or sCD4). Figure 5A shows that α-defensin-2 effectively blocked the binding of gp120 to CD4 irrespective of the protein that was immobilized on plastic, thus confirming a direct interaction with both molecules. Quantitative assessment of α-defensin-2–mediated interference with gp120/CD4 binding showed an IC50 of 1.78 μM (Figure 5B).
Inhibition of gp120/CD4 binding by α-defensin-2. (A) Interference of α-defensin-2 with the reciprocal binding of CD4 and gp120BaL in ELISA. The plates were coated with sCD4 at 5 μg/mL and gp120BaL at 2 μg/mL, respectively; α-defensin-2 (7.3 μM) was preincubated with either sCD4 or gp120BaL immobilized to plastic and then removed by washing before the addition of the respective ligands (gp120BaL or sCD4, both at 1 μg/mL) to the liquid phase. Binding of gp120BaL was revealed using mAb 2G12 and binding of sCD4 using mAb DB81, which binds more efficiently to CD4 when it is complexed with gp120 (S. Burastero and P.L., manuscript in preparation). (B) Competition of α-defensin-2 with recombinant HIV-1 gp120BaL for binding to plastic-immobilized sCD4 in ELISA. The plates were coated with sCD4 at 5 μg/mL; α-defensin-2 was added prior to Leu3a or gp120BaL and kept in the wells throughout the reaction period. Error bars indicate SD of mean values obtained from 3 repeated assays.
Inhibition of gp120/CD4 binding by α-defensin-2. (A) Interference of α-defensin-2 with the reciprocal binding of CD4 and gp120BaL in ELISA. The plates were coated with sCD4 at 5 μg/mL and gp120BaL at 2 μg/mL, respectively; α-defensin-2 (7.3 μM) was preincubated with either sCD4 or gp120BaL immobilized to plastic and then removed by washing before the addition of the respective ligands (gp120BaL or sCD4, both at 1 μg/mL) to the liquid phase. Binding of gp120BaL was revealed using mAb 2G12 and binding of sCD4 using mAb DB81, which binds more efficiently to CD4 when it is complexed with gp120 (S. Burastero and P.L., manuscript in preparation). (B) Competition of α-defensin-2 with recombinant HIV-1 gp120BaL for binding to plastic-immobilized sCD4 in ELISA. The plates were coated with sCD4 at 5 μg/mL; α-defensin-2 was added prior to Leu3a or gp120BaL and kept in the wells throughout the reaction period. Error bars indicate SD of mean values obtained from 3 repeated assays.
α-defensins downmodulate cell-surface expression of CD4
To further investigate the mechanisms of α-defensin–mediated inhibition of HIV-1–induced fusion, we tested the expression of the primary cellular receptors used by HIV-1 (ie, CD4, CCR5, and CXCR4) using a unique CD4+ T-cell clone, PM1,37 which naturally expresses both CCR5 and CXCR4 and is thereby sensitive to HIV-1 isolates of different coreceptor usage phenotype. Figure 6A shows that α-defensin-2 induced a strong reduction of binding of the anti-CD4 mAb Leu3a (mean fluorescence intensity [MFI] reduction from 6 independent experiments, 73.4% ± 7.5%). By contrast, with the exception of CD26 and CD184 (CXCR4), which were slightly decreased (mean ± SD MFI reduction from 3 independent experiments, 19% ± 2.1% and 26.8% ± 9.5%, respectively), none of the other surface markers tested (CD2, CD45RO, CD46, CD195 [CCR5], HLA-DR) showed a reduced expression. Similar data (not shown) were obtained with synthetic α-defensin-1. The decrease in CD4 expression after treatment with α-defensins was confirmed using other cell types, such as IL-2–activated PBMCs and different CD4+ T-cell lines (including NIH-3T3 mouse fibroblasts stably transfected with human CD4 either alone or in combination with CCR5 or CXCR4), as well as with a panel of different mAbs directed to different domains of CD4 (not shown). As seen in the HIV-1 envelope-mediated fusion assay, α-defensin-2 was slightly more effective than α-defensin-1 in downmodulating CD4 expression; however, neither protein had any effect on CD4 expression when used at concentrations below 3.5 μM (data not shown).
Effect of α-defensin-2 on cell-surface protein expression: induction of CD4 downmodulation. Flow cytometry analysis of the surface expression of CD2, CD4 (Leu3a), CD26, CD45RO, CD46, HLA-DR, CCR5 (2D7), and CXCR4 (44717.111) in PM1 cells. The cells were preincubated for 60 minutes with or without an equimolar mixture of α-defensins 1 and 2 (total concentration, 14.7 μM) in the absence of serum; then the appropriate mAbs were added for cell-surface staining. Solid histograms represent the expression of the indicated markers; empty profiles indicate the background fluorescence signal obtained using irrelevant, isotype-matched mAbs. The numbers denote the MFI for the indicated marker. The histograms shown are representative of 4 to 6 independent experiments with similar results. (B) Time-course analysis of the effect of α-defensin-2 on binding of the anti-CD4 mAb Leu3a and OKT4 to live and formaldehyde-fixed PM1 cells. The cells were incubated without (empty bars) or with α-defensin-2 (14.7 μM) for 20 minutes (light gray bars) or 120 minutes (dark gray bars) and then stained with the mAbs. Fixed cells (treated with 2% paraformaldehyde at 4°C for 30 minutes) were washed twice and incubated for 120 minutes with α-defensins before antibody staining (black bars). Error bars indicate SD of mean values obtained from 3 repeated assays.
Effect of α-defensin-2 on cell-surface protein expression: induction of CD4 downmodulation. Flow cytometry analysis of the surface expression of CD2, CD4 (Leu3a), CD26, CD45RO, CD46, HLA-DR, CCR5 (2D7), and CXCR4 (44717.111) in PM1 cells. The cells were preincubated for 60 minutes with or without an equimolar mixture of α-defensins 1 and 2 (total concentration, 14.7 μM) in the absence of serum; then the appropriate mAbs were added for cell-surface staining. Solid histograms represent the expression of the indicated markers; empty profiles indicate the background fluorescence signal obtained using irrelevant, isotype-matched mAbs. The numbers denote the MFI for the indicated marker. The histograms shown are representative of 4 to 6 independent experiments with similar results. (B) Time-course analysis of the effect of α-defensin-2 on binding of the anti-CD4 mAb Leu3a and OKT4 to live and formaldehyde-fixed PM1 cells. The cells were incubated without (empty bars) or with α-defensin-2 (14.7 μM) for 20 minutes (light gray bars) or 120 minutes (dark gray bars) and then stained with the mAbs. Fixed cells (treated with 2% paraformaldehyde at 4°C for 30 minutes) were washed twice and incubated for 120 minutes with α-defensins before antibody staining (black bars). Error bars indicate SD of mean values obtained from 3 repeated assays.
To address the question of whether the reduced expression of CD4 induced by α-defensins was due to bona fide receptor modulation or epitope occupancy, a time-course analysis was performed after exposure of PM1 cells to synthetic α-defensin-2. Two mAbs that recognize distinct immunoglobulin-like domains of the glycoprotein were used: Leu3a (specific for the CDR2 region of D1) and OKT4 (specific for D3 and D4). As illustrated in Figure 6B, α-defensin-2 induced a rapid loss of Leu3a binding at 20 minutes, with a further reduction at 120 minutes. By contrast, OKT4 binding was only moderately decreased at the early time point but was markedly reduced at 120 minutes. These results suggested that the Leu3a epitope, unlike the OKT4 epitope, was directly masked by binding of α-defensin-2. This hypothesis was confirmed in experiments with formaldehyde-fixed cells, which showed that α-defensin-2 significantly reduced the binding of Leu3a but had no effect on the binding of OKT4, confirming the results obtained by competition ELISA with plastic-immobilized recombinant sCD4 (Figure 3B). The progressive loss of OKT4 reactivity in living cells, but not in formaldehyde-fixed cells, clearly demonstrated that the CD4 molecule was progressively downmodulated from the cellular surface after treatment of live cells with α-defensin-2.
Discussion
Several recent reports have shown that α-defensins act as natural inhibitors of HIV-1 infection,6,15,23,24 but the mechanisms of such inhibition are still incompletely understood. In this study, we provide evidence that α-defensins inhibit the earliest stages of the HIV-1 infectious cycle as shown by blockade of envelope-mediated cell fusion, a process that closely mimics fusion between the viral and cellular membranes during the virus-entry process. The physiologic relevance of this observation was suggested by the fact that α-defensins effectively inhibited fusion mediated not only by laboratory strains but also by clinical HIV-1 isolates of different genetic background and biologic phenotype. Moreover, we formally proved a direct and specific binding of α-defensins to the CD4 receptor as well as to the external HIV-1 envelope glycoprotein, gp120, demonstrating for the first time that α-defensins interfere with the reciprocal binding of gp120 and CD4.
Our results are in agreement with previous binding studies with both θ- and α-defensins using surface plasmon resonance6 but differ from the conclusion of other investigators who reported that α-defensins do not interfere with the HIV-1 entry step.23 Such conclusion was based on parallel testing of recombinant α-defensin-1 and AMD3100, an HIV-entry inhibitor that targets the CXCR4 coreceptor. While AMD3100 inhibited HIV-1 replication only if added at the time of virus infection, inhibition by α-defensins was observed both when added at 0 and 2 hours after infection. However, this evidence is only indirect and does not permit us to formally exclude inhibition at the entry level. Rather, it is consistent with a dual mechanism of α-defensin–mediated HIV-1 inhibition acting both at the virus entry and postentry levels. By contrast, the experimental approaches used in the present study permitted us to focus exclusively on the earliest steps of the viral infectious cycle, formally proving that α-defensins inhibit the virus docking to the target cell. Thus, our findings add ground to the hypothesis that α-defensins may antagonize HIV-1 infection at least at 2 distinct levels24 : at the mucosal surface and in absence of serum, they neutralize HIV-1 virions by preventing envelope-receptor interaction and blocking virus entry; after the virus has entered the cells and irrespective of the presence of serum, α-defensins suppress HIV replication presumably by interfering with intracellular signaling events.
We also showed that exposure of CD4+ T cells to α-defensins leads to a dramatic down-regulation of the CD4 receptor as well as to a slight reduction of expression of the CXCR4 coreceptor. Although other investigators have reported a lack of effects of α-defensins on CD4 and CXCR4 expression,24 the experimental conditions used in their study were markedly different from ours. First, α-defensins were used at about 1.5 μM, while we found that concentrations below 3.5 μM are insufficient for inducing CD4 downmodulation; second, α-defensins were incubated with the cells for a prolonged time (16 hours), while we observed that after 18 hours the expression of CD4 and CXCR4 tends to return to baseline levels, most likely due to receptor recycling38 associated with α-defensin degradation and/or aggregation. The ability of α-defensins to downmodulate the cell-surface expression of CD4 may have important implications for the potency and duration of the antiviral effects of α-defensins secreted in vivo. Moreover, modification of CD4 and CXCR4 expression may represent a novel mechanism of immunomodulation by α-defensins, because these 2 molecules are physiologically relevant receptors constitutively expressed on the surface of both T cells and mononuclear phagocytes. Other immunomodulatory effects of α-defensins have been described, including a direct chemotactic activity on naive T cells and immature dendritic cells11 as well as indirect effects mediated by up-regulation of CC-chemokine release,26 enhanced transcription of the CXC-chemokine IL-8,39 and increased production of proinflammatory cytokines such as TNF-α and IL-1β.40
Although the variety of molecular targets of α-defensins might suggest a nonspecific binding activity, we were able to demonstrate specificity both at the intramolecular and intermolecular levels. Using a panel of mAbs of defined epitope specificity, we identified selected regions of CD4 and gp120 that interact with α-defensins. Consistent with their ability to block the binding of gp120 to CD4, α-defensins effectively competed with mAbs that recognize the D1 region of the CD4 glycoprotein, which includes the gp120-binding site.41 On gp120, we observed competition with mAbs mapping both to the CD4- and to the coreceptor-binding site but no competition with mAbs to the V3 loop and to the heavily glycosylated region recognized by mAb 2G12.42 In addition to this intramolecular binding specificity, we demonstrated that α-defensins failed to exert significant effects on a panel of other cell-surface proteins expressed by CD4+ T cells, including CD2, CD26, CD45RO, CD46, CCR5, and HLA-DR. How can we reconcile the binding specificity of α-defensins with their ability to interact with a variety of molecules? This apparent paradox may be resolved in light of recent studies that documented a lectinlike activity of α- and θ-defensins.6 Our data are consistent with this model, because both CD4 and gp120 are glycoproteins. However, it has to be emphasized that we failed to observe a generalized and unselective binding of glycoproteins, as further proved by the lack of interaction with the 2G12 epitope of gp120, which is essentially formed by carbohydrate moieties.42 Interestingly, retrocyclin, which shares a high degree of homology with α-defensins,10 also binds to CD4 and gp120 with very high affinity, but it does not apparently interfere with the reciprocal binding of the 2 molecules.9 Likewise, another HIV-inhibitory lectin, cyanovirin-N (CV-N), inhibits envelope-mediated membrane fusion by binding to CD4 and gp120; nonetheless, mapping studies with mAbs indicated that CV-N does not occlude or alter the CD4-binding site or CD4-induced epitopes but inhibits the binding of mAb 2G12 to gp120.43 These findings suggest that although α-defensins share several characteristics with other lectins that inhibit HIV-1 entry, they possess a peculiar binding fingerprint, most likely through the recognition of one or more specific carbohydrate motifs. The distribution of such motifs will dictate the molecular binding spectrum and, thereby, the biologic activities of α-defensins.
The protective role of α-defensins in vivo in the course of HIV-1 infection is still debated. These antibiotic peptides are produced at high levels by neutrophils, which represent the predominant leukocyte population in the vaginal epithelium.44 Indeed, high concentrations of α-defensins are present in the mucous plug that occludes the uterine cervix.45 Neutrophils release massive quantities of α-defensins upon degranulation in response to cytokines or, as recently reported, CC chemokines.46 Recent studies have reported elevated levels of CC chemokines in the genital tracts of women who are exposed to HIV-1 and yet remain uninfected.47 Furthermore, α-defensins may play an important role as natural antagonists of HIV-1 entry at the intestinal mucosal level. The gut is increasingly recognized as a major site of early HIV replication: in the gut-associated lymphoid tissue, where a substantial proportion of the CD4+ T cells in the body reside,48 a massive depletion of CD4+ T cells occurs during acute HIV-1 infection. Inflammation causes the infiltration of neutrophils by transmigration across intestinal epithelia in response to chemotactic signals,49,50 which in turn release large amounts of α-defensins. Moreover, the epithelium of the intestinal tract expresses human β-defensins (hBD) 1 to 4, and hBD-2 and -3 were recently shown to be strongly induced upon HIV-1 exposure7 and to suppress both R5 and X4 HIV-1 replication.7,51 The remaining 2 human defensins (HD), HD-5 and HD-6, are expressed in the intestinal Paneth cells and probably contribute to innate defense mechanisms of the intestinal mucosal surface.52 Thus, it is conceivable that α-defensins exert anti-HIV activity mainly at the mucosal level and at sites of active inflammation, where they can reach considerable levels, rather than in blood, where they are diluted and rapidly inactivated by plasmatic proteins. It has been estimated that 106 neutrophils contain approximately 5 μg α-defensins, which translates into a cellular concentration above 10 mg/mL.53 This means that levels of α-defensins in the micromolar range may be easily be reached in the extracellular milieu in the vicinity of activated neutrophils, indicating that the hypothesis that α-defensins may block HIV-1 entry by inhibiting the binding of the virus to its cellular receptor is physiologically relevant.
Altogether, the unique biologic properties of α-defensins are likely to result in a complex modulation of both HIV replication and immune-cell functions, which may influence the natural history of HIV infection. In vivo studies will be important to elucidate the immunologic and clinical relevance of α-defensin secretion in different anatomic sites during the course of HIV infection or in vaccinated individuals as well as in other infectious or inflammatory disorders.
Authorship
Contribution: L.F. designed and performed research, analyzed data, and wrote the paper; F.S., M.T., and L.V. performed research; and P.L. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paolo Lusso, Unit of Human Virology, Department of Biological and Technological Research (DIBIT), San Raffaele Scientific Institute, 20132 Milan, Italy; e-mail: paolo.lusso@hsr.it.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Italian Ministry of Health (ISS AIDS Project 2004 and ICAV 2004) (P.L.) and the European Union (Projects Eurovac I and II) (P.L.). The authors thank Dr E. A. Berger for vaccinia constructs, Dr G. Scarlatti for primary HIV-1 isolates, Dr E. M. Fenyo for a primary HIV-1 isolate, and the NIH ARRRP for providing multiple reagents and viral isolates.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal