Abstract
Liver gene transfer is a highly sought goal for the treatment of inherited and infectious diseases. Lentiviral vectors (LVs) have many desirable properties for hepatocyte-directed gene delivery, including the ability to integrate into nondividing cells. Unfortunately, upon systemic administration, LV transduces hepatocytes relatively inefficiently compared with nonparenchymal cells, and the duration of transgene expression is often limited by immune responses. Here, we investigated the role of innate antiviral responses in these events. We show that administration of LVs to mice triggers a rapid and transient IFNαβ response. This effect was dependent on functional vector particles, and in vitro challenge of antigen-presenting cells suggested that plasmacytoid dendritic cells initiated the response. Remarkably, when LVs were administered to animals that lack the capacity to respond to IFNαβ, there was a dramatic increase in hepatocyte transduction, and stable transgene expression was achieved. These findings indicate that, even in the setting of acute delivery of replication-defective vectors, IFNs effectively interfere with transduction in a cell-type–specific manner. Moreover, because disabling a single component of the innate/immune network was sufficient to establish persistent xenoantigen expression, our results raise the hope that the immunologic barriers to gene therapy are less insurmountable than expected.
Introduction
Lentiviral vectors (LVs) are a promising candidate system for therapeutic gene transfer. Because of their capacity to transduce nondividing cells and stably integrate a gene expression cassette of relatively large size and complexity, LVs have significant potential for achieving long-term expression of a therapeutic molecule. Several groups, including our own, have carried out studies using LV for in vivo gene delivery in rodents.1-5 Efficient gene transfer to the liver could be achieved; however, hepatocytes, which were the main target of the therapy, were transduced at a relatively low efficiency compared with nonparenchymal cells. At a low vector dose, this effect was particularly pronounced. While a high frequency of Kupffer cells (KCs) were found to be vector positive, only a small fraction of hepatocytes were transduced.
Interestingly, by increasing the concentration of injected vector a threshold is reached in which hepatocyte transduction becomes dose responsive, and improved hepatocyte gene transfer is achieved. This may be due to the requirement for saturating the particle-clearance systems of the sinusoid-lining cells in blood-filtering organs.6 Nonetheless, a better understanding of the rate-limiting factors in transduction would help to improve both the dose-effect relationship and risk-benefit ratio of systemic LV administration.
A high incidence of transgene-specific immunity has also been observed in studies using LVs for in vivo gene delivery.1-5 This response resulted in elimination of transduced cells and/or neutralization of the transgene product, and ultimately negated the benefits of gene transfer. The role of transgene expression within antigen-presenting cells (APCs) of the hematopoietic system has been identified as a factor contributing to the development of antitransgene immunity.1 However, the effect of innate immune responses on LV-mediated gene transfer has not been examined in detail.
Recent studies have investigated the consequences of exposing human dendritic cells (DCs) in culture to wild-type HIV, the parent virus of many LVs.7,8 These works indicate that HIV activates a subset of DCs, plasmacytoid DCs (pDCs), through engagement of toll-like receptor 7 (TLR7), a pattern-recognition receptor (PRR) for single-stranded RNA (ssRNA). In response to HIV, pDCs produce high levels of IFNαβ, potent antiviral cytokines, which can lead to their own maturation, as well as the maturation of other DC subsets.8,9 This, in turn, can activate the adaptive immune system. Since LVs are derived from HIV and contain ssRNA by necessity, LVs may also trigger a similar innate immune response. If this were to occur in vivo, it could contribute to the induction of the adaptive immune response that results in clearance of transduced cells.
The studies of HIV, although informative, were carried out in vitro, and do not necessarily reflect the in vivo conditions of a systemically administered LV. In addition, LVs are nonreplicating, hybrid vectors. They lack all viral accessory proteins associated with virulence and pathogenesis, and have been pseudotyped by the envelope of an unrelated virus. This affects their biodistribution, target cell binding, and entry. Thus, the aforementioned studies do not necessarily provide insight into the potential consequences of LV-mediated activation of the innate immune system.
To address these issues, we have undertaken studies to monitor the innate host response following in vivo LV delivery to mice. Our results provide new findings indicating that LVs activate an IFNαβ response early after administration, which plays a major role in limiting the efficiency and stability of gene transfer.
Materials and methods
Vector production
Third-generation LVs were produced by Ca3PO4 transfection into 293T cells. Supernatants were collected, passed through a 0.22-μm filter, and purified by ultracentrifugation as described.10 Titer was estimated on 293T cells, and vector particles were measured by HIV-1 Gag p24 antigen immunocapture (NEN Life Science Products, Boston, MA). Vector infectivity was calculated as the ratio between titer and particle. For concentrated vesicular stomatitis virus (VSV) pseudotyped vector, titer ranged from 0.7 to 1.5 × 1010 transducing units (TU)293T/mL, and particles ranged from 90 to 150 μg/mL p24. For concentrated gp64 pseudotyped vector, the titer was 109 TU/mL, and the particles were 100 μg/mL p24.
In vivo vector administration
Balb/c, C57BL/6, 129sv, and Nu/Nu mice were purchased from Charles River Laboratories (Milan, Italy), and IFNαβ receptor knockout mice (IFNαβR−/−) were purchased from B&K Universal Limited (Grimston, United Kingdom). Mice were maintained in specific pathogen–free conditions. Vector administration was carried out by tail-vein injection. All animal procedures were performed according to protocols approved by the Hospital San Raffaele Institutional Animal Care and Use Committee.
Quantitative PCR and RT-PCR
DNA from cells and tissues was extracted by using the Blood & Cell Culture DNA Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA from cells was extracted by using Tri Reagent (Sigma, St Louis, MO), according to the manufacturer's instructions. Vector copies per genome (C/G) were quantified by quantitative polymerase chain reaction (Q-PCR) using the primer and probe set previously described.1 Copies per genome (C/G) were calculated by the following equation: (ng LV/ng endogenous DNA) × (no. of LV integrations in the standard curve). Reverse transcription (RT) was carried out on 2 μg total RNA from the livers and spleens of treated mice using the random hexamers protocol of the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions using the primer/probe set previously described.1 All reactions were carried out in triplicate in an ABI Prism 7900 (Applied Biosystems, Weiterstadt, Germany).
Cytokine expression analysis
The RNase protection assay for quantitation of mRNA was performed as previously described.11 The mouse interleukin-1α (mIL-1α), mIL-1β, mIL-2, mIL-3, mIL-4, mIL-5, mIFN-γ, mTNF-α, mTNF-β, 2′5′ oligoadenylate synthetase (OAS), and mL32 subclones in pGEM-4 transcription vector were described in a previous report.12 Mouse serum or cell supernatants were analyzed at indicated time points and tested by enzyme-linked immunosorbent assay (ELISA) for the presence of murine IFNα (PBL Biomedical, Piscataway, NJ) and by cytokine bead array for TNF-α.
Immunohistochemistry
For immunofluorescence, tissues were prepared as previously described13 and stained with the indicated antibodies. Images were visualized with a Zeiss Axioskop2 microscope using triple laser confocal microscopy with a Zeiss Plan-Neofluar 20×/0.5 numerical aperture objective lens and a Zeiss W-PI 10 × 0.23 objective lens as eyepiece (Zeiss, Arese, Italy). Images were acquired using a Radiance 2100 camera (Bio-Rad, Segrate, Italy) and LaserSharp 2000 acquisition software (Bio-Rad). Morphometric analysis was used to determine the frequency of green fluorescent protein (GFP)–positive hepatocytes. Confocal images were imported into ImageJ software (http://rsb.info.nih.gov/ij/). Three random liver fields were measured per mouse. We scored the total number of TOPRO-3–positive large nuclei, which were taken to be hepatocytes, to determine the total number of hepatocytes per field. Since most hepatocytes are binucleated, caution was taken to prevent double scoring of single hepatocytes. GFP-positive hepatocytes were scored based on 488:515 excitation-emission signal, and morphology to distinguish them from nonparenchymal cells. Frequencies were calculated based on the counting of a minimum of 1000 hepatocytes per mouse.
Plasmacytoid DCs and CD11b+ splenocyte isolation
Spleens were excised from Balb/c mice and incubated with collagenase D to obtain a unicellular solution. CD11b splenocytes and pDCs were isolated by commercially available kit according to the manufacturer's instructions (no. 130-049-601 and no. 130-091-263, respectively; Miltenyi Biotec, Bergisch Gladbach, Germany). Per experimental analysis, 5 to 6 spleens were used.
IFN-γ ELISPOT assay
GFP-specific IFN-γ–secreting cells were enumerated by ELISA as previously described.14 Briefly, 5 × 104 magnetically sorted splenic CD8+ T cells (Miltenyi Biotec) or total intrahepatic leukocytes were plated in enzyme-linked immunospot (ELISPOT) plates coated with anti–IFN-γ capture antibody (2.5 μg/mL, R46A2; BD Bioscience, San Jose, CA) in the presence of IL-2 (50 U/mL; BD) and 1 × 105 irradiated (30 Gy [3000 Rad]) wild-type EL-4 or EL-4.eGFP. After 42 hours, IFN-γ–producing cells were detected by anti–IFN-γ detection antibody. Spots were counted by the Eli.Scan system (A.EL.VIS, Hannover, Germany).
Results
LV entry and RT occurs rapidly after intravenous administration, but vector DNA is quickly lost
Balb/c mice were intravenously injected via tail vein with 10 μg of HIV-1 Gag p24 equivalents of VSV glycoprotein pseudotyped third-generation LV (VSV.LV)–encoding enhanced GFP under the control of the ubiquitously expressed phosphoglycerate kinase (PGK) promoter. This vector dose was chosen based on previous experience, which suggested that at this concentration the threshold for achieving significant hepatocyte transduction starts to be overcome.
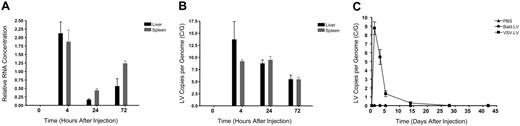
Q-PCR analysis was used to follow transduction kinetics. At 4 hours after injection high levels of LV genomic RNA and proviral DNA could be detected within the liver and spleen (Figure 1A-B). In contrast, no vector DNA or RNA could be detected at the same and later time points in animals injected with a control LV, which was assembled without a viral envelope (bald.LV). The inability to detect vector sequences from mice treated with bald.LV suggests that in VSV.LV-treated animals the RNA and DNA content was not from extracellular vector particles or plasmid DNA trapped within the organs, but indeed due to vector particles that had undergone cell entry and RT. An additional Q-PCR analysis, using primers and a probe specific for plasmid backbone sequences, did not detect significant levels of transfer plasmid (data not shown), and thus further confirmed that the detected vector DNA within the liver and spleen was the result of LV transduction.
Kinetic profile of LV content within the liver and spleen following systemic injection. (A) Quantification of LV RNA from the liver (▪) and spleen (⊡) of VSV.LV-treated Balb/c mice. Quantitative RT-PCR (Q-RT-PCR) was used to measure GFP RNA. All samples were normalized to murine GAPDH. Values are presented as the means ± SEM for 3 mice per group. (B) Quantification of LV DNA from the liver (▪) and spleen (⊡) of VSV.LV-treated Balb/c mice. Spleen and liver samples were analyzed at the indicated time point by Q-PCR to measure LV copies per genome (C/G). All samples were normalized to murine β-actin. Values are presented as the means ± SEM for 3 mice per group. (C) Analysis of vector persistence in the liver. Q-PCR analysis was performed as in panel B on liver samples from mice treated with PBS (▴) or 10 μg of VSV.LV (▪) or bald.LV (•).
Kinetic profile of LV content within the liver and spleen following systemic injection. (A) Quantification of LV RNA from the liver (▪) and spleen (⊡) of VSV.LV-treated Balb/c mice. Quantitative RT-PCR (Q-RT-PCR) was used to measure GFP RNA. All samples were normalized to murine GAPDH. Values are presented as the means ± SEM for 3 mice per group. (B) Quantification of LV DNA from the liver (▪) and spleen (⊡) of VSV.LV-treated Balb/c mice. Spleen and liver samples were analyzed at the indicated time point by Q-PCR to measure LV copies per genome (C/G). All samples were normalized to murine β-actin. Values are presented as the means ± SEM for 3 mice per group. (C) Analysis of vector persistence in the liver. Q-PCR analysis was performed as in panel B on liver samples from mice treated with PBS (▴) or 10 μg of VSV.LV (▪) or bald.LV (•).
By 24 hours, RNA content had sharply declined, but by 72 hours, RNA levels were again elevated, indicative of vector-mediated transcription. Interestingly, these transduction kinetics were consistent with those reported for HIV infection in vitro,15 and indicate that cellular entry and RT of the vector occurs rapidly following in vivo administration.
Vector DNA content, although initially high, began to diminish soon after RT. By 72 hours after injection, vector content had declined 3-fold from peak detection levels within the liver (Figure 1C). The mechanism responsible for this initial loss of vector DNA was unclear, however; following this early and rapid decline, there was a delayed, but near complete clearance of LV DNA. This second phase was expected, as it is well established that intravenous delivery of a ubiquitously expressed LV results in immune-mediated destruction of transduced cells, and ultimately, the loss of vector DNA.
In vivo administration of LV activates an IFNαβ response
To measure the innate response to LV, we used a RNase protection assay (RPA) to monitor changes in the hepatic and splenic expression of cytokines and cytokine-responsive genes known to be produced by activated cells of the innate immune system.16 RPA provides a sensitive method for detecting changes in the transcriptional profile of multiple genes within a tissue. The liver and spleen are the most well transduced tissues following intravenous injection of LVs, and are major targets of systemic gene therapy as well as important sites of innate host protection.
Balb/c and C57BL/6 mice were treated with 10 μg of LV. Within 4 hours of vector administration there was a more then 8-fold induction of OAS, a downstream product of IFNαβ signaling, and a specific marker of IFNα and IFNβ expression12 in the liver and spleen of both groups of mice (Figures 2, S1 [found on the Blood website; see the Supplemental Materials link at the top of the online article). Induction of IFNα was also found by direct measurement within the serum of treated mice (Figure 2C). Concomitant with the IFNαβ response, there was a strong up-regulation in the expression of TNF-α, a potent inflammatory cytokine, although only a modest up-regulation of IL-1α and IL-1β, 2 other inflammatory cytokines. OAS expression began to decline by 24 hours after injection, and by 72 hours OAS returned to pretreatment levels, indicating resolution of the IFNαβ response. Overall, these results demonstrate that in vivo administration of LV rapidly triggers a strong but transient IFNαβ response.
Monitoring the innate immune response following in vivo LV administration. Balb/c mice were treated with 10 μg of LV via intravenous injection. The cytokine profile of the liver (A) and spleen (B) was measured by RPA at the indicated times. Three mice per group are shown. Quantitative phosphor imaging analysis of the RPA was carried out, and the results are presented as the means ± SEM. All samples were normalize to L32 and presented as fold induction compared with PBS-treated controls. ▪ indicates OAS; •, TNF-α; ▴, IL-1α; and ⊡, IL-1β. (C) Circulating IFNα levels. Serum samples were analyzed at the indicated time for IFNα expression by ELISA. Values are presented as the means ± SEM for 3 mice per group.
Monitoring the innate immune response following in vivo LV administration. Balb/c mice were treated with 10 μg of LV via intravenous injection. The cytokine profile of the liver (A) and spleen (B) was measured by RPA at the indicated times. Three mice per group are shown. Quantitative phosphor imaging analysis of the RPA was carried out, and the results are presented as the means ± SEM. All samples were normalize to L32 and presented as fold induction compared with PBS-treated controls. ▪ indicates OAS; •, TNF-α; ▴, IL-1α; and ⊡, IL-1β. (C) Circulating IFNα levels. Serum samples were analyzed at the indicated time for IFNα expression by ELISA. Values are presented as the means ± SEM for 3 mice per group.
Induction of the IFNαβ response is dependent on LV infection
To improve vector titer and broaden cell tropism, the VSV envelope has routinely been used for pseudotyping LVs.17,18 The high transduction efficiency of this vector, in particular for cells of the innate immune system such as DCs and macrophages, may be responsible for the robust induction of IFNαβ. Alternatively, the VSV envelope may be acting as a PRR ligand and triggering the innate response.
To determine whether VSV.G was responsible for the observed IFNαβ response, we pseudotyped the LV.PGK.GFP vector with the gp64 glycoprotein envelope from the baculovirus Autographa californica multinuclear polyhedrosis virus (gp64.LV). Previous studies have demonstrated that LV can be pseudotyped with the gp64 envelope, and that these vectors may even have a reduced tropism for hematopoietic cells.19,20
Since the transduction efficiency of a vector, particularly for hematopoietic-lineage cells, may influence the innate stimulatory capacity of that vector, we initially set out to evaluate the transduction profile of gp64.LV. Balb/c mice were administered 15 μg of gp64.LV by IV, and killed at 4 hours and 7 days after injection. A higher vector dose was used for these experiments because of the lower infectivity (TU/p24 ratio) of gp64.LV compared with VSV.LV. Indeed, even at the higher dose, transduction levels were still lower with gp64.LV than those obtained with VSV.LV. Nonetheless, gp64.LV was able to mediate significant levels of gene transfer (Figure 3A-B) in both the liver (0.7 ± 0.07 C/G) and spleen (0.9 ± 0.2 C/G). Confocal fluorescence microscopy analysis found that GFP expression was confined predominantly to hepatocytes in the liver and stromal cells in the spleen (Figure 3B). However, a fraction of GFP-positive cells in both the liver and spleen were found to costain for the hematopoietic-specific marker CD45, and thus indicates that gp64.LV also transduces hematopoietic cells, albeit to a lesser extent than VSV.LV.
LV-mediated induction of innate immunity is dependent on vector infectivity. (A) Balb/c mice were injected via tail vein with PBS, 10 μg VSV.LV, or 15 μg gp64.LV. Spleen and liver samples were analyzed at 4 hours after injection by Q-PCR to measure LV copies per genome (C/G). All samples were normalized to murine β-actin. Values are presented as the means ± SEM for 3 mice per group. □ indicates PBS; ▪, VSV.LV; and ⊡, gp64.LV mice. (B) Confocal immunofluorescence microscopy analysis of liver and spleen sections from mice administered gp64.LV 7 days earlier. Fixed frozen sections were costained with anti-GFP (green) antibodies to identify transduced cells, and anti-CD45 (red) antibodies to identify hematopoietic lineage cells. Arrows indicate selected cells that costain for both GFP and CD45; these cells appear yellow. TO-PRO-3 (blue) was used to stain nuclei. Scale bars equal 200 μm. (C) The cytokine profile, as measured by RPA, of the liver (left) and spleen (right) 4 hours after injection with the indicated LV (n=3 per group). Analysis was carried out as described in Figure 2. HI.LV indicates heat-inactivated LV; bald.LV, envelope-negative LV. Asterisks indicate t-test significance value of treatment group compared with bald.LV (*P < .05; **P < .005; ***P < .001).
LV-mediated induction of innate immunity is dependent on vector infectivity. (A) Balb/c mice were injected via tail vein with PBS, 10 μg VSV.LV, or 15 μg gp64.LV. Spleen and liver samples were analyzed at 4 hours after injection by Q-PCR to measure LV copies per genome (C/G). All samples were normalized to murine β-actin. Values are presented as the means ± SEM for 3 mice per group. □ indicates PBS; ▪, VSV.LV; and ⊡, gp64.LV mice. (B) Confocal immunofluorescence microscopy analysis of liver and spleen sections from mice administered gp64.LV 7 days earlier. Fixed frozen sections were costained with anti-GFP (green) antibodies to identify transduced cells, and anti-CD45 (red) antibodies to identify hematopoietic lineage cells. Arrows indicate selected cells that costain for both GFP and CD45; these cells appear yellow. TO-PRO-3 (blue) was used to stain nuclei. Scale bars equal 200 μm. (C) The cytokine profile, as measured by RPA, of the liver (left) and spleen (right) 4 hours after injection with the indicated LV (n=3 per group). Analysis was carried out as described in Figure 2. HI.LV indicates heat-inactivated LV; bald.LV, envelope-negative LV. Asterisks indicate t-test significance value of treatment group compared with bald.LV (*P < .05; **P < .005; ***P < .001).
To measure the innate immune response to gp64.LV, RPA was carried out (Figure 3C). Within 4 hours of injection, there was a 4-fold induction of OAS in both the liver and spleen, and a 14-fold induction of TNF-α expression in the liver. Thus, the IFNαβ response to LV is not specific to the VSV envelope, but likely is a function of other vector components.
To rule out the possibility that contaminants, such as plasmid DNA or endotoxins, which can be carried over from vector production, were directly responsible for the activation of the IFNαβ response, we repeated the experiments with 10 μg of VSV.LV that was heat-inactivated by incubation at 65°C (HI.LV). In mice treated with HI.LV, we did not observe significant induction of innate cytokines (Figures 3C,S1). Since heat inactivation does not diminish the adjuvant activity of endotoxins or CpG immunostimulatory sequences, these findings rule out contaminants as a source of IFNαβ induction in response to LV administration.
A second control vector, bald.LV, which was not pseudotyped with a viral envelope, was administered to Balb/c and C57BL/6 mice. As noted, vector RNA and proviral DNA could not be detected in mice treated with 10 μg of bald.LV, and shows that the early stages of vector transduction does not occur with this vector. Here again, we did not observe induction of IFNαβ, as measured by OAS expression (Figures 3C,S1), and thus suggests that cell entry is required for triggering the observed response.
Overall, these results demonstrate that activation of the IFNαβ response is dependent on functional vector particles and accompanies productive infection.
pDCs, not macrophages or myeloid DCs, produce high levels of IFNα in response to LVs
Having established that LV transduction triggers IFNαβ expression in vivo, we next set out to establish the cellular origin of the response. Initial activation of the IFNαβ response is largely mediated by the innate immune system. Most important in the context of viral infections are monocytes/macrophages, myeloid DCs, pDCs, and natural killer (NK) cells, which, through their unique ability to express particular PRRs, are able to recognize viral components, such DNA and RNA, and trigger an antiviral response.21
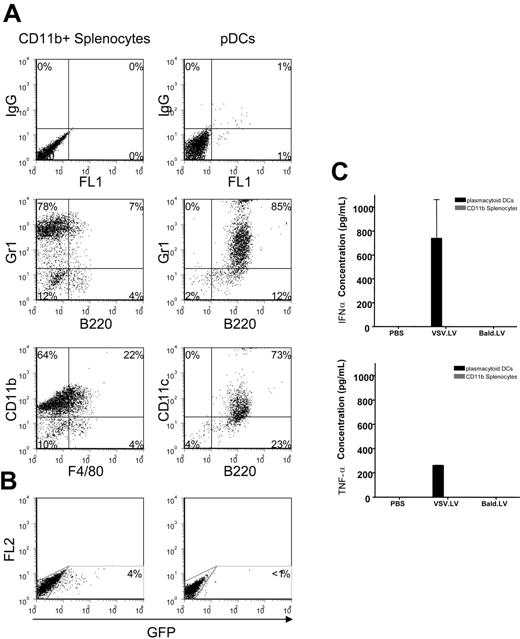
To determine which cells of the innate immune system were responding to LV, we monitored expression of IFNα and TNF-α from splenocytes following exposure to the vector. Cells were isolated from the spleen of Balb/c mice by magnetic sorting, and fluorescence-activated cell sorter (FACS) analysis was used to determine cellular identity (Figure 4A). Splenocytes purified by CD11b-sorting included CD11b+Gr-1+F4/80−B220− myeloid DCs and CD11b+Gr-1+F4/80+B220− macrophages. Cells isolated by CD3, CD19, CD11b, and CD49b depletion, followed by B220-positive selection, were predominately pDCs, as determined by their CD11c+Gr-1+B220+ phenotype.
pDCs, but not CD11b+ splenocytes, are activated by LVs. (A) Phenotype analysis of isolated splenocyte populations. FACS acquisition of magnetic-sorted splenocytes was carried out to determine the phenotype of the isolated cells. Per analysis, 5 to 6 spleens were pooled (n=3). Representative plots are shown. (B) Cells from panel A were exposed to 600 ng/mL p24 of the indicated LV. After LV administration (5 days), FACS analysis was performed to measure GFP expression. (C) Supernatants from transduced cells were collected at 48 hours, and ELISA was used to measure IFNα and TNF-α concentrations. Values are presented as the means ± SEM (n=3).
pDCs, but not CD11b+ splenocytes, are activated by LVs. (A) Phenotype analysis of isolated splenocyte populations. FACS acquisition of magnetic-sorted splenocytes was carried out to determine the phenotype of the isolated cells. Per analysis, 5 to 6 spleens were pooled (n=3). Representative plots are shown. (B) Cells from panel A were exposed to 600 ng/mL p24 of the indicated LV. After LV administration (5 days), FACS analysis was performed to measure GFP expression. (C) Supernatants from transduced cells were collected at 48 hours, and ELISA was used to measure IFNα and TNF-α concentrations. Values are presented as the means ± SEM (n=3).
Cell were exposed to either PBS or 600 ng p24/mL of VSV.LV or bald.LV, a concentration that corresponds to approximately 10% of the vector dose used in vivo. Higher vector doses were not used because of the possible confounding effect of VSV-mediated toxicity to the cells. Supernatants were collected at 48 hours after exposure. As shown in Figure 4C, pDCs produced high levels of IFNα and TNF-α in response to VSV.LV, but not to bald.LV. In contrast, LV did not trigger cytokine production from CD11b+ splenocytes. Interestingly, FACS analysis found that less than 1% of pDCs were GFP positive (Figure 4B), indicating that transduction is not required for triggering pDC production of IFNα. This may explain how gp64.LV, which has a low infectivity for hematopoietic cells, could trigger an IFNαβ response. Overall, our results are consistent with those recently reported for wildtype HIV,7,22 and suggest that the IFNαβ response seen in vivo following LV administration may be primarily mediated by pDCs.
The type I IFN response limits the efficiency and stability of LV-mediated gene transfer
IFNαβs have a broad range of antiviral properties.23 By triggering a cell to express the double-stranded RNA (dsRNA)–dependent protein kinase (PKR), Mx proteins, or OAS, IFNs can inhibit transcription, block translation, and promote the degradation of dsRNA. IFNαβ can also play a role in the initiation of an adaptive immune response by inducing DC maturation,24 a critical event for DC-mediated T-cell activation.21 Many of the antiviral activities of the IFNαβs, however, act during viral replication. LVs though, are nonreplicating vectors, which are acutely administered as a single bolus, and so transduction may not be affected by a developing antiviral state. In addition, because the IFNαβ response to LVs is self-limiting, it was also unclear whether transient induction would have an effect on the stability of gene transfer.
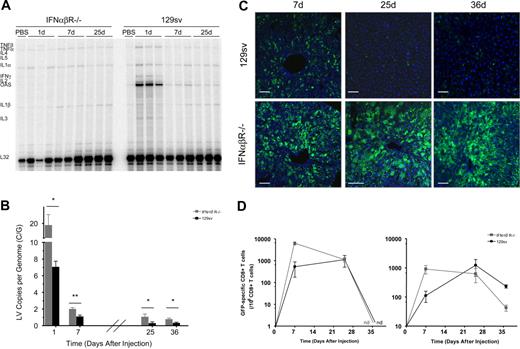
To address these questions, we treated IFNαβR−/− mice and strain-matched controls (129sv) with 15 μg of VSV.LV via tail-vein injection (n = 6/group/time point). RPA analysis was performed, and, as expected, in the absence of IFNαβ signaling LV did not induce OAS expression (Figure 5A). Interestingly, we also did not observe up-regulation of TNF-α expression in the liver of LV-treated IFNαβR−/− mice, while there was induction of TNF-α in normal mice. These results indicate that production of TNF-α, in response to LV, is mediated by IFNαβ, and not directly induced by the vector.
The IFNαβ response mediates reduced transduction efficiency and plays a role in immune-mediated vector clearance. IFNαβR−/− and wild-type strain-matched controls (129sv) were treated with 15 μg VSV.LV (n=6/group/time point). (A) Cytokine profile of the liver and spleen (not shown) were measured by RPA. Analysis was carried out as described in Figure 2. (B) Q-PCR analysis of LV DNA content within the liver and spleen of wild-type (▪) and IFNαβR−/− (⊡) mice at 24 hours after injection. All samples were normalized to murine β-actin. Values are presented as the means ± SEM. Asterisks indicate t-test significance values of wild-type compared with IFNαβR−/− mice (*P <.05; **P < .005). (C) Confocal immunofluorescence analysis of liver sections at the indicated time. Fixed frozen sections were costained with anti-GFP (green) antibodies to identify transduced cells. TO-PRO-3 (blue) was used to stain nuclei. Images are representative of 6 mice analyzed. Scale bars equal 75 μm. (D) ELISPOT analysis of the frequency of GFP-specific IFN-γ secreting CD8+ T cells from the liver (left) and spleen (right) of treated mice. Results are presented as the means ± SEM (n=3/group/time point). ND indicates not detected.
The IFNαβ response mediates reduced transduction efficiency and plays a role in immune-mediated vector clearance. IFNαβR−/− and wild-type strain-matched controls (129sv) were treated with 15 μg VSV.LV (n=6/group/time point). (A) Cytokine profile of the liver and spleen (not shown) were measured by RPA. Analysis was carried out as described in Figure 2. (B) Q-PCR analysis of LV DNA content within the liver and spleen of wild-type (▪) and IFNαβR−/− (⊡) mice at 24 hours after injection. All samples were normalized to murine β-actin. Values are presented as the means ± SEM. Asterisks indicate t-test significance values of wild-type compared with IFNαβR−/− mice (*P <.05; **P < .005). (C) Confocal immunofluorescence analysis of liver sections at the indicated time. Fixed frozen sections were costained with anti-GFP (green) antibodies to identify transduced cells. TO-PRO-3 (blue) was used to stain nuclei. Images are representative of 6 mice analyzed. Scale bars equal 75 μm. (D) ELISPOT analysis of the frequency of GFP-specific IFN-γ secreting CD8+ T cells from the liver (left) and spleen (right) of treated mice. Results are presented as the means ± SEM (n=3/group/time point). ND indicates not detected.
LV content was measured in the liver and spleen of treated animals by Q-PCR (Figures 5B,S2B). At 24 hours after injection, IFNαβR−/− mice had almost 3-fold higher vector content within the liver compared with normal mice (19 ± 5 and 7 ± 0.8 C/G, respectively). This marked difference, early after vector administration, suggests that the IFNαβ response negatively impacts the initial steps of LV transduction.
To further investigate this phenomenon, the GFP expression profile of the liver and spleen was analyzed (Figures 5C,S2A). As seen in Figure 5C, there was a striking difference in the pattern of GFP expression between the 2 treatment groups within the liver. In normal mice, at the dose tested, vector expression occurred predominately within KCs. Only a small fraction of hepatocytes were GFP positive, while in IFNαβR−/− mice, both hepatocytes and nonparenchymal cells were well transduced and expressed the transgene. Since vector DNA content was also found to be higher in IFNαβR−/− mice, even at 24 hours after injection, our data would suggest that the effects of IFNαβ are on vector transduction, and not on transcription or translation of the transgene.
In normal 129sv mice, by day 36, vector content in the liver dropped below 0.3 C/G (Figure 5B). GFP-positive cells were found in some mice, however, at low frequency, and predominately within nonparenchymal cells. In contrast, the LV content within the liver of IFNαβR−/− mice remained above 0.7 C/G up to 36 days. Importantly, GFP expression was maintained in a large fraction of hepatocytes. This unexpected finding indicates that vector-transduced cells persist in IFNαβR−/− mice.
To better understand the immunologic status of GFP in the 2 treatment groups, ELISPOT analysis for IFN-γ–producing cells was used to monitor the frequency of GFP-responsive CD8+ T cells in both the liver and spleen (Figures 5E,S2C). In untreated controls, as expected, no positive cells were found, whereas in LV.PGK.GFP-treated 129sv mice, GFP-specific CD8+ T cells were detected. Interestingly, we also found GFP-specific CD8+ T cells in LV.PGK.GFP-treated IFNαβR−/− mice, but by 36 days after injection, the frequency of GFP-specific CD8+ T cells declined to less than 0.005% of total CD8+ T cells in the spleen, and were undetectable in the liver. These findings indicate that priming of GFP-specific CD8+ T cells occurs in IFNαβR−/− mice; however, because vector content and transgene expression persisted, the primed cells do not appear to be capable of mediating productive vector clearance.
In normal mice, transgene expression is progressively down-regulated and ultimately cleared by the adaptive immune response. Thus, it is difficult to distinguish any direct effect of the IFNαβ response on the pattern of liver cell transduction, since transduced hepatocytes may quickly be cleared by a cellular immune response. To directly assess the influence of the IFNαβ response on LV transduction, we compared nude and IFNαβR−/− mice injected with a dose-matched concentration of LV (n = 3/group). Similar to normal mice, GFP expression in the livers of nude mice was predominately observed in KCs, as determined by costaining with the macrophage-specific marker F4/80 (Figure 6A). Morphometric analysis revealed that 6% ± 0.6% of hepatocytes were GFP positive in the nude mice, whereas in the IFNαβR−/− mice, 37% ± 3% of hepatocytes were GFP positive (Figure 6B). This dramatic disparity strongly suggests that the IFNαβ response has a more profound impact on vector transduction than the adaptive immune response.
The IFNαβ response, and not the adaptive immune system, affects the liver transduction profile of LVs. (A) Confocal immunofluorescence analysis of the liver from nude (Nu/Nu; n=3) and IFNαβR−/− (n=6) mice treated with 15 μg VSV.LV 3 weeks earlier. Fixed frozen sections were costained with anti-GFP (green) and anti-F4/80 (red) antibodies to identify transduced cells and KCs, respectively. TO-PRO-3 (blue) was used to stain nuclei. Scale bars equal 75 μm. (B) Quantitative analysis of the frequency of GFP-positive hepatocytes from LV-treated Nu/Nu and IFNαβR−/− mice. Results are presented as the means ± SEM counts performed on 3 random sections from individual mice.
The IFNαβ response, and not the adaptive immune system, affects the liver transduction profile of LVs. (A) Confocal immunofluorescence analysis of the liver from nude (Nu/Nu; n=3) and IFNαβR−/− (n=6) mice treated with 15 μg VSV.LV 3 weeks earlier. Fixed frozen sections were costained with anti-GFP (green) and anti-F4/80 (red) antibodies to identify transduced cells and KCs, respectively. TO-PRO-3 (blue) was used to stain nuclei. Scale bars equal 75 μm. (B) Quantitative analysis of the frequency of GFP-positive hepatocytes from LV-treated Nu/Nu and IFNαβR−/− mice. Results are presented as the means ± SEM counts performed on 3 random sections from individual mice.
Discussion
Here, we report that intravenous administration of late-generation LVs in mice induces a rapid and transient IFNαβ response in both the liver and spleen. This supports previous studies carried out with human cells in vitro, which demonstrated that HIV and LV can trigger activation of DCs.7,8,25 Our work extends these findings by providing insight into the effect of this response on LV gene transfer. Unexpectedly, we found that IFNαβ strongly inhibits transduction efficiency, specifically within the liver, and contributes to immune-mediated clearance of transduced cells.
Induction of the IFNαβ response occurred within 4 hours of LV injection, independent of the envelope pseudotype, and was concomitant with the presence of reverse-transcribed vector DNA. IFN release did not occur when mice were injected with noninfectious bald and HI.LV particles. Thus, these results strongly point to a mechanism of innate activation by the vector, which requires cell entry. This is an interesting finding since the innate immune system has the capacity to recognize many pathogen-associated molecular patterns (PAMPs) without being infected by the pathogen.26
Several intracellular and endosomal PRRs have been identified, including TLR7 and TLR9, which recognize ssRNA and unmethylated CpG DNA, respectively.27-32 Both HIV and LV carry genetic information as ssRNA and enter the cell through an endosome-mediated pathway. Thus, both have the potential to trigger intracellular PRRs. Since TLR7 is restricted to endosomes, and primarily expressed in pDCs, this would explain our findings that an infectious LV particle is required for innate activation, and that pDCs but not other hematopoietic cells respond to LV. Indeed, Bhardwaj and colleagues have carried out detailed studies demonstrating that HIV activates pDCs through TLR7 signaling.7 However, it has been shown that endogenous RT occurs within the viral particle itself;33 thus, we cannot rule out the possibility that TLR9 and/or an alternate DNA PRR31,32 is also triggered by LV. Indeed, preliminary work from our lab indicates that a TLR7/9 antagonist is not sufficient to prevent LV from inducing an IFNαβ response in vivo (B.D.B. and G.S., unpublished observations, March 2006). Thus, further investigation will be required to identify additional elements within the vector and/or target cells that are involved in triggering the innate host response, with the hope that future strategies can be developed to prevent this response.
Our results indicate that the IFNαβ response has a profound effect on LV-mediated gene transfer. In the absence of IFNαβ signaling, we observed a dramatic increase in the efficiency of liver transduction, with more than 35% of hepatocytes transduced after a single vector administration. It is not clear why transduction of hepatocytes is affected by IFNαβ. One explanation may be related to tissue architecture and LV biodistribution. DCs and KCs are among the first cells to be exposed to the vector, and may be the most efficient at uptaking vector particles.6,34 IFNαβ release by these cells may occur before hepatocyte transduction is complete. As such, IFNαβ signaling, or a cytokine induced by IFNαβ, may have the opportunity to affect the hepatocytes into an antiviral state that prevents transduction from being completed.
Only a few studies of wild-type HIV have examined the effects of IFNαβs on the early steps of viral infection.35-37 In T cells, IFNα was found to interfere with HIV provirus formation, although the specific mechanism mediating the effect was not identified.35 Our results, which show that by 24 hours after injection, well before the development of an adaptive immune response, there was a 2.7-fold differential in vector content between the livers of normal and IFNαβR−/− mice, consistent with a mechanism of interference in the early stages of transduction. It has recently been shown that IFNαβ signaling can induce expression of several retroviral restrictive factors, including Trim5α and APOBEC3G, and suggests a possible mechanism for IFNαβ to negatively impact LV transduction.38,39 The observation that hepatocyte transduction by LV can also be greatly improved above a particular dose threshold would be consistent with a mechanism of restriction involving a saturatable factor, such as Trim5α or APOBEC3G. In interpreting our findings, however, we should also take into account the differences in expression and antiviral activity of members of these gene families among different species. Moreover, since IFNαβs have a broad range of effects on target cells,23 more detailed investigation will need to be carried out to specifically address this complex phenomenon.
Vector content and transgene expression persisted for more than 5 weeks after injection in all LV-treated IFNαβR−/− mice. This occurred despite expansion of GFP-specific CD8+ T cells. IFNαβR−/− mice are capable of clearing a viral infection through an adaptive immune response,40-42 and thus T cells do not have an absolute requirement for IFNαβR signaling in order to be primed and to carry out their effector function. There was some decrease in vector content over time. However, since GFP-expressing cells persisted after contraction of the GFP-specific CD8+ T-cell population, it appears that immune-mediated vector clearance was not realized. Persistence of transgene-expressing cells may be due, in part, to the weak immunogenicity of GFP in the 129sv mouse background. Nonetheless, since vector content was significantly lower in the normal mice compared with the IFNαβR−/− mice, a role for IFNαβ in the decline is indicated.
Although we found that IFNαβ was not required for T-cell activation, it may be important in determining the functionality of the responding T cells. IFNαβs can mediate some increase in the survival capacity of activated T cells, although they do not directly stimulate proliferation,43 and the killing capacity of CD8+ T cells has been shown to be affected by IFNαβ.44 In the absence of IFNαβ signaling, the frequency of virus-specific CD8+ T cells with the capacity to express granzyme B is reduced by half, and cytotoxic T lymphocyte (CTL) function is severely diminished.44 This effect may be due to a continuous need for IFNαβR signaling by responding T cells, which cannot occur in IFNαβR−/− mice. However, as recently shown, it may also be due to a requirement for IFNαβ signaling at the time of CD8+ T-cell activation.45,46
From the latter perspective, our data suggests that the transient IFNαβ response triggered shortly after administration of LV plays the pivotal role in setting in motion the processes leading to the generation of antigen-specific, functionally competent T cells. The apparent lack of redundant pathways which compensate for the absence of IFNαβR signaling in our study may be due to the unique setting of LV gene therapy. LV particles are short-lived, replication-deficient, and stripped of most known virulence factors. They are acutely administered to healthy tissues and neither the viral antigens nor other signals derived from injured cells are thought to persist in the transduced tissue. In such attenuated settings, and contrary to the infection by a wild-type pathogen, disabling only 1 component of the innate/immune network may suffice to tilt the balance of the response from antigen clearance to antigen tolerance.
An alternative or complementary explanation is that the improved transduction efficiency of LV, due to the absence of IFNαβ, played a role in preventing immune-mediated vector clearance. High antigen doses can have a tolerizing effect.47 This was initially shown in an lymphocytic choriomeningitis virus (LCMV) model of infection, where increasing the initial viral dose resulted in more rapid exhaustion of antiviral CTLs and enabled viral persistence.48 Evidence from this model suggests that one of the functions of the IFNαβ response is to reduce initial viral loads, through its antiviral activity, and thereby prevent CTL exhaustion.41 Similarly, by preventing IFNαβ signaling, LV transduction dramatically increased, resulting in higher antigen levels, which may have prevented a productive CTL response.
The work presented here provides the first detailed in vivo analysis of the innate host response to LV. We found that LV induces a strong IFNαβ response that reduces transduction efficiency and plays a major role in preventing stable gene transfer. Our results indicate that the minimum requirement for stimulating this response is an infectious LV particle, and intimates gene delivery itself as the culprit. Nonetheless, because the response is transient, the development of strategies to prevent it, such as neutralizing IFNαβ antibodies or IFNαβR or PRR antagonists, which would require only temporary administration, should serve to improve the effectiveness and stability of LV-mediated gene transfer. For the treatment of diseases such as the hemophilias, where efficient and long-term gene transfer is desired, this could overcome a major barrier to successful gene therapy.
Authorship
Contribution: B.D.B. designed and performed research, analyzed data, and wrote the paper; G.S. designed and performed research, analyzed data, and edited the paper; A.A., E.H., L.S.S., and A.Z. performed research and data analysis; M.G.R. and L.G.G. designed research, analyzed data, and edited the paper; and L.N. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
B.D.B. and G.S. contributed equally to this work.
Correspondence: Luigi Naldini, San Raffaele Telethon Institute for Gene Therapy, Via Olgettina 58, 20132 Milan, Italy; e-mail: naldini.luigi@hsr.it.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The authors would like to thank Matteo Iannacone for advice and assistance.
This work was supported by grants from Telethon and the Italian Ministry of Scientific Research (L.N.). B.D.B. is the recipient of a Natural Science and Engineering Research Council of Canada fellowship.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal