Abstract
Natural killer (NK) cells contribute to host immunity, including tumor surveillance, through the production of interferon gamma (IFN-γ). Although there is some knowledge about molecular mechanisms that induce IFN-γ in NK cells, considerably less is known about the mechanisms that reduce its expression. Here, we investigate the role of the Hlx transcription factor in IFN-γ production by NK cells. Hlx expression is induced in monokine-activated NK cells, but with delayed kinetics compared to IFN-γ. Ectopic Hlx expression decreases IFN-γ synthesis in primary human NK cells and IFN-γ promoter activity in an NK-like cell line. Hlx protein levels inversely correlate with those of STAT4, a requisite factor for optimal IFN-γ transcription. Mechanistically, we provide evidence indicating that Hlx overexpression accelerates dephosphorylation and proteasome-dependent degradation of the active Y693-phosphorylated form of STAT4. Thus, Hlx expression in activated NK cells temporally controls and limits the monokine-induced production of IFN-γ, in part through the targeted depletion of STAT4.
Introduction
Innate immunity is characterized by the ability of immune cells to rapidly detect an invading pathogen and to restrict its dissemination while targeting compromised host cells for elimination. This complex task is achieved in part through the production of soluble cytokines and chemokines by natural killer (NK) cells.1 NK cells elaborate interferon gamma (IFN-γ) in response to stimulation with monokines, particularly IL-12 in combination with IL-15, IL-18, or IL-1β.2 IFN-γ signals through a heterodimeric receptor and STAT1 to promote the maturation and activation of monocytes, leading to improved antigen presentation and the establishment of macrophage effector functions.3
The importance of IFN-γ is illustrated by naturally occurring mutations in IFN-γ receptor subunits and STAT1 in humans, who are highly susceptible to systemic disease after mycobacterial infection or vaccination with bacillus Calmette-Guerin.4 In addition, mice lacking IFN-γ responsiveness exhibit increased incidence of tumors, consistent with a role for IFN-γ in tumor surveillance by immune cells.5 Although the beneficial roles of IFN-γ are unquestionable, excessive IFN-γ can be detrimental to the host. In fact, high levels of IFN-γ are implicated in the etiology of inflammatory bowel disease.6 In addition, unabated IFN-γ production leads to enhanced apoptosis of hematopoietic stem cells and impaired NK cell development.7 Therefore, it is not surprising that IFN-γ synthesis is subject to stringent control in vivo, with multiple checkpoints including transcriptional control.8 Indeed, numerous trans-acting factors have been implicated as positive regulators of IFN-γ gene expression8 ; however, the negative regulation of this process is far less understood at a molecular level.
The H2.0-like homeobox 1 (HLX1, HB24, Hlx) gene encodes a highly conserved putative homeobox transcription factor9 that is preferentially expressed by T-helper 1 (TH1) polarized CD4+ T cells.10,11 Ectopic expression of Hlx during TH2 differentiation leads to increased IFN-γ mRNA and protein expression.10–12 In this study, we report that Hlx is a negative regulator of IFN-γ production by NK cells and that its inhibitory function is achieved at least in part through the proteasomal degradation of STAT4, a key transcription factor for IFN-γ mRNA synthesis.13,14
Materials and methods
Human NK cell isolation and cell lines
Murine NK cell culture
Hlx+/− mice were maintained on an FVB/N background. Dams from Hlx+/− breedings were killed at E13.5, and fetal livers were isolated. DNA was extracted from embryonic tissue for genotyping (Extract-N-Amp Tissue PCR Rapid Genotyping Kit; Sigma, St Louis, MO).17 Fetal liver cells were cultured in 96-well, round-bottom plates (104 cells/well) in DMEM with 10% FBS (Invitrogen, Carlsbad, CA), antibiotic/antimycotic, glutamine, pegylated rat stem cell factor (100 ng/mL; Amgen, Thousand Oaks, CA), murine IL-7 (10 ng/mL; Peprotech, Rocky Hill, NJ), human Flt-3 ligand (100 ng/mL; Amgen), and human IL-2 (1000 IU/mL; Roche, Basel, Switzerland). Successful derivation of NK cells was confirmed cytometrically with NK1.1-APC and DX5-PE, CD19-biotin, CD14-biotin, CD3-biotin, Ter119-biotin, and streptavidin-PerCP-Cy5.5 (BD PharMingen, San Diego, CA).
Retroviral infection of primary NK and NK-92 cells
The entire Hlx open-reading frame, containing its native start and stop codons, was amplified from human leukocyte cDNA and introduced into pCR2.1-TOPO (Invitrogen). Forward and reverse primers for PCR were modified at their 5′ ends with BamHI and EcoRI sites, respectively. After bidirectional sequencing, the Hlx cDNA was mobilized to the PINCO retroviral transfer vector as a BamHI-EcoRI fragment. Alternatively, for IFN-γ reporter assays, Hlx was subcloned into pcDNA3.1 (Invitrogen). Retrovirus was produced, and cells were infected and purified as described.16
Monokine stimulation of NK cells
Human NK cells were stimulated at 2 million/mL in RPMI medium containing 10% FBS and IL-12 (10 ng/mL; Genetics Institute, Cambridge, MA), IL-15 (100 ng/mL; Amgen), or IL-18 (50 ng/mL; R&D Systems, Minneapolis, MN). Before stimulation at 1 million/mL, NK-92 cells were washed extensively with PBS and rested for 24 hours in RPMI containing 10% FBS in the absence of IL-2. Murine NK cells were stimulated at 2 million/mL in DMEM containing 10% FBS with the cytokines murine IL-12 (10 ng/mL; Genetics Institute) and human IL-15 (3 μg/mL; Amgen). For proteasomal inhibition, MG-132 (Calbiochem), epoxomicin (Sigma), and lactacystin were reconstituted according to the manufacturer's instructions and incubated at 20 μM final concentration with rested PINCO-Hlx–infected NK-92 cells for 1 hour at 37°C before IL-12/IL-18 was added for the indicated times. To inhibit protein translation, cycloheximide (Sigma) was reconstituted in 100% ethanol and incubated at 20 μg/mL final concentration with rested PINCO-Hlx–infected NK-92 cells for 30 minutes at 37°C before IL-12/IL-18 was added for the indicated times.
Detection of IFN-γ and Y693 pSTAT4
QRT-PCR
Total RNA was extracted and reverse transcribed.18 Sequences of all QRT-PCR primers and probes are available on request. QRT-PCR reactions were performed and analyzed by the ΔCt method after normalizing to internal 18S control reactions, as described.20 Results—expressed as the mean ± SEM of triplicate wells—were the n-fold difference of transcript levels in an 18S-normalized sample compared with calibrator cDNA. Calibrator cDNA was unstimulated NK (Figure 1A), unstimulated CD56dim NK (Figure 1C), or unstimulated PINCO NK-92 (Figure 3C).
Detection of Hlx protein and immunoblotting
Hlx protein was detected using affinity-purified rabbit antisera raised against a GST-Hlx fusion protein (Abgent, San Diego, CA). To construct the GST-Hlx fusion protein, a human Hlx cDNA fragment corresponding to amino acids 333 to 488 was PCR amplified and cloned into the BamHI and EcoRI sites of pGEX-6P-1 (Amersham Biosciences, Freiburg, Germany). Affinity-purified Hlx antiserum was used at 1:1000 dilution. Immunoblotting was performed with whole cell lysates18 or nuclear and cytosolic protein.21 Antibodies were anti–pSTAT4 Y693 (1:5000 mouse IgG2b monoclonal; BD PharMingen), anti–total STAT4 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), anti–β-actin (1:1000; Santa Cruz Biotechnology), anti-Ku70 (1:1000; Santa Cruz Biotechnology), and anti-Brg1 (1:1000).
Reporter assays
Five million DERL-7 were transfected by nucleofection (Amaxa, Cologne, Germany) using Solution V and DNA as follows: 5 μg empty pGL3-Basic (Promega, Madison, WI) or the IFN-γ firefly luciferase vector (a kind gift of Howard Young, National Cancer Institute22 ), 5 μg pcDNA3.1-Hlx or empty pcDNA3.1 vector (Invitrogen), 50 ng TK-renilla luciferase vector (pRL-TK; Promega), and program O-17. After electroporation, cells were immediately placed in 10% RPMI containing IL-12/IL-18 and were incubated for 6 hours at 37°C. Cells were subsequently washed with ice-cold PBS and lysed in 140 μL 1 × passive lysis buffer (Promega), and luminescence was analyzed with the dual-luciferase reporter system (Promega). All assays were performed in triplicate, and all firefly luciferase values were normalized for transfection efficiency by subtracting the renilla luciferase activity derived from the cotransfected pRL-TK plasmid. The activity of the pGL3 basic reporter vector alone was subtracted from that of the vector with the IFN-γ promoter, and the mean ± SD of the triplicate values of the difference is shown.
Statistical analysis
Data were analyzed using a Student 2-tailed t test. P values below .05 were considered statistically significant.
Results
Hlx expression is induced in monokine-activated NK cells
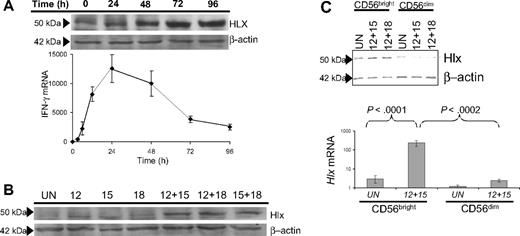
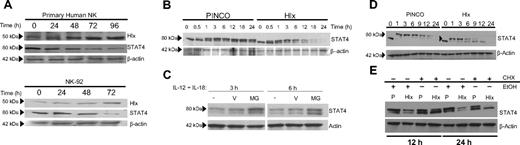
We first investigated the relationship between Hlx protein and IFN-γ mRNA levels in primary human NK cells over time after treatment with IL-12 and IL-18 (IL-12/IL-18). Hlx protein expression was readily detectable by immunoblotting in resting NK cells, increased on IL-12/IL-18 treatment, peaked at 72 hours, and was maintained after 96 hours of stimulation (Figure 1A, upper panel). In contrast, IFN-γ mRNA expression—measured by quantitative reverse transcription–polymerase chain reaction (QRT-PCR)—peaked at 24 hours and subsequently declined (Figure 1A, lower panel). Maximal induction of Hlx expression required costimulation with different cytokines (eg, IL-12/IL-18) (Figure 1B) that, as reported,2 synergistically promoted high IFN-γ production. Treatment with individual monokines led to modest or undetectable changes in Hlx protein levels (Figure 1B), suggesting that increased Hlx expression may have a role in the regulation of IFN-γ production.
Monokine-dependent Hlx expression by human CD56bright NK cells is delayed with respect to IFN-γ transcription. (A) Delayed induction of Hlx protein with respect to IFN-γ production in primary NK cells. (upper) Total NK cells were stimulated with IL-12/IL-18 for indicated time points, and Hlx protein levels were analyzed by immunoblotting. β-Actin levels were analyzed to ensure equal loading (n = 4 experiments). (lower) In parallel, IFN-γ mRNA levels were analyzed by QRT-PCR. Mean ± SEM from 4 separate experiments is shown. (B) Maximal Hlx protein induction requires monokine costimulation. Total NK cells were stimulated for 72 hours, as indicated, and Hlx and β-actin protein levels were analyzed by immunoblotting (n = 3 experiments). (C) Preferential induction of Hlx protein in the CD56bright NK subset. (upper) FACS-purified unstimulated NK subsets were lysed directly (UN) or after stimulation with the indicated monokine combinations (72 hours). Lysates were analyzed for Hlx and β-actin protein by immunoblotting (n = 3 experiments). (lower) FACS-purified NK subsets were lysed immediately for RNA (UN) or stimulated with IL-12/IL-15 for 12 hours before lysis. Hlx mRNA levels were analyzed by QRT-PCR. Mean ± SEM from 5 separate donors is shown.
Monokine-dependent Hlx expression by human CD56bright NK cells is delayed with respect to IFN-γ transcription. (A) Delayed induction of Hlx protein with respect to IFN-γ production in primary NK cells. (upper) Total NK cells were stimulated with IL-12/IL-18 for indicated time points, and Hlx protein levels were analyzed by immunoblotting. β-Actin levels were analyzed to ensure equal loading (n = 4 experiments). (lower) In parallel, IFN-γ mRNA levels were analyzed by QRT-PCR. Mean ± SEM from 4 separate experiments is shown. (B) Maximal Hlx protein induction requires monokine costimulation. Total NK cells were stimulated for 72 hours, as indicated, and Hlx and β-actin protein levels were analyzed by immunoblotting (n = 3 experiments). (C) Preferential induction of Hlx protein in the CD56bright NK subset. (upper) FACS-purified unstimulated NK subsets were lysed directly (UN) or after stimulation with the indicated monokine combinations (72 hours). Lysates were analyzed for Hlx and β-actin protein by immunoblotting (n = 3 experiments). (lower) FACS-purified NK subsets were lysed immediately for RNA (UN) or stimulated with IL-12/IL-15 for 12 hours before lysis. Hlx mRNA levels were analyzed by QRT-PCR. Mean ± SEM from 5 separate donors is shown.
Because combination monokine treatment (eg, IL-12/IL-18 or IL-12/IL-15) strongly stimulates IFN-γ production by CD56bright but not CD56dim human NK cells,2,20 we investigated whether the expression of Hlx was restricted to a specific CD56 NK subset. Indeed, though Hlx mRNA and protein levels were comparable in resting CD56 NK subsets, induction of Hlx mRNA and protein expression occurred preferentially in IL-12/IL-18– and IL-12/IL-15–stimulated CD56bright NK cells (Figure 1C). Thus, Hlx expression is induced by monokine costimulation and enhanced within the CD56bright NK subset; however, higher levels of Hlx correlated with decreased IFN-γ production by NK cells.
Hlx negatively regulates IFN-γ production in monokine-activated NK cells
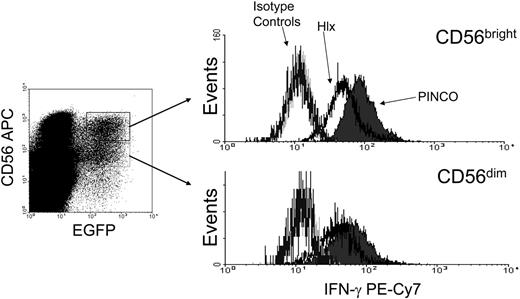
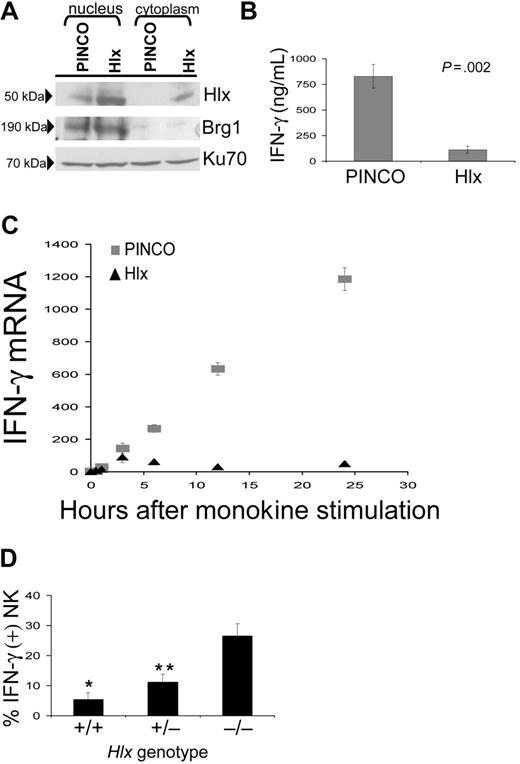
Based on the kinetics of Hlx expression relative to IFN-γ transcription, it is conceivable that Hlx acts as a negative regulator of IFN-γ production in CD56bright NK cells. Thus, IFN-γ production was assessed by intracellular staining in IL-12/IL-18–stimulated primary human NK cells previously transduced with the EGFP (enhanced green fluorescence protein)–expressing PINCO-Hlx or empty PINCO retrovirus (Figure 2). Interestingly, a significant decrease in the proportion of IFN-γ+ cells was observed in the Hlx-expressing EGFP+CD56bright NK cells (CD56bright PINCO-Hlx compared with CD56bright PINCO; P < .02; n = 3 experiments) and in the CD56dim fraction (CD56dim PINCO-Hlx compared with CD56dim PINCO; P < .006; n = 3 experiments). In similar experiments, human NK-92 cells, which as reported have a phenotype that resembles the CD56bright NK cell subset,23 were infected with the Hlx or the empty retrovirus and were EGFP-sorted. As expected, anti-Hlx Western blots performed on subcellular fractions from EGFP+ PINCO– and EGFP+ PINCO-Hlx–transduced NK-92 cell lysates showed that Hlx was indeed overexpressed and primarily localized in the nuclear compartment (Figure 3A). Note that Brg1 and Ku70 were detected as controls for purity of the nuclear fraction and equal loading, respectively (Figure 3A). Consistent with the data obtained with primary CD56bright NK cells, Hlx overexpression led to a marked decrease in IFN-γ protein levels in IL-12/IL-18–stimulated NK-92 cells after 24 hours of stimulation (Figure 3B). Moreover, levels of IFN-γ mRNA, albeit similar in unstimulated vector- and Hlx-transduced NK-92 cells, dramatically increased over time after IL-12/IL-18 stimulation of vector-transduced cells but not in NK-92 cells overexpressing Hlx (Figure 3C). Thus, our data not only indicated that sustained Hlx expression impairs the ability of NK cells to produce IFN-γ in response to monokine costimulation, they also suggested that a monokine-induced increase of Hlx level is most likely required to temporally control the expression and, therefore, the production of IFN-γ by activated NK cells. Indeed, Hlx-deficient murine NK cells show enhanced IFN-γ mRNA and protein production after in vitro monokine costimulation with IL-12 and IL-15 compared with wild-type and heterozygous control NK cells (Figure 3D and data not shown).
Hlx inhibits IFN-γ production by primary CD56bright NK. PINCO- or PINCO-Hlx–infected primary human NK cells were stimulated for 24 hours with IL-12/IL-18 and were subjected to staining for CD56 and IFN-γ. (left) Infected cells were gated on CD56brightEGFP+ or CD56dimEGFP+. (right) IFN-γ staining in each population compared with isotype controls. Results shown are representative of 3 experiments (CD56bright PINCO-Hlx compared with CD56bright PINCO; P < .02).
Hlx inhibits IFN-γ production by primary CD56bright NK. PINCO- or PINCO-Hlx–infected primary human NK cells were stimulated for 24 hours with IL-12/IL-18 and were subjected to staining for CD56 and IFN-γ. (left) Infected cells were gated on CD56brightEGFP+ or CD56dimEGFP+. (right) IFN-γ staining in each population compared with isotype controls. Results shown are representative of 3 experiments (CD56bright PINCO-Hlx compared with CD56bright PINCO; P < .02).
Hlx inhibits IFN-γ mRNA and protein expression in NK-92 cells. (A) Overexpression of Hlx protein in NK-92 cells. FACS-purified NK-92 cells transduced with PINCO or PINCO-Hlx were subjected to nucleocytoplasmic fractionation and immunoblotting for Hlx. Enrichment of nuclear protein was confirmed by Brg1 staining. Equal loading was confirmed by Ku70 staining. (B) Inhibition of IFN-γ production by Hlx. PINCO- or PINCO-Hlx–infected NK-92 cells were stimulated with IL-12/IL-18, and supernatants were harvested after 24 hours for IFN-γ ELISA. Shown are the mean ± SEM from 5 separate experiments. (C) Time course of IFN-γ mRNA in PINCO-compared with PINCO-Hlx–transduced NK-92 in response to IL-12/IL-18. Shown are results of 1 of 4 representative experiments; error bars represent SD of triplicate PCR reactions. (D) Increased IFN-γ production by Hlx−/− NK in response to monokine costimulation. Fetal liver–derived NK of indicated genotypes were subjected to 24-hour stimulation with IL-12 and IL-15, followed by intracellular staining for IFN-γ. Mean ± SEM percentages of IFN-γ+ NK1.1+ cells from 9 experiments with NK cells derived from 9 or more livers of each genotype are shown. *P < .001 Hlx+/+ compared with Hlx−/−. **P < .005 Hlx+/− compared with Hlx−/−).
Hlx inhibits IFN-γ mRNA and protein expression in NK-92 cells. (A) Overexpression of Hlx protein in NK-92 cells. FACS-purified NK-92 cells transduced with PINCO or PINCO-Hlx were subjected to nucleocytoplasmic fractionation and immunoblotting for Hlx. Enrichment of nuclear protein was confirmed by Brg1 staining. Equal loading was confirmed by Ku70 staining. (B) Inhibition of IFN-γ production by Hlx. PINCO- or PINCO-Hlx–infected NK-92 cells were stimulated with IL-12/IL-18, and supernatants were harvested after 24 hours for IFN-γ ELISA. Shown are the mean ± SEM from 5 separate experiments. (C) Time course of IFN-γ mRNA in PINCO-compared with PINCO-Hlx–transduced NK-92 in response to IL-12/IL-18. Shown are results of 1 of 4 representative experiments; error bars represent SD of triplicate PCR reactions. (D) Increased IFN-γ production by Hlx−/− NK in response to monokine costimulation. Fetal liver–derived NK of indicated genotypes were subjected to 24-hour stimulation with IL-12 and IL-15, followed by intracellular staining for IFN-γ. Mean ± SEM percentages of IFN-γ+ NK1.1+ cells from 9 experiments with NK cells derived from 9 or more livers of each genotype are shown. *P < .001 Hlx+/+ compared with Hlx−/−. **P < .005 Hlx+/− compared with Hlx−/−).
Hlx represses IFNG promoter activity in NK cells
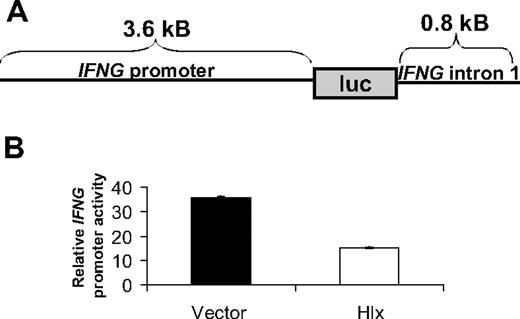
Because Hlx overexpression in NK-92 cells resulted in decreased IFN-γ mRNA levels and because Hlx is a homeobox transcription factor, we reasoned that Hlx may regulate the transcriptional activity of the IFNG promoter in NK cells. To begin to test this hypothesis, we sought to determine whether Hlx could inhibit IFNG promoter activity in vivo. We made use of luciferase reporter assays in the DERL-7 NK-like cell line,24 which is easily transduced by electroporation.15,25 DERL-7 was electroporated with a previously characterized full-length human IFNG promoter-firefly luciferase reporter construct containing an approximately 3.6-kb IFNG 5′ flanking sequence and an 0.8-kB enhancer element from the first intron of IFNG following the luciferase open-reading frame (Figure 4A). 22,26 After electroporation, cells were treated with IL-12/IL-18 for 6 hours. After normalizing for transfection efficiency, we determined that Hlx overexpression repressed IFNG promoter activity (Figure 4B). However, on subsequent deletion analysis of the 3.6-kb IFNG promoter and the 0.8-kb intron 1 enhancer element within the construct, we were unable to map Hlx repressive activity to any particular region (data not shown). Thus, the mechanism of IFNG promoter regulation by Hlx is likely complex and dependent on multiple cis-regulatory elements.
Hlx inhibits IFNG promoter activity in DERL-7 cells. (A) The human IFNG luciferase construct used in this study consisted of 3.6 kB of a 5′ flanking sequence upstream of the transcriptional start site, the firefly luciferase open-reading frame, and 0.8 kB of the intron 1 enhancer element. (B) DERL-7 cells with Hlx or empty vector were transfected with the IFNG luciferase construct, and luciferase activity was measured after 6-hour stimulation with IL-12/IL-18. Mean ±SD of 1 of 3 representative experiments is shown.
Hlx inhibits IFNG promoter activity in DERL-7 cells. (A) The human IFNG luciferase construct used in this study consisted of 3.6 kB of a 5′ flanking sequence upstream of the transcriptional start site, the firefly luciferase open-reading frame, and 0.8 kB of the intron 1 enhancer element. (B) DERL-7 cells with Hlx or empty vector were transfected with the IFNG luciferase construct, and luciferase activity was measured after 6-hour stimulation with IL-12/IL-18. Mean ±SD of 1 of 3 representative experiments is shown.
Hlx promotes proteasome-dependent STAT4 degradation after monokine stimulation
We further hypothesized that Hlx might negatively regulate IFN-γ mRNA levels by inhibiting the function or activity of a positive regulator of IFN-γ production. STAT4 is a known trans-activator of the IFN-γ promoter in NK cells.13,14 STAT4 is activated by tyrosine phosphorylation at tyrosine residue 693 (Y693) by Jak2/Tyk2 kinases within seconds of IL-12 stimulation, resulting in its rapid nuclear translocation and direct association with multiple cis-regulatory elements within the IFN-γ promoter.26–28 In IL-12/IL-18–stimulated primary human NK or NK-92 cells, we observed that the increase of Hlx expression was accompanied by a decrease in total STAT4 protein levels (Figure 5A), suggesting that Hlx may negatively regulate IFN-γ gene transcription by suppressing STAT4 expression. Thus, levels of total STAT4 protein were also evaluated in NK-92 cells transduced with Hlx or empty retrovirus, EGFP-sorted and stimulated with IL-12/IL-18. Before stimulation, Hlx- and vector-transduced NK-92 cells expressed similar levels of unphosphorylated STAT4 (lower, faster-migrating band at 0 hour; Figure 5B). Similarly, both populations demonstrated comparable induction of Y693 pSTAT4 levels after IL-12/IL-18 stimulation (upper, slower-migrating band at 0.5 hours; Figure 5B). However, Hlx overexpression inhibited long-term monokine-induced STAT4 activation as levels of Y693 pSTAT4 progressively decreased over time in Hlx-expressing NK-92 cells but not in vector-transduced NK-92 cells (Figure 5B). In parallel experiments, we did not observe an effect of Hlx on total and tyrosine-phosphorylated STAT1 and STAT5, indicating that Hlx exerts its effect specifically on STAT4. We also found that retroviral expression of Hlx in primary NK reduced the proportion of Y693 pSTAT4+ cells after IL-12/IL-18 treatment (from 78.0% ± 5.4% in vector-transduced cells to 67.1% ± 5.4% in Hlx-transduced cells; n = 5 independent experiments; P < .005).
Hlx overexpression leads to proteasome-dependent degradation of STAT4. (A) Endogenous Hlx levels increased as total STAT4 levels declined. Primary human NK or NK-92 cells were harvested at the indicated time points after IL-12/IL-18 stimulation, and Hlx, STAT4, and β-actin protein levels were determined by immunoblotting. (n = 4 experiments in primary NK, n = 2 experiments in NK-92). (B) Hlx overexpression decreased total and Y693 pSTAT4 levels in NK-92 after IL-12/IL-18 treatment. FACS-purified PINCO- or Hlx-expressing NK-92 cells were harvested at indicated time points after IL-12/IL-18 stimulation. Total STAT4 and β-actin levels were assessed by immunoblotting (n = 4 experiments). (C) Proteasome inhibition rescued loss of STAT4 in NK-92 cells overexpressing Hlx. Cells were preincubated for 1 hour with 20 mM MG-132 (MG), an equal amount of DMSO vehicle, or medium alone (—), followed by IL-12/IL-18 treatment for the indicated times. Total protein was harvested, and total STAT4 and β-actin levels were assessed by immunoblotting (n = 3 experiments). (D) Diminished Y693 pSTAT4 levels in Hlx-overexpressing NK-92 cells despite inhibition of new protein synthesis. PINCO- or Hlx-expressing NK-92 cells were preincubated for 30 minutes with 10 μM CHX, followed by IL-12/IL-18 treatment for the indicated times. Total cellular protein was harvested, and total STAT4 and β-actin levels were assessed by immunoblotting (the arrowhead indicates the position of the 80 kDa marker). (E) Comparison of CHX compared with ethanol (EtOH) carrier effects on STAT4 expression in PINCO- (P) and Hlx-expressing NK-92 after IL-12/IL-18 stimulation for the indicated times (n = 3 experiments).
Hlx overexpression leads to proteasome-dependent degradation of STAT4. (A) Endogenous Hlx levels increased as total STAT4 levels declined. Primary human NK or NK-92 cells were harvested at the indicated time points after IL-12/IL-18 stimulation, and Hlx, STAT4, and β-actin protein levels were determined by immunoblotting. (n = 4 experiments in primary NK, n = 2 experiments in NK-92). (B) Hlx overexpression decreased total and Y693 pSTAT4 levels in NK-92 after IL-12/IL-18 treatment. FACS-purified PINCO- or Hlx-expressing NK-92 cells were harvested at indicated time points after IL-12/IL-18 stimulation. Total STAT4 and β-actin levels were assessed by immunoblotting (n = 4 experiments). (C) Proteasome inhibition rescued loss of STAT4 in NK-92 cells overexpressing Hlx. Cells were preincubated for 1 hour with 20 mM MG-132 (MG), an equal amount of DMSO vehicle, or medium alone (—), followed by IL-12/IL-18 treatment for the indicated times. Total protein was harvested, and total STAT4 and β-actin levels were assessed by immunoblotting (n = 3 experiments). (D) Diminished Y693 pSTAT4 levels in Hlx-overexpressing NK-92 cells despite inhibition of new protein synthesis. PINCO- or Hlx-expressing NK-92 cells were preincubated for 30 minutes with 10 μM CHX, followed by IL-12/IL-18 treatment for the indicated times. Total cellular protein was harvested, and total STAT4 and β-actin levels were assessed by immunoblotting (the arrowhead indicates the position of the 80 kDa marker). (E) Comparison of CHX compared with ethanol (EtOH) carrier effects on STAT4 expression in PINCO- (P) and Hlx-expressing NK-92 after IL-12/IL-18 stimulation for the indicated times (n = 3 experiments).
Interestingly, at later time points, we observed that ectopic Hlx expression not only interfered with STAT4 activation, it also significantly reduced the level of unphosphorylated STAT4 protein (Figure 5B). This reduction in total STAT4 protein levels led us to test whether Hlx overexpression is associated with a decrease in STAT4 mRNA; however, no difference in STAT4 transcript levels was detectable by QRT-PCR in Hlx compared with vector-infected NK-92 cells during time-course experiments in IL-12/IL-18 (n = 4 experiments; data not shown). Alternatively, it has been reported that Y693 pSTAT4 undergoes proteasome-dependent degradation in T lymphocytes after IL-12 treatment.29,30 We therefore attempted to reverse the loss of Y693 pSTAT4 protein through the application of MG-132, an inhibitor of the 26S proteasome subunit. Indeed, MG-132 rescued the loss of Y693 pSTAT4 after short-term (3 or 6 hours) treatment of Hlx-expressing NK-92 cells with IL-12/IL-18 (Figure 5C). Similar results were obtained with the additional proteasome inhibitors epoxomicin and lactacystin (data not shown). Interestingly, proteasome inhibition also rescued expression of the unphosphorylated STAT4 protein, particularly after 6-hour stimulation with IL-12/IL-18 (Figure 5C). The E3 ubiquitin ligase SLIM has been shown to catalyze polyubiquitylation of Y693 pSTAT4 in vitro and to attenuate STAT4 protein expression in vivo.30 Therefore, we sought to determine whether SLIM protein levels were altered by Hlx overexpression by immunoblotting, but no change in SLIM expression was observed (data not shown).
To confirm that the loss of STAT4 protein was caused by decreased protein stability, new protein synthesis was inhibited using cycloheximide (CHX), and the kinetics of STAT4 degradation were monitored in NK-92 expressing Hlx compared with empty vector. As expected, in the presence of CHX, Hlx overexpression resulted in decreased Y693 pSTAT4 levels beginning after 6 hours of IL-12/IL-18 stimulation compared with empty vector (Figure 5D). Surprisingly, however, CHX treatment resulted in an accumulation of unphosphorylated STAT4 in cells expressing Hlx and empty vector but with more rapid kinetics with Hlx overexpression (Figure 5D). Thus, it appears that Hlx overexpression accelerates the dephosphorylation of Y693 STAT4 and that its subsequent degradation is reliant on new protein synthesis. To confirm these findings were specific to CHX treatment, we treated Hlx and vector expressing NK-92 with CHX or ethanol carrier for 12 hours or 24 hours and repeated the STAT4 Western blot (Figure 5E). Again, we found that the inhibitory effect of Hlx overexpression on Y693 pSTAT4 levels was insensitive to CHX treatment. Furthermore, we confirmed the stabilizing influence of CHX treatment on the levels of unphosphorylated STAT4, regardless of Hlx or vector expression.
Discussion
In this study, we found that Hlx is a novel negative regulator of IFN-γ production in NK cells after monokine costimulation. The delayed kinetics of Hlx protein induction with respect to IFN-γ itself are consistent with a model in which Hlx serves as a feedback inhibitor of IFN-γ synthesis in activated CD56bright NK. This Hlx-mediated negative feedback pathway is likely important in vivo, given the potentially deleterious effects of unchecked IFN-γ production on the host.6,7 The process of Hlx protein induction after monokine costimulation is likely complex. Indeed, Hlx regulation must occur at transcriptional and posttranscriptional levels because parallel mRNA and protein expression studies have established long lags between monokine costimulation and the onset of Hlx mRNA expression (approximately 12 hours; data not shown) and between the onset of detectable Hlx mRNA and appreciable Hlx protein expression (Figure 1A). In future studies, it will be insightful to analyze the HLX promoter itself and to determine the direct influence of monokines on its activity and the potential effects of NK-derived cytokines that have been shown to inhibit IFN-γ production, such as transforming growth factor-β and TWEAK,15,31 on Hlx mRNA and protein expression.
In addition, we identified the STAT4 transcription factor as a key molecular target of Hlx action in NK cells. Multiple lines of evidence have established the role of STAT4 as a requisite, direct transactivator of the IFNG promoter in NK cells after monokine stimulation.13,14,27 Constitutive STAT4 activation is strongly implicated in autoimmune disease, including rheumatoid arthritis and inflammatory bowel disease.6,32 The inhibitory effect of Hlx on STAT4 levels identifies Hlx as a potentially relevant molecular target to exploit in autoimmunity. Ultimately, the physiological and pathophysiological roles of Hlx in the attenuation of STAT4 transcriptional activity and IFN-γ production must be determined in vivo.
Mechanistically, we found that Hlx promotes the degradation of Y693 pSTAT4. Previous work has identified SLIM as an E3 ubiquitin ligase with substrate specificity toward Y693 pSTAT4.30 However, this study did not explore the possibility that Y693 pSTAT4 undergoes dephosphorylation before its degradation by the proteasome. In our study, through the use of CHX, we implicated Hlx specifically in the dephosphorylation of pSTAT4 Y693 (Figure 4E), and we demonstrated that subsequent degradation of unphosphorylated STAT4 requires new protein synthesis. These findings suggest that the process of Y693 pSTAT4 degradation is complex, consisting of a CHX-insensitive dephosphorylation step and a CHX-sensitive degradation step. The specific role of Hlx in Y693 pSTAT4 dephosphorylation remains unclear. Although the Hlx protein structure lacks sequence homology to known tyrosine phosphatase domains, it is conceivable that Hlx may recruit a protein tyrosine phosphatase to DNA-bound Y693 pSTAT4 dimers or tetramers. In addition, it is possible that Hlx may associate directly with DNA through its homeodomain and may displace DNA-bound Y693 pSTAT4 multimers, rendering them more susceptible to dephosphorylation and degradation. Moreover, we have not fully rejected the hypothesis that Hlx functions as a transcription factor and that it is in fact the transcriptional target(s) of Hlx, translated before CHX and monokine treatment, that trigger Y693 pSTAT4 dephosphorylation after IL-12/IL-18 stimulation. Indeed, it is conceivable that SLIM is one such target of Hlx.
Besides identifying a role for STAT4 in Hlx-dependent regulation of IFN-γ, our work identifies the IFNG promoter as a potential direct target of Hlx transcriptional repression. However, in preliminary experiments, we were unable to identify the specific region required for Hlx repression (data not shown). This difficulty may be attributable to a complex requirement for multiple cis-regulatory motifs throughout the IFNG promoter for Hlx repression of IFN-γ to occur. Therefore, in subsequent work, it will be essential to distinguish the effects of Hlx on STAT4 from its direct effect, if any, at the IFNG promoter. Stimulation of Hlx overexpressing NK-92 cells with IL-2 or IL-15, alone or in combination with IL-18, did not influence IFN-γ mRNA or protein levels compared with cells infected with empty retroviral vector (data not shown). Because STAT4 is activated minimally under these conditions (data not shown), this finding is consistent with a model in which the effects of Hlx are mediated primarily by STAT4. STAT4 is required for IFN-γ production in response to IL-12,13,14 making it prohibitive to explore the consequence of Hlx overexpression in STAT4-deficient NK cells on IFN-γ production after IL-12/IL-18 treatment or to determine the effect of Hlx on the activity of IFNG promoter constructs lacking STAT4 enhancer elements. Given these challenges, it may be insightful instead to perform a detailed structure–function analysis of Hlx itself so as to identify Hlx mutants that maintain IFN-γ repression without affecting STAT4 levels and vice versa.
Our interest in Hlx was prompted by published accounts of its role as a positive regulator of IFN-γ production in CD4+ T lymphocytes.10–12 Although the data presented here are not consistent with this role in NK cells, considerable caution should be exercised in comparing this work with the T-cell studies given the distinctly different mechanisms of IFN-γ production in NK and CD4+ T-cell populations. Although NK cells produce IFN-γ within minutes of monokine stimulation,20 CD4+ T cells must undergo a process of polarization, entailing cell division and active chromatin remodeling at the IFNG locus, before IFN-γ transcription can occur.33 Ectopic expression of Hlx during TH2 polarization of CD4+ T cells has been shown to promote IFN-γ production, but Hlx expression during TH1 polarization and in fully TH1- and TH2-polarized CD4+ T cells reportedly does not influence IFN-γ synthesis.10–12 These findings have led to the hypothesis that Hlx functions in a discrete window during early TH-cell differentiation to promote IFN-γ production.10–12
Interestingly, recent work from the Zheng34 laboratory clearly implicates Hlx as a negative regulator of the IL-4 receptor alpha chain (IL-4Rα). Because IL-4 is required for normal TH2 differentiation, Hlx deficiency results in impaired production of TH2 cytokines, and this phenotype can be reversed by ectopic retroviral expression of IL-4Rα.34 Apart from promoting TH2 differentiation, IL-4 is a potent inhibitor of IFN-γ production by T cells.35 Therefore, by decreasing the responsiveness of a CD4+ T cell to IL-4 during early TH2 polarization, Hlx may indirectly promote the production of IFN-γ by this lymphocyte population. If the effect of Hlx on IFN-γ is attributed entirely to IL-4Rα regulation, it will be important in the future to determine whether the loss of IL-4Rα expression eliminates the increase in IFN-γ seen on ectopic Hlx expression in TH2 cells. In addition, it will be interesting to determine whether Hlx influences IL-4Rα expression by NK cells given that IL-4 plays a very different role in NK cells than in T cells, namely to costimulate IFN-γ production.36,37
Thus, it is likely that Hlx serves different roles in the T and NK lineages, and the extent of its function may be limited to discrete developmental windows. It is not without precedent that transcription factors perform divergent functions in multiple lineages. GATA-3 is one transcriptional regulator shown to have apparently opposing functions in IFN-γ regulation by NK and CD4+ T cells.38 Alternatively, one can envision multiple scenarios that could account for opposing Hlx activities in CD4+ T and NK cells, such as posttranslational modification of Hlx or unique expression of Hlx interacting proteins in lineage-specific fashion. It is anticipated that more detailed studies of the Hlx protein and its interactions will elucidate its mechanism of action in T- and NK-cell lineages in the future.
Authorship
Contribution: B.B. designed and performed the research and wrote the manuscript. T.L.H. performed the research. A.G.F. designed the research. B.W.B. performed the research. J.Y designed the research. R.T. designed the research. H.C.M. collected the data. M.L.C. performed the research. M.A. performed the research. D.M.B. performed the research. A.L. analyzed the data. D.J. analyzed the data. D.P. designed the research. M.D.B. designed the research. M.A.C. designed the research and edited the original and revised versions of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael A. Caligiuri, A458 Starling-Loving Hall, 320 W 10th Ave, Columbus, OH 43210; e-mail: michael.caligiuri@osumc.edu.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Cancer Institute grants P30 CA16059, CA68458, and CA95426 (M.A.C.) and by National Institutes of Health grants DK02791 and DK61219 (M.D.B.).
We thank Dr Howard Young (National Cancer Institute, Frederick, MD) for providing the IFNG promoter luciferase construct; Dr Michael Grusby (Harvard University) for the generous donation of SLIM antisera, and Dr Denis Guttridge (The Ohio State University) and Dr Wei-ping Zheng (University of Rochester) for helpful advice.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal