Abstract
The identification of alternative splice variants of Survivin that possess distinct functions from those originally identified for the main Survivin isoform has greatly increased the complexity of our understanding of the role of Survivin in different cells. Previous functional studies of the Survivin splice variants have been performed almost exclusively in cancer cells. However, Survivin has increasingly been implicated in other normal physiologic and pathophysiologic processes, including angiogenesis. In this study, we dissect the involvement of Survivin ΔEx3 in angiogenesis. We show by confocal microscopy that a pool of endothelial Survivin ΔEx3 is localized to membrane ruffles. We also demonstrate that Survivin ΔEx3 is the Survivin splice variant responsible for modulating angiogenesis in vitro, in tube formation assays, and in vivo, in an in vivo angiogenesis assay. Our data indicate that Survivin ΔEx3 may regulate angiogenesis via several mechanisms including cell invasion, migration, and Rac1 activation. Our findings identify a novel pathway regulating angiogenesis through Survivin ΔEx3 and a novel mechanism for Rac1 activation during angiogenesis. In conclusion, our results provide new insights into the regulation of endothelial cell homeostasis and angiogenesis by the Survivin proteins.
Introduction
Angiogenesis is a key process required in numerous physiologic and pathophysiologic processes, including wound healing, peripheral vascular disease, myocardial ischemia, and neoplasia. It is a complex, multistep process in which new microvessels develop from endothelial cells of preexisting blood vessels. Endothelial cells must respond to a number of extracellular signals during vascular assembly.1
Survivin is an inhibitor of apoptosis protein (IAP) involved in blocking the pathway that leads to programmed cell death induced by a variety of death stimuli.2,3 In addition to its antiapoptotic function, Survivin also acts as a chromosomal passenger protein (CPP), ensuring the proper alignment of chromosomes during mitosis and allowing for equal and complete cell division.4-6 Transcription from the SURVIVIN gene locus yields at least 5 described alternative splice variants that, in addition to Survivin, have been designated Survivin 2B,7 Survivin ΔEx3,7 Survivin 3B,8 and Survivin 2α.9 It has been demonstrated that some of these isoforms have subcellular localization patterns that could be associated with unique functional properties.9-11 The localization of Survivin 3B has not yet been described. The overwhelming majority of published reports on Survivin focus on its involvement in cancer, or its relevance in angiogenesis in the context of cancer. Fewer reports have examined the function of Survivin in normal development.12-15 In addition, most studies have not assessed the function of the individual Survivin splice variants in either normal cells or in cancer. Preliminary reports suggest that Survivin and Survivin ΔEx3 have antiapoptotic properties,7 Survivin 2B has proapoptotic properties,7 and Survivin 2α may act as a natural dominant negative of Survivin's antiapoptotic activities.9 No function has yet been ascribed to Survivin 3B. The potential role of the Survivin variants in angiogenesis is unknown.
Here, we demonstrate that Survivin ΔEx3 is highly expressed in vascular endothelial cells; that it is necessary for activation of the small GTPase Rac1 during endothelial tube formation; and that it is required for in vivo endothelial cell invasion. Thus, we provide substantial evidence that Survivin ΔEx3 plays a prominent role in the important physiologic and pathophysiologic process of angiogenesis.
Materials and methods
Patients
Tissue samples obtained from pediatric patients with a diagnosis of medulloblastoma, ependymoma, choroid plexus tumors, or various angiomas were selected from the Columbus Children's Hospital (CCH) pathology database based on availability of paraffin-embedded tumor material. All patients were treated at CCH between 1992 and 2005. Approval was obtained from the Columbus Children's Research Institute institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki.
Tissue microarray (TMA)
Formalin-fixed, paraffin-embedded tissue samples were obtained from nonneoplastic tissue, from complete autopsy cases performed at CCH. Regions within each tissue were selected by a pathologist (D.R.B.) to ensure the use of viable and well-preserved tissue. Tissues included cerebral cortex, pituitary, kidney, thyroid, adrenal, thymus, bone marrow, skin, cortical bone, colon, stomach, aortic region, liver, small intestine, lymph node, peripheral nerve, esophagus, lung, spleen, larynx, duodenum, urinary bladder, heart, ovary, skeletal muscle, tongue, and testes. Cores of tissue were punched from a “donor” block (punch diameters = 2 mm) and arrayed into a “recipient” paraffin block. The TMA was produced using a Manual Tissue Arrayer-1 (MTA-1; Beecher Instruments, Sun Prairie, WI).
Immunohistochemistry
Paraffin-embedded tissue samples (medulloblastoma, ependymoma, or angiomas) were sectioned at a thickness of 4 to 5 μm on a Leica microtome (Leica, Bannockburn, IL) and placed on charged slides. Heat antigen retrieval was performed on a pressure cooker with Antigen Retrieval Citra (Biogenex, San Ramon, CA) according to the manufacturer's instructions. Blocking was performed with Powerblock (Biogenex) for 10 to 15 minutes. The samples were incubated at 4°C overnight with either the antiSurvivin (1:200; Novus Biologicals, Littleton, CO), anti–Survivin ΔEx3 antibody (1:400; Abcam, Cambridge, MA), or anti-PP1 antibody (PP1 [E-9], sc-7482, 1:25; Santa Cruz Biotechnology, Santa Cruz, CA). Immunohistochemistry was performed using the avidin-biotin method with reagents from Biocare Medical (Concord, CA). Color development was achieved by applying peroxidase substrate chromagen AEC (Dako Cytomation, Carpinteria, CA). Slides were counterstained with Mayer hematoxylin and mounted in aqueous mounting solution. Standard operating procedures were used for hematoxylin and eosin staining, and staining with antibodies to CD31 and CD34 using a Ventana autostainer (Ventana, Tucson, AZ). Images were acquired with a Leica microscope at ×40, ×100, or ×400 magnification (5×/0.15, 10×/0.3, and 40×/0.75 objective lenses), and processed with Adobe Photoshop (Adobe Systems, San Jose, CA).
Western blotting
Protein extracts from an ependymoma, normal brain ependyma, and HeLa cells transfected with myc-tagged Survivin ΔEx3 were made, as previously described.16 One hundred and fifty micrograms of protein extract from ependymoma and normal brain ependyma and 10 μg of protein extract from transfected HeLa cells were separated on an 18% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto a nitrocellulose membrane. The membrane was immunoprobed as previously described.11 To demonstrate Survivin up-regulation by VEGF treatment in human umbilical vein endothelial cells (HUVECs), protein lysates were prepared from cells treated with VEGF or mock treated. Fifty micrograms of protein extract was separated in a 4% to 20% Precise Tris-HEPES-SDS Pre-cast gel (Pierce Biotechnology, Rockford, IL) and transferred to PVDF membrane (Bio-Rad, Hercules, CA). The membrane was immunoprobed as previously described.
Cells
Human umbilical vein endothelial cells (HUVECs), kindly provided by Dr Veela Mehta, were cultured in Medium 200 (including growth supplements) (Cascade Biologicals, Portland, OR) at 37°C, 5% CO2.
Immunofluorescence
Cells, grown on glass coverslips, were washed in PBS and fixed in freshly prepared 4% (wt/vol) paraformaldehyde for 30 minutes at room temperature. The cells were then permeabilized with 0.2% Triton-X100 for 30 minutes before being processed for indirect immunofluorescence using anti–Survivin ΔEx3 polyclonal antibody (Abcam), followed by anti–rabbit biotin and NeutrAvidin Texas Red or NeutrAvidin fluorescein (Molecular Probes, Carlsbad, CA). Other markers included Golgi marker antibody GM130 (BD Pharmingen, San Diego, CA), Mitotracker CMXRos (Molecular Probes), or anti-PP1 antibody (Santa Cruz Biotechnology). All incubations were performed at room temperature for 1 hour in PBS containing 1% BSA. After each incubation, the cells were washed in PBS. Coverslips were mounted onto slides using VectaShield (Vector Labs, Burlingame, CA). Cells were visualized with a Zeiss LSM510-META confocal laser-scanning microscope (Zeiss, Thornwood, NY), using a C-Apochromat × 63/1.2 corr water immersion lens, and processed using the acquisition software provided with the microscope.
Plasmids and transfections
The construction of GFP–Survivin ΔEx3 and myc-tagged Survivin ΔEx3 have been previously described.10 Survivin ΔEx3–specific shRNA was constructed by cloning the targeting sequence consisting of sequence at the join of exons 2 and exon 4 of the SURVIVIN gene locus into pSUPER.17 SH3 shRNA was constructed by cloning the targeting sequence consisting of a 19-nt region within exon 3 (which is absent from Survivin ΔEx3 mRNA). The construction and cloning of the other Survivin shRNAs used in this study have been previously reported.18 Transfections were performed using Effectene transfection reagent (Qiagen, Valencia, CA) at a 1:10 DNA/Effectene ratio, according to the manufacturer's instructions.
PP1 siRNA transfection of HUVECs
Pan-PP1 siRNA was obtained from Santa Cruz Biotechnology and consists of a pool of 6 20- to 25-nucleotide siRNAs designed to knock down PP1 gene expression. For siRNA transfection, 60 pmol pan-PP1 siRNA diluted in siRNA transfection medium (Santa Cruz Biotechnology) was added to 6 μL siRNA transfection reagent (Santa Cruz Biotechnology) diluted in an equal volume of siRNA transfection medium. The mixture was allowed to incubate at room temperature for 30 minutes, and further diluted in siRNA transfection medium before adding to HUVECs, in a 6-well plate. After 8 hours of incubation, the medium was supplemented with full growth medium for a further 48 hours of incubation, before analysis by immunofluorescence.
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction (PCR) was performed as previously described using Taqman probes using an ABI 7700 (Applied Biosystems, Foster City, CA). Each PCR reaction was performed in triplicate, and the average result was used for calculations. Ribosomal RNA was used as an internal control for each sample. Results from each sample were compared with control-treated RNA as a calibrator, using the 2−ΔΔCT calculation method (ABI).
Antibody treatment of cells
HUVE cells were preincubated with 10 μg/mL rabbit polyclonal anti–Survivin ΔEx3 or normal rabbit IgG (Santa Cruz Biotechnology) for 15 minutes at 37°C. To block VEGF receptors, anti-VEGFR1 (Flt-1) or anti-VEGFR2 (Flk1/KDR) monoclonal antibodies (R&D Systems, Minneapolis, MN) were preincubated with HUVECs at 50 ng/mL for 30 minutes at 37°C. The antibodies were maintained at 10 μg/mL and 50 ng/mL, respectively, for any subsequent experimental steps.
Apoptosis and proliferation assays
Apoptosis was evaluated by fluorescence-activated cell sorting (FACS) analysis using fluorescein isothiocyanate–conjugated annexin V assay (Roche Diagnostics, Indianapolis, IN). For proliferation, cells were pulse labeled with 10 μM 5-bromo 2′deoxyuridine (Sigma-Aldrich, St Louis, MO) for 1 hour at 37°C. The cells were fixed in ice-cold 70% ethanol for 20 minutes, followed by a denaturation step in 2 N HCl for 20 minutes and a neutralization step with 0.1 M sodium borate for 2 minutes, all at room temperature. The sample was then subjected to immunostaining with an anti-BrdU monoclonal antibody, or an isotype-matched IgG, conjugated with fluorescein isothiocyanate (eBioscience, San Diego, CA) and analyzed by flow cytometry.
MTT assay
MTT assay was performed to assess cell proliferation/viability of HUVECs. We used the TACS-MTT assay according to the manufacturer's description (R&D Systems), in 96-well plates. Experiments were performed in triplicate. Mean and standard deviation for 3 independent experiments are shown.
Caspase 3 assay
Active caspase 3 was assayed with the Caspase 3/7-GLO assay (Promega, Madison, WI) according to the manufacturer's instructions, in 96-well plates. Experiments were performed in triplicate. Means and standard deviation for 3 independent experiments are shown. For comparison of caspase activity, we designated the level of caspase 3 activity in negative control cells as 100% and expressed the level of caspase 3 activity in the experimental sample relative to the negative control.
In vitro tube formation assay
Reduced growth factor basement membrane extract (Trevigen, Gaithersburg, MD) was plated evenly in a 24-well plate, and incubated at 37°C for 30 minutes before adding HUVECs (5 × 104 cells/well). Tube formation was studied over 48 hours and photographs of representative × 100 fields were taken at 0, 4, 16, 24, and 48 hours. Endothelial cells were quantified by counting branches from each endothelial cell and the length of the tube formed using SPOT Advanced software (Diagnostic Instruments, Sterling Heights, MI) at 4 and 16 hours. Where indicated, cells were preincubated with anti–Survivin ΔEx3 polyclonal antibody or control IgG for 15 minutes at 37°C before plating or anti-VEGFR1 (Flt-1) or anti-VEGFR2 (Flk1/KDR) monoclonal antibodies for 30 minutes at 37°C. Supplementation with VEGF was achieved by addition of soluble VEGF (Biosource International, Carlsbad, CA) to the culture medium at a concentration of 20 ng/mL for 24 hours and was present throughout the experimental period at the same concentration. Rac1 inhibition was achieved by addition of In Solution Rac1 inhibitor (Calbiochem, San Diego, CA) to the culture medium at a final concentration of 100 μM.
Statistical analysis
Statistical analysis was performed using 2-tailed Student t test. A P value of .05 or less was considered significant.
Directed in vivo angiogenesis assay (DIVAA)
The DIVAA assay was obtained from Trevigen and performed, with modifications, as originally described.19 Briefly, sterile 0.15 cm × 1 cm–long semiclosed surgical silicone tubing (angioreactors) was filled with 18 μL high-concentration basement membrane extract with or without angiogenic factors (Trevigen), containing 50 μg/mL anti–Survivin ΔEx3 or rabbit IgG control, at 4°C. The angioreactors were then inverted and incubated at 37°C for 1 hour to allow gel formation. The angioreactors were then implanted subcutaneously into the dorsal flank of 6- to 8-week-old athymic nude female mice.
Endothelial cell invasion assay
FITC-labeled Griffonia lectin (FITC-lectin; Trevigen), an endothelial cell selective reagent, was used to quantify invading endothelial cells into the angioreactor. Briefly, the DIVAA angioreactor was collected from the mouse and the basement membrane extract mix recovered and digested with dispase (Trevigen) for 1 hour at 37°C. Cell pellets and insoluble fractions were collected by centrifugation at 5000g for 2 minutes at room temperature, and washed 3 times with PBS. After the last wash, the cell pellets were resuspended in 25 μg/mL FITC-lectin and incubated at 4°C overnight. The stained pellets were centrifuged and washed with cold PBS, and the final pellet was resuspended and relative fluorescence determined in a spectrofluorometer (Applied Biosystems). The mean relative fluorescence ± SD for 3 replicate assays was determined.
Rac1 pull-down assay
To evaluate Rac1 activation during HUVEC tube formation on basement membrane extract, cell extracts were prepared at 4 hours after plating. Cells were washed twice with PBS and lysed at 4°C in MLB (25 mM HEPES, pH 7.5, 150 mM NaCl, 1% Igepal CA-630, 10 mM MgCl2, 1 mM EDTA, and 2% glycerol). The resultant protein samples were precleared with GST-agarose (Cell Signaling, Charlottesville, VA) for 10 minutes at 4°C. The samples were centrifuged at 16 000g for 2 minutes at 4°C and the supernatant was removed. Active Rac1 was immunoprecipitated from the experimental samples by adding PAK1-PBD agarose (Cell Signaling) at 4°C for 1 hour. The bound fraction was separated in an SDS–polyacrylamide gel, and the presence of Rac1 in the bound fraction was detected by Western blotting using primary anti-Rac1 antibody (clone 23A8; Cell Signaling). The amount of activated Rac1 was normalized against total Rac1 in the corresponding lysate, as previously described.20 Integrated density values of the bands were measured using a Syngene Chemi Genius2 BioImaging system and analyzed with Syngene GeneTools software (Syngene, Frederick, MD).
Results
(1) Survivin ΔEx3 is expressed in vascular endothelium
The known dual function of Survivin as an IAP and a mitotic regulator combined with our previous findings that ectopic expression of Survivin ΔEx3 can direct Survivin to unique intracellular compartments11 prompted us to examine the endogenous expression pattern of Survivin ΔEx3.
To this end, we evaluated the localization of Survivin ΔEx3 in 30 formalin-fixed, paraffin-embedded pediatric brain tumor tissues using a recently available, commercial, polyclonal antibody raised against the peptide SWLPWIEASGRS, unique to Survivin ΔEx3. In this analysis, Survivin ΔEx3 localized to areas of the tissue that were distinct from that of the Survivin protein. While Survivin itself was expressed in more than 50% of tumor cells as previously shown,16 Survivin ΔEx3 was not detected within the tumor cells themselves but was exclusively located within the vascular endothelial cells of the tumor (Figure 1A-B). Although previous studies suggested that Survivin ΔEx3 is also expressed within tumor cells,16 as determined by quantitative PCR analysis of RNA isolated from a tumor cell line or from a human tumor biopsy, the protein analyses performed here did not reveal prominent expression within tumor cells. One potential explanation for these differences could be that the protein levels within the tumor cells were below the threshold of detection by the immunohistochemical assay used in the current study. An alternative explanation could be that the PCR analyses used in the former studies were sensitive enough to detect the expression of Survivin ΔEx3 within the vascular endothelium of the human tumor tissue samples. Human tumors are highly vascular and thus whole tumor biopsies would be expected to contain a significant amount of vascular cells. To substantiate the current observations showing expression of Survivin ΔEx3 in vascular endothelium, we expanded our immunohistochemical evaluation to include a wide range of tissues.
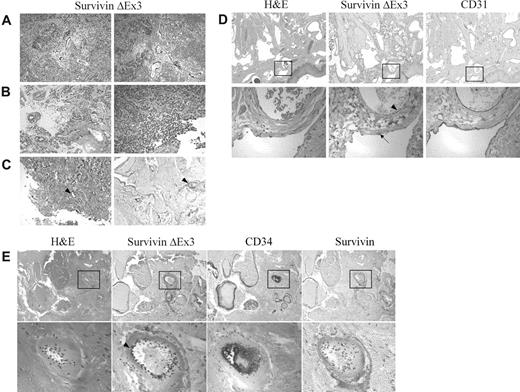
Immunolocalization of Survivin ΔEx3 in brain tumors and angiomas. Formalin-fixed paraffin-embedded tissues were immunostained with antibodies against Survivin, Survivin ΔEx3, CD31, or CD34. Survivin ΔEx3 localized to the vascular endothelium of the brain tumor (A-B) and angioma tissues (C-E arrowhead). Vascular endothelial cells staining positive for Survivin ΔEx3 were also positive for CD31 (D) and CD34 (E). Survivin ΔEx3 was not visualized within the lymphatic endothelium (D, arrow). Survivin was not detected in angioma tissues (E). The tissues represented include (A) medulloblastoma, (B) ependymoma, (C) capillary venous angioma, (D) lymphangioma, and (E) pyogenic granuloma.
Immunolocalization of Survivin ΔEx3 in brain tumors and angiomas. Formalin-fixed paraffin-embedded tissues were immunostained with antibodies against Survivin, Survivin ΔEx3, CD31, or CD34. Survivin ΔEx3 localized to the vascular endothelium of the brain tumor (A-B) and angioma tissues (C-E arrowhead). Vascular endothelial cells staining positive for Survivin ΔEx3 were also positive for CD31 (D) and CD34 (E). Survivin ΔEx3 was not visualized within the lymphatic endothelium (D, arrow). Survivin was not detected in angioma tissues (E). The tissues represented include (A) medulloblastoma, (B) ependymoma, (C) capillary venous angioma, (D) lymphangioma, and (E) pyogenic granuloma.
Different types of angiomas, including capillary-venous hemangioma, cavernous (venous) hemangioma, lymphangioma, arteriovenous malformation, and cystic hygroma, were evaluated for both Survivin and Survivin ΔEx3 expression by immunohistochemical staining. Importantly, none of these tissues expressed Survivin at detectable levels (Figure 1E). By contrast, Survivin ΔEx3 was prominently expressed in the vascular endothelium of most vessels examined including capillaries and venous and arterial blood vessels within all subtypes of angiomas (Figure 1C-E arrowhead). The endothelial cell markers CD31 (Figure 1D) and CD34 (Figure 1E) also localized to the same regions of the angiomas as Survivin ΔEx3, which included the vascular endothelial cells of venous and arterial blood vessels, as well as smaller capillaries. In contrast to vascular endothelial cells, lymphatic endothelial cells did not express Survivin ΔEx3 at detectable levels (Figure 1D arrow). This demonstrates a clear and specific association of Survivin ΔEx3 with the vascular endothelium, and supports a role for Survivin ΔEx3 in these cells.
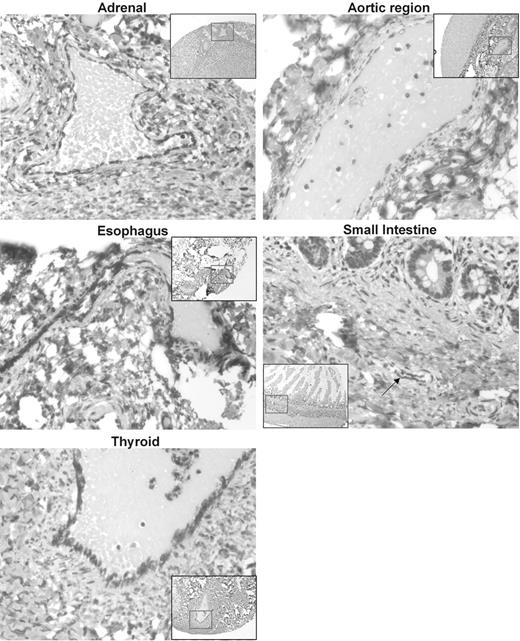
To further characterize the expression of Survivin ΔEx3 within vascular endothelial cells, we constructed a tissue microarray from a wide spectrum of normal human tissue containing normal vasculature. Survivin ΔEx3 was expressed in the cytoplasm of vascular endothelial cells present in areas of all tissues examined, as shown in Figure 2.
Immunolocalization of Survivin ΔEx3 in normal tissues. A tissue microarray (TMA) was constructed with normal healthy tissues from a variety of organs. TMA slides were immunostained with an antibody against Survivin ΔEx3. Survivin ΔEx3 localized to the cytoplasm of vascular endothelial cells of blood vessels within all tissues examined including the adrenal cortex, aortic region, esophagus, thyroid, and small intestine, shown here. Arrow indicates blood vessel within small intestine.
Immunolocalization of Survivin ΔEx3 in normal tissues. A tissue microarray (TMA) was constructed with normal healthy tissues from a variety of organs. TMA slides were immunostained with an antibody against Survivin ΔEx3. Survivin ΔEx3 localized to the cytoplasm of vascular endothelial cells of blood vessels within all tissues examined including the adrenal cortex, aortic region, esophagus, thyroid, and small intestine, shown here. Arrow indicates blood vessel within small intestine.
To validate the specificity of the Survivin ΔEx3 antibody used in the immunohistochemistry experiments, we performed Western blotting using protein extracts isolated from human ependymoma tumors, normal brain ependyma, and HeLa cells transfected with a myc-tagged Survivin ΔEx3 construct. A single protein band, migrating at the expected molecular weights of endogenous Survivin ΔEx3 or myc-tagged Survivin ΔEx3, was visualized exclusively in the lanes containing the human tumor samples and the transfected HeLa cells, but not the normal brain ependyma (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). No other bands were detected on these blots. Due to the high degree of similarity of the peptide used to raise the Survivin ΔEx3 antibody and a portion of the human protein phosphatase 1 protein (PP1) (Figure S2), we performed further validation of the specificity of the Survivin ΔEx3 antibody. Immunohistochemical analysis of angioma samples using a PP1 antibody or the Survivin ΔEx3 antibody demonstrated that while Survivin ΔEx3 exclusively stains the cytoplasmic compartment of vascular endothelial cells within the angiomas, the PP1 antibody stains within the nucleus and the cytoplasm of these cells, as well as in these regions of other cells within the tissue (Figure S3). Additionally, HUVE cells were treated with either a pan-PP1 siRNA or Survivin ΔEx3–specific shRNAs. These cells were then stained with either an anti-PP1 antibody or the anti–Survivin ΔEx3 antibody to demonstrate that Survivin ΔEx3 expression is maintained, following treatment with PP1 siRNA (Figure S4). By contrast, treatment with the Survivin ΔEx3 shRNA resulted in loss of staining by the Survivin ΔEx3 antibody, with no appreciable difference in staining by the PP1 antibody (Figure S4). These data provide substantial evidence that the Survivin ΔEx3 antibody is specific for Survivin ΔEx3.
(2) Subcellular localization of Survivin ΔEx3
The high levels of expression of Survivin ΔEx3 within the cytoplasm of vascular endothelial cells prompted us to further evaluate its subcellular localization pattern. We previously reported the expression of tagged Survivin ΔEx3 in both the nucleus and mitochondria of malignant cells11 ; Song et al later also described an additional nucleolar localization for ectopically expressed Survivin ΔEx3 in cervical cancer.21 The endogenous localization of Survivin ΔEx3 has not previously been described, largely due to the lack of specific antibodies to this protein. We investigated the localization of Survivin ΔEx3 in cultured HUVE cells by confocal microscopy using a 3-step immunocytochemistry procedure (“Materials and methods”). In contrast to Survivin, Survivin ΔEx3 localized diffusely to the cytoplasm during mitosis and was not associated with the DNA at the kinetochore, or present in the midbody/cleavage furrow at cytokinesis (Figure 3A). In quiescent cells, Survivin ΔEx3 localized within the cytoplasm, in large punctate cytoplasmic structures that colocalized in part with a Golgi apparatus marker (GM130), but not with the mitochondrial marker Mitotracker (Figure 3B). In a smaller proportion of cells, Survivin ΔEx3 was also observed in the region of the plasma membrane and the actin cytoskeleton. This staining was at the cell periphery, in the spreading edge, in structures that resemble membrane ruffles and lamellipodia (Figure 3B). This finding suggests a potential function for Survivin ΔEx3 in endothelial cell spreading and migration.
Subcellular localization of Survivin ΔEx3 in HUVE cells. HUVE cells were seeded on glass coverslips and immunostained with antibodies against Survivin ΔEx3, alpha tubulin, or GM130, or the mitochondrial-specific dye Mitotracker, as described in “Materials and methods.” (A) In dividing cells, Survivin ΔEx3 (red) was localized in the cytoplasm, and did not colocalize with alpha-tubulin (green) at any stage of mitosis (note absence from the midbody/cleavage furrow [arrows]). (B) In resting cells, Survivin ΔEx3 (green) partially colocalized with the Golgi apparatus marker GM130 (red; colocalization resulting in yellow pixels, arrows), but did not colocalize with Mitotracker (red). In a number of cells, Survivin ΔEx3 also localized to the actin cytoskeleton (green, arrow) as well as to the spreading edge of the cell, resembling a membrane ruffle (green, arrow). Nuclear localization was also observed in a minority of cells (arrow; nucleus was pseudocolored red for better contrast, and in all other images the nucleus is depicted in blue from Hoechst staining). Scale bar represents 20 μm.
Subcellular localization of Survivin ΔEx3 in HUVE cells. HUVE cells were seeded on glass coverslips and immunostained with antibodies against Survivin ΔEx3, alpha tubulin, or GM130, or the mitochondrial-specific dye Mitotracker, as described in “Materials and methods.” (A) In dividing cells, Survivin ΔEx3 (red) was localized in the cytoplasm, and did not colocalize with alpha-tubulin (green) at any stage of mitosis (note absence from the midbody/cleavage furrow [arrows]). (B) In resting cells, Survivin ΔEx3 (green) partially colocalized with the Golgi apparatus marker GM130 (red; colocalization resulting in yellow pixels, arrows), but did not colocalize with Mitotracker (red). In a number of cells, Survivin ΔEx3 also localized to the actin cytoskeleton (green, arrow) as well as to the spreading edge of the cell, resembling a membrane ruffle (green, arrow). Nuclear localization was also observed in a minority of cells (arrow; nucleus was pseudocolored red for better contrast, and in all other images the nucleus is depicted in blue from Hoechst staining). Scale bar represents 20 μm.
(3) Disruption of Survivin ΔEx3 in vascular endothelial cells does not induce apoptosis or inhibit proliferation
Multiple investigators have demonstrated a role for Survivin in inhibiting apoptosis in vascular endothelial cells.22-25 Down-regulation of Survivin by antisense oligonucleotides and siRNAs results in an increase in apoptosis in these cells.26 In this work, we examined the effects of Survivin ΔEx3 on apoptosis and proliferation in vascular endothelial cells by treatment with a Survivin ΔEx3-specific shRNA, targeting the exon 2–exon 4 boundary, which does not target any other isoform except Survivin ΔEx3 (Figure 5A). Treatment with this shRNA resulted in a 30% to 50% reduction of Survivin ΔEx3 transcripts, as assayed by Taqman real-time reverse-transcription (RT)–PCR (not shown), and an appreciable reduction in Survivin ΔEx3 protein (Figure S5). As one control for these experiments, we also treated cells with a combination of shRNAs that target all known Survivin splice variants, with the exception of Survivin 2α. The levels of transcript reduction observed after treatment with these shRNAs was on average 60% for Survivin, Survivin 2B, and Survivin ΔEx3 (not shown). Apoptosis was evaluated using annexin V and proliferation was assessed by pulse labeling with 5-bromo 2′deoxyuridine (BrdU).
A small (mean = 4%), statistically insignificant increase in apoptosis was observed when cells were treated with the Survivin ΔEx3–specific shRNA, compared with a scrambled shRNA control, suggesting Survivin ΔEx3 does not play a significant role in protecting endothelial cells from apoptosis (Figure 4A). From this experiment alone, we cannot exclude the possibility that the observed minimal effect on apoptosis could be related to a compensatory up-regulation of Survivin, as Survivin has been previously shown to protect endothelial cells from apoptosis.24 To provide further evidence for the negligible effect of Survivin ΔEx3 shRNA on inducing cell death, we performed assays for active caspase 3 using the caspase 3/7 GLO assay. We observed a mean 5.4% increase in caspase 3 activity in cells treated with Survivin ΔEx3 shRNA relative to scrambled control. This small increase correlates with the percent increase in apoptosis observed by the annexin V assay and was not statistically significant (P = .31) (Figure 4B). No significant changes in proliferation were observed, suggesting Survivin ΔEx3 does not play a significant role in regulating cell growth (Figure 4C). Treatment of HUVE cells with a combination of shRNAs that target all Survivin splice variants, with the exception of Survivin 2α, resulted in a statistically significant increase in apoptosis (mean = 32.5%), as has been previously demonstrated26 (Figure 4A).
Disruption of Survivin ΔEx3 does not induce apoptosis or affect proliferation in HUVE cells. HUVE cells were cotransfected with scrambled shRNA, Survivin ΔEx3 shRNA, or combination shRNA along with pHcRed. Red fluorescent cells were separated by FACS at 48 hours. (A) Cells were assayed for apoptosis by staining with annexin V. (B) Cells were grown in a 96-well plate and assayed for active caspase 3 with caspase 3/7 GLO. The y-axis represents the percentage of caspase 3 activity, relative to control. (C) Cells were pulsed with 10 μM BrdU for 1 hour, and staining was performed as described in “Materials and methods,” using an anti–BrdU-FITC or an IgG isotype-FITC antibody. Both apoptosis and proliferation were analyzed by flow cytometry. Experiments were performed in triplicate, and data are shown for a representative sample set. Error bars are the standard error of the mean of three replicates per sample.
Disruption of Survivin ΔEx3 does not induce apoptosis or affect proliferation in HUVE cells. HUVE cells were cotransfected with scrambled shRNA, Survivin ΔEx3 shRNA, or combination shRNA along with pHcRed. Red fluorescent cells were separated by FACS at 48 hours. (A) Cells were assayed for apoptosis by staining with annexin V. (B) Cells were grown in a 96-well plate and assayed for active caspase 3 with caspase 3/7 GLO. The y-axis represents the percentage of caspase 3 activity, relative to control. (C) Cells were pulsed with 10 μM BrdU for 1 hour, and staining was performed as described in “Materials and methods,” using an anti–BrdU-FITC or an IgG isotype-FITC antibody. Both apoptosis and proliferation were analyzed by flow cytometry. Experiments were performed in triplicate, and data are shown for a representative sample set. Error bars are the standard error of the mean of three replicates per sample.
(4) Interference with Survivin ΔEx3 inhibits vascular tube formation in vitro
Due to its prominent expression in the vascular endothelium in both normal and neoplastic tissues and its localization to the plasma membrane and actin cytoskeleton, we postulated that Survivin ΔEx3 could play a functional role in the growth of new vessels. To test this hypothesis, we used 2 different methodologies to disrupt Survivin ΔEx3 activity in an in vitro tube formation assay. These included down-regulating Survivin ΔEx3 mRNA with shRNA and inhibiting Survivin ΔEx3 protein with the anti–Survivin ΔEx3 antibody.
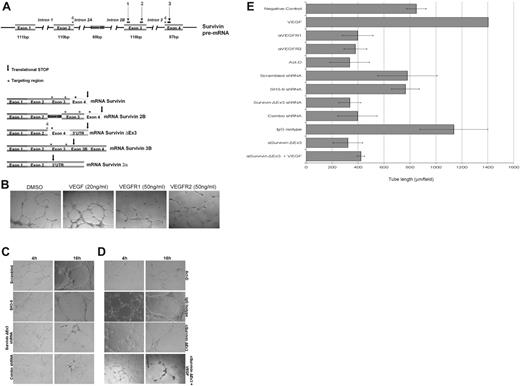
To down-regulate transcription, we used the aforementioned Survivin ΔEx3–specific shRNA. A scrambled shRNA and a cocktail of 3 shRNAs targeting all Survivin isoforms, except Survivin 2α, were used as controls (Figure 5A). HUVECs were transfected with the shRNA plasmids in combination with a plasmid encoding eGFP. The cells were sterile sorted by FACS to isolate the population of cells that was successfully transfected, based on eGFP fluorescence. This enriched population was plated on basement membrane extract (BME) with complete growth factor medium to support in vitro tube formation. As shown in Figure 5C, treatment with the shRNA cocktail or Survivin ΔEx3–specific shRNA resulted in an impaired ability to form tubes on BME compared with treatment with either a scrambled shRNA or a Survivin shRNA not targeting Survivin ΔEx3 (SH3-9). The difference was most marked between 4 and 16 hours after plating. At this point, a tubular network of interconnecting branches was well established in all control samples; however, few contacts between cells, with shorter protrusions and few branches, were observed in the cells treated with the shRNA cocktail and Survivin ΔEx3–specific shRNAs.
Disruption of Survivin ΔEx3 inhibits vascular tube formation in vitro. (A) Schematic representation of the Survivin gene structure and shRNA targeting regions. Colored boxes represent exons from the SURVIVIN gene. The 5 identified splice variants are represented, and the targeting region is shown as an asterisk above the area targeted. The arrow represents the location of the translational stop in each of the mRNAs. Numbers 1 to 3 consist of the combination of shRNAs previously described,12 whereas number 4 represents the Survivin ΔEx3–specific shRNA. (B) HUVE cells were treated with VEGFR1 or VEGFR2 blocking antibodies (50 ng/mL) or supplemented with VEGF (20 ng/mL); (C) HUVECs were transfected with scrambled shRNA, non–Survivin ΔEx3 shRNA (SH3-9), Survivin ΔEx3 shRNA, or a combination of 3 Survivin shRNAs; or (D) HUVECs were treated with Survivin ΔEx3 antibody (10 μg/mL), Survivin ΔEx3 antibody (10 μg/mL) plus VEGF (20 ng/mL), isotype control antibody (10 μg/mL), or actinomycin-D (7.5 μg/mL) and plated on growth factor–reduced BME in 96-well plates. For all conditions, tube formation was assayed at 4 and 16 hours and quantitative data were collected at the 16-hour time point by measuring the length of the tubules formed under an inverted light microscope at ×100 magnification (40×/0.75 objective lens). Quantitative data shown in panel E are mean ± SE of 5 replicates per sample.
Disruption of Survivin ΔEx3 inhibits vascular tube formation in vitro. (A) Schematic representation of the Survivin gene structure and shRNA targeting regions. Colored boxes represent exons from the SURVIVIN gene. The 5 identified splice variants are represented, and the targeting region is shown as an asterisk above the area targeted. The arrow represents the location of the translational stop in each of the mRNAs. Numbers 1 to 3 consist of the combination of shRNAs previously described,12 whereas number 4 represents the Survivin ΔEx3–specific shRNA. (B) HUVE cells were treated with VEGFR1 or VEGFR2 blocking antibodies (50 ng/mL) or supplemented with VEGF (20 ng/mL); (C) HUVECs were transfected with scrambled shRNA, non–Survivin ΔEx3 shRNA (SH3-9), Survivin ΔEx3 shRNA, or a combination of 3 Survivin shRNAs; or (D) HUVECs were treated with Survivin ΔEx3 antibody (10 μg/mL), Survivin ΔEx3 antibody (10 μg/mL) plus VEGF (20 ng/mL), isotype control antibody (10 μg/mL), or actinomycin-D (7.5 μg/mL) and plated on growth factor–reduced BME in 96-well plates. For all conditions, tube formation was assayed at 4 and 16 hours and quantitative data were collected at the 16-hour time point by measuring the length of the tubules formed under an inverted light microscope at ×100 magnification (40×/0.75 objective lens). Quantitative data shown in panel E are mean ± SE of 5 replicates per sample.
To disrupt Survivin ΔEx3 protein, HUVE cells were pretreated with a Survivin ΔEx3 antibody or rabbit IgG control before plating on BME for in vitro tube formation. As additional controls, we treated cells with soluble VEGF, anti-VEGFR1,27,28 or anti-VEGFR228 antibodies or with actinomycin-D,29 known inducers and inhibitor of in vitro angiogenesis, respectively. Tube formation was quantified by counting the number of branches and tube length from 5 representative fields per replicate, in a double-blinded fashion, at the 16-hour time point. Whereas cells treated with soluble VEGF showed an increase in tube formation (Figure 5B), cells treated with Survivin ΔEx3 antibody, actinomycin-D (Figure 5D), or VEGFR1 or VEGFR2 antibodies (Figure 5B) showed similar abnormalities to those observed following Survivin ΔEx3 shRNA–specific treatments (Figure 5C); inhibition of tube formation was by approximately half, as demonstrated by quantification of tube length (Figure 5E). Since VEGF transcriptionally up-regulates Survivin (as previously shown30 and Figure S5), and, therefore, the other splice forms regulated by this promoter, including Survivin ΔEx3, we also evaluated the effects of VEGF in cells treated with Survivin ΔEx3 antibody. Tube length was only marginally increased (Figure 5D-E), suggesting that the addition of VEGF cannot fully compensate for inhibition of Survivin ΔEx3. Statistical comparisons of tube length for the various conditions are shown in Table S1. Taken together, these data suggest that Survivin ΔEx3 is required for the early steps in endothelial tube formation on BME.
(5) Survivin ΔEx3 regulates Rac1 activation during endothelial tube formation
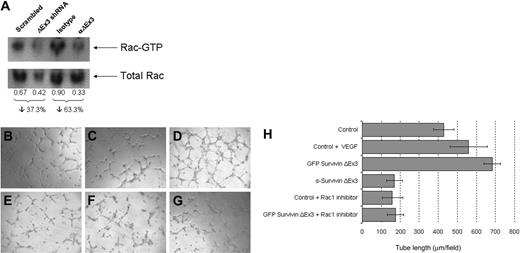
Rac1 is a well-characterized small GTPase of the Rho subfamily that is known to regulate endothelial morphogenesis and capillary assembly.31 In an attempt to delineate the mechanism by which interference with Survivin ΔEx3 impairs endothelial tube formation in vitro, we investigated whether Survivin ΔEx3 is required for the activation of Rac1 during endothelial tube formation on BME. Rac1 activity in HUVE cells seeded on BME was measured by GST-PBD pull-down assay.32 Down-regulation of Survivin ΔEx3 was achieved by treatment with Survivin ΔEx3–specific shRNA, and Survivin ΔEx3 protein inhibition was achieved by using a Survivin ΔEx3–specific antibody. We assessed Rac1 activation at 4 hours after plating on BME, a time point at which we observed obvious defects in tube formation using both methodologies. In these experiments, Rac1 activation was decreased by 37% in shRNA-treated cells, compared with cells treated with a scrambled shRNA control. A decrease of 63% was observed in cells treated with a Survivin ΔEx3 antibody, compared with an IgG control (Figure 6A). These observations temporally coincided with the marked reduction in tube formation when treated cells were plated on BME for 4 hours, suggesting Survivin ΔEx3 has an effect on Rac1 activation during angiogenesis.
Survivin ΔEx3 is involved in Rac1 activation during endothelial tube formation. (A) HUVECs plated onto BME were analyzed for Rac1 activation at the tube formation stage (4 hours) by GST-PBD pull-down assays. The amount of active Rac1 was normalized against total Rac1 in the corresponding lysate, and the integrated density values of the bands were measured using Syngene GeneTools analysis software. (B-H) HUVE cells were plated on growth factor–reduced BME in 96-well plates supplemented with VEGF (20 ng/mL) or Rac1 inhibitor (100 μM), untransfected, or after transfection with eGFP Survivin ΔEx3. (B) Control; (C) control + VEGF; (D) GFP Survivin ΔEx3; (E) Survivin ΔEx3 antibody (10 μg/mL); (F) control + Rac1 inhibitor; and (G) GFP Survivin ΔEx3 + Rac1 inhibitor. Tube formation was assayed at 4 hours after plating on BME, and quantitative data were collected by measuring the length of the tubules formed under an inverted light microscope at ×100 magnification (40×/0.75 objective lens). Quantitative data shown in panel H are mean ± SE of 5 replicates per sample.
Survivin ΔEx3 is involved in Rac1 activation during endothelial tube formation. (A) HUVECs plated onto BME were analyzed for Rac1 activation at the tube formation stage (4 hours) by GST-PBD pull-down assays. The amount of active Rac1 was normalized against total Rac1 in the corresponding lysate, and the integrated density values of the bands were measured using Syngene GeneTools analysis software. (B-H) HUVE cells were plated on growth factor–reduced BME in 96-well plates supplemented with VEGF (20 ng/mL) or Rac1 inhibitor (100 μM), untransfected, or after transfection with eGFP Survivin ΔEx3. (B) Control; (C) control + VEGF; (D) GFP Survivin ΔEx3; (E) Survivin ΔEx3 antibody (10 μg/mL); (F) control + Rac1 inhibitor; and (G) GFP Survivin ΔEx3 + Rac1 inhibitor. Tube formation was assayed at 4 hours after plating on BME, and quantitative data were collected by measuring the length of the tubules formed under an inverted light microscope at ×100 magnification (40×/0.75 objective lens). Quantitative data shown in panel H are mean ± SE of 5 replicates per sample.
To further substantiate the involvement of Rac1 in Survivin ΔEx3–mediated angiogenesis, we used a commercial Rac1 inhibitor in tube formation assays on BME. HUVECs were transfected with eGFP Survivin ΔEx3 and sterile sorted by FACS to isolate the population of cells that was successfully transfected. This enriched population was plated on BME with complete growth factor medium to support in vitro tube formation. To disrupt Survivin ΔEx3 protein, HUVE cells were pretreated with a Survivin ΔEx3 antibody before plating on BME. Supplementation with VEGF (20 ng/mL) or cell-permeable Rac1 inhibitor (100 μM) was achieved by adding these to the culture medium upon plating on BME. Tube formation was quantified by counting the number of branches and tube length from 5 representative fields per replicate, in a double-blinded fashion, at the 4-hour time point. The addition of GFP Survivin ΔEx3 increased tube length significantly at 4 hours (Figure 6D,H), when compared with control (Figure 6B,H). The magnitude of increased tube length was comparable with that observed after treatment with VEGF alone at this time point (Figure 6C,H). There were no significant differences in tube length between control and GFP Survivin ΔEx3 at the 16-hour time point; however, the overall tube length was increased in both conditions over time (not shown). These observations suggest that Survivin ΔEx3 likely plays a specific role in the initial stages of vascular tube formation. Inhibition of the Rac1 pathway by a Rac1 inhibitor resulted in a dramatic reduction in tube length in both control and GFP Survivin ΔEx3–transfected cells. This effect was similar to that observed when cells were inhibited with the Survivin ΔEx3 antibody (Figure 6E,H). The effect observed was not due to cytotoxicity of the inhibitor, as determined by an MTT viability assay on the HUVE cells (Figure S6). The Survivin ΔEx3–dependent increase in tube formation at the 4-hour time point, abrogated by treatment with a Rac1 inhibitor (Figure 6G-H), further substantiates the role of Rac1 in Survivin ΔEx3-mediated angiogenesis.
(6) Interference with Survivin ΔEx3 inhibits directed in vivo angiogenesis
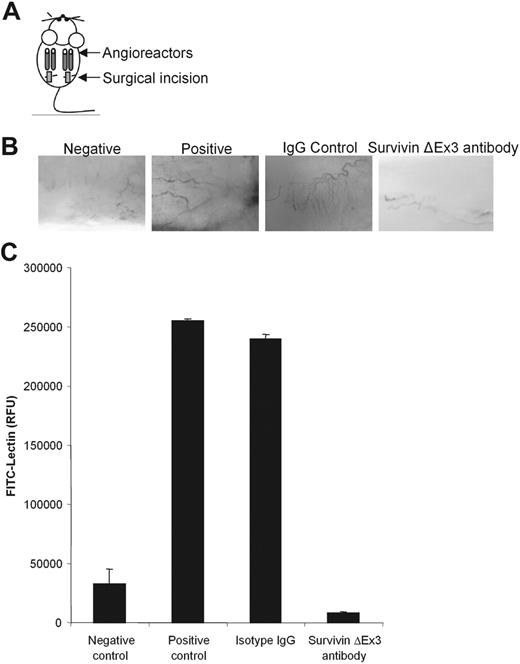
To substantiate the in vitro observations showing that Survivin ΔEx3 is required for angiogenesis, we performed a directed in vivo angiogenesis assay (DIVAA).19 This assay provides a quantitative assessment of angiogenic responses to the inhibition of Survivin ΔEx3. Angioreactors filled with extracellular matrix premixed with or without angiogenic factors (heparin, FGF-2) containing either normal rabbit IgG (experimental control) or Survivin ΔEx3–specific antibody (experimental sample) were implanted subcutaneously into the dorsal flank of athymic nude mice (Figure 7A). At a predetermined time point known to produce significant vascular growth (11 days after implantation), angioreactors were removed. Results at this time point showed prominent vascular growth in the control samples, as demonstrated by invasion of the angioreactor by multiple, branched capillaries (Figure 7B). By contrast, vascularization of the angioreactor was virtually absent in the Survivin ΔEx3 antibody-treated sample, similar to that observed for the negative control angioreactor (Figure 7B). This result suggests that inhibiting Survivin ΔEx3 can directly suppress endothelial cell invasion into the angioreactor.
Disruption of Survivin ΔEx3 results in a diminished angiogenic response in vivo. (A-C) DIVAA was performed as previously described. Angioreactors (A) were prefilled with extracellular matrix with or without the angiogenic factors heparin and FGF-2 (negative and positive controls) or containing angiogenic factors and supplemented with IgG control or Survivin ΔEx3 antibody, then implanted into the dorsal flanks of athymic nude mice. (B) After 11 days, the angioreactors were removed and photographed under a dissecting microscope to establish blood vessel invasion. (C) Quantification of angiogenic responses was performed after recovery of cell pellets from the angioreactors following dispase digestion and staining with FITC-lectin. Results represent the mean fluorescence ± SE from 4 replicates.
Disruption of Survivin ΔEx3 results in a diminished angiogenic response in vivo. (A-C) DIVAA was performed as previously described. Angioreactors (A) were prefilled with extracellular matrix with or without the angiogenic factors heparin and FGF-2 (negative and positive controls) or containing angiogenic factors and supplemented with IgG control or Survivin ΔEx3 antibody, then implanted into the dorsal flanks of athymic nude mice. (B) After 11 days, the angioreactors were removed and photographed under a dissecting microscope to establish blood vessel invasion. (C) Quantification of angiogenic responses was performed after recovery of cell pellets from the angioreactors following dispase digestion and staining with FITC-lectin. Results represent the mean fluorescence ± SE from 4 replicates.
Fluorescein-labeled Griffonia simplicifolia lectin-1 (FITC-lectin), an endothelial cell selective marker, was used to quantify the endothelial cell responses to inhibition of Survivin ΔEx3.19 Endothelial cells within the angioreactors were dispersed from the matrix by dispase digestion and stained with FITC-lectin, as described. The mean relative fluorescence intensity of cells isolated from the Survivin ΔEx3 antibody-treated angioreactor was 28-fold lower than that observed for the control IgG angioreactor (Figure 7C). This effect was more significant than the differences observed between the negative control sample lacking angiogenic modulators and the positive control sample containing angiogenic modulators (8-fold difference), representing a significantly higher level of endothelial cell invasion when Survivin ΔEx3 is not inhibited (Figure 7C). Furthermore, a 4-fold decrease in mean intensity in the Survivin ΔEx3 antibody-treated angioreactor was observed compared with the negative control angioreactor, lacking angiogenic modulators. These data suggest that inhibition of Survivin ΔEx3 in vivo significantly reduces endothelial cell invasion, resulting in dramatically impaired angiogenesis.
Discussion
The previously understood functional roles of Survivin have greatly increased in complexity since the identification of alternative splice variants that possess distinct functions from those originally identified for the main Survivin isoform. Studies of the Survivin splice variants have been performed almost exclusively with cancer cells; however, Survivin itself has been increasingly implicated in other physiologic and pathophysiologic processes, including angiogenesis.22-26 Previous studies have shown that the main isoform of Survivin is expressed at higher levels in the endothelial cells of newly formed blood vessels within tumors33 as well as within injured or ischemic brain.34 Functional studies have not yet addressed the roles of the individual splice variants in angiogenesis.
One of the early findings linking Survivin to angiogenesis came from work using VEGF. VEGF is one of the strongest proangiogenic factors and it is known to induce Survivin transcription in vascular endothelial cells.30 Increased expression of Survivin in newly formed microvessels within the injured brain of a mouse model for stroke has been demonstrated.34 Mice with heterozygous deficiency for survivin also had a reduced vessel density and decreased extent of neovascularization.34 One report using antisense oligonucleotides and siRNA to Survivin in endothelial cells showed that down-regulation of Survivin led to growth inhibition, increased apoptosis, and decreased capillary formation.26 However, in this work, based on the siRNA design, all Survivin isoforms would have been targeted, eliminating the possibility of evaluating the contribution of the individual Survivin splice variants. Disruption of Survivin using antisense and dominant-negative mutants that affect all of the Survivin variants resulted in a decrease in new tumor formation and in tumor angiogenesis.33 The identity of the specific Survivin variant(s) responsible for proangiogenic properties was unclear from these experiments.
To assess the potential involvement of Survivin ΔEx3 in angiogenesis, we evaluated the protein expression pattern of Survivin ΔEx3 in sections of both normal and tumor-derived tissues and detected a prominent association of Survivin ΔEx3 with vascular endothelial cells. Survivin ΔEx3 localized to the vascular endothelium in a variety of tissues, including malignant tumors, proliferative benign angiomas, and diverse normal organs. The striking and specific expression of Survivin ΔEx3 within vessels led to our investigation of a functional role for Survivin ΔEx3 in angiogenesis. To this end, we functionally disrupted the expression of Survivin ΔEx3, using both shRNA-specific targeting and antibody-mediated inhibition, in both cultured vascular endothelial cells and in an in vivo model of angiogenesis. Disruption of Survivin ΔEx3 resulted in an impaired ability of vascular endothelial cells to form tubes on BME that was of similar magnitude to that observed in cells treated with antibodies to VEGF receptors. This dramatic effect was only marginally overcome by the addition of VEGF to the medium. In the in vivo angiogenesis assay, vascularization was inhibited following addition of the Survivin ΔEx3 antibody. In support of these results, exogenous expression of Survivin ΔEx3 led to increased vascularization that was quantitatively similar to that observed following the addition of VEGF. The sum of these results implicates Survivin ΔEx3 as a direct mediator of angiogenesis.
The cellular mechanism of angiogenesis involves endothelial cell spreading, migration, and regulation of cell-cell and cell-matrix adhesion, all of which require significant reorganization of the actin cytoskeleton. Ruffles, lamellipodia, and focal adhesions are complex structures that regulate cell spreading and migration. Investigation of the intracellular localization of Survivin ΔEx3 in endothelial cells led to the identification of a subcellular pool of the protein that localized within the plasma membrane consistent with membrane ruffles.
The Rho family of proteins (Rac, Rho, cdc42), members of the ras superfamily of small GTPases, has been identified as a key regulator in the formation of membrane ruffles, together with the organization of actin cytoskeleton in many cell types.35,36 Rho family GTPases are involved in mitogenic pathways that control the progression of the G1-phase of the cell cycle. Rac1, specifically, has been reported to stimulate transcription of cyclin D137,38 and, together with cdc42, to promote pRb phosphorylation.39 This, in turn, results in the release of the E2F transcriptional activators that further induce transcription from the Survivin promoter.40 Results here, indicating that Survivin ΔEx3 localizes to cell margins, the site of GTPase activity, suggest the potential involvement of some of these proteins. Demonstration that Survivin ΔEx3 affects Rac1 activation during vascular tube formation in vitro and that inhibition of endothelial Survivin ΔEx3 greatly impairs endothelial cell invasion and migration in vivo specifically supports the involvement of Survivin ΔEx3 through Rac1 in this process. Demonstration that a Rac1 inhibitor suppresses an increase in tube formation following ectopic expression of Survivin ΔEx3 further suggests that Survivin ΔEx3 acts through Rac1 in angiogenesis. Together, these data strongly suggest that Survivin ΔEx3, through Rac1, is a key regulator of locomotory processes within the vascular endothelial cell that lead to cell spreading and migration.
In summary, this study demonstrates that the member of the inhibitor of apoptosis protein family, Survivin ΔEx3, is involved in angiogenesis. Here, we identify a new pathway in which Survivin ΔEx3 mediates activation of the small GTPase Rac1 and regulates angiogenesis. We also describe a novel functional role for Survivin ΔEx3 that is distinct from Survivin in angiogenesis. In conclusion, our results provide new insight into the regulation of endothelial cell homeostasis and angiogenesis by the Survivin proteins.
Authorship
Contribution: H.C. designed and performed research, participated in data analysis and interpretation, and participated in writing the paper; J.R.F. performed research, data analysis, and interpretation; D.R.B. provided key tools, and participated in data analysis and interpretation; M.P.H. performed research; R.A.A. designed research, contributed key tools and reagents, participated in data analysis and interpretation, and participated in writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rachel A. Altura, Columbus Children's Research Institute, 700 Children's Dr, Rm WA5021, Columbus, OH 43205; e-mail: alturar@ccri.net.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by Hope Street Kids Foundation (H.C. and R.A.A.); NIH T32 HD043003-02 (J.R.F.); Jessica Antonina Kass Memorial Endowment Fund (D.R.B.); and NICHD P30HD34615-05 (R.A.A.).
We would like to thank Dr Veela Mehta for providing HUVE cells, Dr Osama El-Assal for assistance with mouse surgeries, Cynthia McAllister for assistance with cell sorting, Florinda Jaynes for assistance with histology, and Robin Bagley for the construction of the TMA. We also thank Phyllis Forrest for compiling pathology reports for this study.

![Figure 3. Subcellular localization of Survivin ΔEx3 in HUVE cells. HUVE cells were seeded on glass coverslips and immunostained with antibodies against Survivin ΔEx3, alpha tubulin, or GM130, or the mitochondrial-specific dye Mitotracker, as described in “Materials and methods.” (A) In dividing cells, Survivin ΔEx3 (red) was localized in the cytoplasm, and did not colocalize with alpha-tubulin (green) at any stage of mitosis (note absence from the midbody/cleavage furrow [arrows]). (B) In resting cells, Survivin ΔEx3 (green) partially colocalized with the Golgi apparatus marker GM130 (red; colocalization resulting in yellow pixels, arrows), but did not colocalize with Mitotracker (red). In a number of cells, Survivin ΔEx3 also localized to the actin cytoskeleton (green, arrow) as well as to the spreading edge of the cell, resembling a membrane ruffle (green, arrow). Nuclear localization was also observed in a minority of cells (arrow; nucleus was pseudocolored red for better contrast, and in all other images the nucleus is depicted in blue from Hoechst staining). Scale bar represents 20 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/4/10.1182_blood-2006-02-003749/4/m_zh80040708300003.jpeg?Expires=1767734010&Signature=bkZuuwzNa0PsrmJW0RXDlZyT~4iGbouvQsvGzs--pm6J~skoR-jErxCtcDdmIg3zOyRTDzw~QDYJKU4K30vAiIKvJUIX0qFk7UyIWpbbltky3rVj2U-yLYCGRAqcTtxYZl5mM7RaBdePXmU4PZNw6iq~d37t0mKBBO1QTBcXVZLlmFrk5wzTsaKj6BdQOiYZZ-jR5K6c1GrT2kcac5xSemwS0-kb5sYRXSymimKK2qJTEPQC0aoHrRA3xtVbrKZQAjS1OKtKI6IlI3xvOt42XF-3VTzCAKO842PJ857Xzo7eOne90hJjl-9ArarOJbRLqsMhAkn8isGmdq1902G4hw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal