Abstract
Posttranslational histone modification by acetylation or methylation regulates gene expression. Here, we investigated the role of the histone lysine methyltransferase MLL for angiogenic functions in human umbilical vein endothelial cells. Suppression of MLL expression by siRNA or incubation with the pharmacologic methyltransferase inhibitor 5′-deoxy-5′-(methylthio)adenosine significantly decreased endothelial-cell migration and capillary sprout formation, indicating that methyltransferase activity is required for proangiogenic endothelial-cell functions. Because the expression of homeodomain transcription factors (Hox) is regulated by MLL, we elucidated the role of Hox gene expression. MLL silencing was associated with reduced mRNA and protein expression of HoxA9 and HoxD3, whereas HoxB3, HoxB4, HoxB5, and HoxB9 were not altered. Overexpression of HoxA9 or HoxD3 partially compensated for impaired migration in MLL siRNA-transfected endothelial cells, suggesting that HoxA9 and HoxD3 both contribute to MLL-dependent migration. As a potential underlying mechanism, MLL siRNA down-regulated mRNA and protein levels of the HoxA9-dependent axon guidance factor EphB4. In contrast, MLL knockdown effects on capillary sprouting were not rescued by HoxA9 or HoxD3 overexpression, indicating that MLL affects additional targets required for 3-dimensional sprout formation. We conclude that MLL regulates endothelial-cell migration via HoxA9 and EphB4, whereas sprout formation requires MLL-dependent signals beyond HoxA9 and HoxD3.
Introduction
MLL (mixed lineage leukemia, ALL-1, HRX) is a chromatin regulator homolog to the Drosophila trithorax gene.1-3 MLL was originally discovered by its involvement in human acute leukemia through chromosomal translocation and fusion to a variety of genes.4 MLL fusion proteins were shown to cause myeloid progenitor immortalization by inducing a characteristic expression profile of homeobox (Hox) transcription factors.5,6 Physiologically, the SET domain of MLL catalyzes the methylation of lysine 4 of histone H3, thereby regulating gene expression.7,8 In particular, native MLL is essential for the epigenetic control of gene expression of a set of homeobox proteins HoxA7, HoxA9, and HoxA10 during ex vivo differentiation of embryonic stem cells9 and for physiologic hematopoiesis.10-12 MLL−/− mice die around dpc 10.5 with aberrant rostrocaudal segmentation,13 or, in a different model targeting exons 12 to 14, at dpc 11.5 to 14.5 with edematous bodies and petechiae.12 Beyond segmental identity and hematopoiesis, MLL is crucial for the maintenance of helper T-lymphocyte function.14 As yet, however, a function of MLL in postnatal angiogenesis has not been explored.
Hox transcription factors play important roles during embryonic development of the cardiovascular system.15-17 Certain members of the Hox family also regulate postnatal angiogenesis and endothelial-cell adhesion and migration. HoxD3 promotes endothelial invasion and migration18 and converts resting endothelium to an angiogenic phenotype by inducing proangiogenic genes including the integrin subunits α5 and β3.19 HoxB3 is required for capillary morphogenesis of preformed vascular sprouts.20 HoxA9, which is up-regulated in endothelial cells in response to angiogenic stimulation,21 is essential for prototypical endothelial gene expression during shear stress-induced endothelial-cell maturation17 as well as for the migratory and tube-forming capacities of mature endothelial cells.22 In common, angiogenic downstream signaling of both HoxB3 and HoxA9 involves the regulation of axon guidance molecules of the ephrin family, the ephrin A1 ligand, and the EphB4 receptor, respectively.20,22
Although a variety of Hox factors are established regulators of different facets of endothelial gene expression and function, the angiogenic relevance of upstream mechanisms that control Hox expression in endothelial cells, however, remain poorly characterized. Although endothelial differentiation appeared intact in MLL−/− embryos,10 the dependency of postnatal endothelial-cell function on the presence of MLL is as yet unknown. Therefore, we investigated the role of MLL for gene expression and angiogenic functions of endothelial cells. Our data show that in addition to its essential role for the control of Hox expression in hematopoiesis, MLL also serves as a coordinate upstream regulator of Hox-regulated angiogenesis.
Materials and methods
Cell culture
Pooled human umbilical vein endothelial cells (HUVECs) were purchased from Cambrex (Verviers, Belgium) and cultured in endothelial basal medium (EBM; Cambrex) supplemented with hydrocortisone, bovine brain extract, gentamicin, amphotericin B, epidermal growth factor, and 10% fetal calf serum (FCS; Gibco, Karlsruhe, Germany) until the third passage. After detachment with trypsin, cells were grown in 6-cm culture dishes for at least 18 hours. 5′-deoxy-5′-(methylthio)adenosine (MTA) was bought from Sigma (Taufkirchen, Germany).
Plasmid constructs and transfection
HUVECs (3.5 × 105 cells/6-cm well) were grown to 60% to 70% confluence and then transfected with 3 μg plasmid DNA. Plasmids included a wild-type human HoxA9 construct (HoxA9 in pcDNA3.1-myc-His) as described previously,22 a wild-type HoxD3 (subcloned into pcDNA 3.1-myc-His, Invitrogen, Karlsruhe, Germany), or pcDNA3.1-myc-His as empty vector control. Transfection was performed using Superfect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
RNA interference
For siRNA-mediated gene knockdown, HUVECs were grown to 60% to 70% confluence and either transfected with Transpass R2 transfection reagent (NEB, Frankfurt, Germany) or GeneTrans II (MoBiTec, Göttingen, Germany) according to the manufacturer's protocol. siRNAs were synthesized by Eurogentec or Qiagen targeting human HoxA9 (5′-ucaacaaagaccgagcaaa-3′) or human MLL (MLL siRNA I: 5′-gcuacugaucuugaaucaa-3′) or a second siRNA oligonucleotide targeted against a different region of the MLL mRNA sequence (MLL siRNA II: 5′-cgaucaaaugcccgccuaa-3′). If not stated otherwise, cells were transfected with MLL siRNA I. A nonrelated scrambled siRNA was used as a control (5′-agcguguagcuagcagagg-3′).
Western blot analysis
For Western blot analysis, HUVECs were lysed in a lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mmol Na3VO4, 1 μg/mL leupeptin, and 1 mM phenylmethylsulfonyl fluoride) for 15 minutes on ice. After centrifugation for 15 minutes at 20 000g (4°C), the protein content of the samples was determined according to the Bradford method. Equal amounts of protein were loaded onto SDS-polyacrylamide gels and blotted onto nitrocellulose or PVDF membranes. Western blots were performed by using antibodies directed against HoxA9 (rabbit polyclonal anti-HoxA9; 1:1500, Upstate Biotechnology, Lake Placid, NY), HoxD3 (rabbit polyclonal anti-HoxD3, 1:1000; Zymed, San Francisco, CA), EphB4 (rabbit polyclonal anti-EphB4, 1:400, Santa Cruz Biotechnology, Heidelberg, Germany), di-methylated histone H3 lysine K4 (anti–di-methyl-Histone H3 (Lys4), rabbit antiserum, 1:5000, Upstate Biotechnology), or tubulin (mouse monoclonal antitubulin, 1:2500, Dianova, Hamburg, Germany).
ChIP
For chromatin immunoprecipitation (ChIP), HUVECs were cross-linked for 10 minutes by directly adding 1% formaldehyde to the culture medium. Afterward, cells were harvested by scraping in ice cold PBS. The fixed cells were lysed for 5 minutes on ice (50 mM Tris, pH 8.0, 2 mM EDTA, 0.1% NP-40, 10% glycerol, protease inhibitor cocktail) to obtain cell nuclei. The nuclei were pelleted and lysed for another 5 minutes on ice (50 mM Tris, pH 8.0, 5 mM EDTA, 0.1% SDS). The chromatin was sheared by sonification (Branson Sonifire 450, Branson, Danbury, CT). Sheared chromatin samples were taken as input control or used for immunoprecipitation with an antibody directed against di-methylated histone H3 lysine K4 (Upstate Biotechnology). Following immunoprecipitation the samples were washed 6 times and either subjected to Western blot analysis or polymerase chain reaction (PCR) using primer pairs directed against promoter or exon regions within the HoxA9 or HoxD3 genes (Table 1)
Oligonucleotides used for RT-PCR, real-time PCR, and ChIP analyses
| . | Sense primer, 5′→3′ . | Antisense primer, 5′→3′ . |
|---|---|---|
| RT-PCR/real-time PCR | ||
| HoxA9 | ||
| RT-PCR | gcccggtgcgctctccttcgc | catcctgcggttctggaaccag |
| Real-time PCR | aaaaacaacccagcgaa | caaatggcatcactcgtc |
| HoxB3 | ctgctctcttcggaggctattc | ctgttgctagtggcactggtagg |
| HoxB4 | gtcccactccgcgtgcaaagag | cagagcgcgtgggcgatctcc |
| HoxB5 | ggcttcgagctgctccctgtcc | ggctgatgtgaagcttcctcatcc |
| HoxB9 | gcacgaagtggccagactcctc | gctggaagtgaggggctaggac |
| HoxD3 | agaaggctgcttactatgaaaaccc | acaagtagcggttgaagtggaattc |
| MLL | cagctatcctctcagatccatc | ccttgtctttccggactttctg |
| EphB4 | cctgctatccctgcacctcttc | ctgatccaatggtgttagagtgg |
| Integrin αV | 5′-gggattttgtcaaggaggattc-3′ | 5′-ccaaagtccttgctgctcttgg-3′ |
| Integrin α5 | gaccagagttacgggactcaactg | cttggtccattgcacagctgtgg |
| Integrin β1 | tgagtgcaaccccaactacactg | gtctggaccagtgggacactctg |
| Integrin β3 | gtgcggcaggtggaggattac | cacgtgtttgtagccaaacatgg |
| Meis1 | ggtacgacgatctaccccattac | gtcattgacagaggagcccatgc |
| GAPDH | ||
| RT-PCR | tcaccatcttccaggagcgagatc | gagaccacctggtgctcagtgtag |
| Real-time PCR | ttggtatcgtggaaggactca | tgtcatcatatttggcaggttt |
| ChIP | ||
| HoxA9 P1 | aatgcgatttggctgcttttttatg | tcaaatctagccttgtctctgtac |
| HoxA9 P2 | gggagacgggagagtacagagac | cgtccagcagaacaataacgcg |
| HoxA9 P3 | caactactacgtggactcgttc | gtgatggtggtggtacaccgc |
| HoxD3 P4 | gtagtggattgtcattgagtggg | cctgcctcccaagttgacaaag |
| HoxD3 P5 | ggttaggctgtttggtgcaggtg | cataggtcagctccctggtctc |
| HoxD3 P6 | cacctccttagtccccctggtc | ctgtggtgaggactgtggtttttg |
| . | Sense primer, 5′→3′ . | Antisense primer, 5′→3′ . |
|---|---|---|
| RT-PCR/real-time PCR | ||
| HoxA9 | ||
| RT-PCR | gcccggtgcgctctccttcgc | catcctgcggttctggaaccag |
| Real-time PCR | aaaaacaacccagcgaa | caaatggcatcactcgtc |
| HoxB3 | ctgctctcttcggaggctattc | ctgttgctagtggcactggtagg |
| HoxB4 | gtcccactccgcgtgcaaagag | cagagcgcgtgggcgatctcc |
| HoxB5 | ggcttcgagctgctccctgtcc | ggctgatgtgaagcttcctcatcc |
| HoxB9 | gcacgaagtggccagactcctc | gctggaagtgaggggctaggac |
| HoxD3 | agaaggctgcttactatgaaaaccc | acaagtagcggttgaagtggaattc |
| MLL | cagctatcctctcagatccatc | ccttgtctttccggactttctg |
| EphB4 | cctgctatccctgcacctcttc | ctgatccaatggtgttagagtgg |
| Integrin αV | 5′-gggattttgtcaaggaggattc-3′ | 5′-ccaaagtccttgctgctcttgg-3′ |
| Integrin α5 | gaccagagttacgggactcaactg | cttggtccattgcacagctgtgg |
| Integrin β1 | tgagtgcaaccccaactacactg | gtctggaccagtgggacactctg |
| Integrin β3 | gtgcggcaggtggaggattac | cacgtgtttgtagccaaacatgg |
| Meis1 | ggtacgacgatctaccccattac | gtcattgacagaggagcccatgc |
| GAPDH | ||
| RT-PCR | tcaccatcttccaggagcgagatc | gagaccacctggtgctcagtgtag |
| Real-time PCR | ttggtatcgtggaaggactca | tgtcatcatatttggcaggttt |
| ChIP | ||
| HoxA9 P1 | aatgcgatttggctgcttttttatg | tcaaatctagccttgtctctgtac |
| HoxA9 P2 | gggagacgggagagtacagagac | cgtccagcagaacaataacgcg |
| HoxA9 P3 | caactactacgtggactcgttc | gtgatggtggtggtacaccgc |
| HoxD3 P4 | gtagtggattgtcattgagtggg | cctgcctcccaagttgacaaag |
| HoxD3 P5 | ggttaggctgtttggtgcaggtg | cataggtcagctccctggtctc |
| HoxD3 P6 | cacctccttagtccccctggtc | ctgtggtgaggactgtggtttttg |
RT-PCR indicates reverse transcription-polymerase chain reaction.
Spheroid-based angiogenesis assay
Endothelial-cell spheroids of defined cell number were generated as described previously.23,24 In brief, 12 hours after transfection HUVECs were suspended in culture medium containing 0.2% (wt/vol) carboxymethylcellulose (Sigma) and seeded in nonadherent round-bottom 96-well plates (Greiner, Frickenhausen, Germany). Under these conditions, all suspended cells contribute to the formation of a single spheroid per well of defined size and cell number (400 cells/spheroid). Spheroids were generated overnight, after which they were embedded into collagen gels. The spheroid containing gel was rapidly transferred into prewarmed 24-well plates and allowed to polymerize (30 minutes), then 100 μL endothelial basal medium with or without human basic fibroblast growth factor (bFGF, 30 ng/mL) was added on top of the gel. After 24 hours, pictures were taken using an Axiovert 100M microscope and a Plan-NEOFLUAR 10×/0.30 objective lens. In vitro capillary sprouting was quantified by measuring the cumulative length using AxioVision Rel 4.4 digital imaging software (Zeiss, Jena, Germany). To obtain a measure of the cumulative sprout length per spheroid, every sprout from 10 to 15 spheroids was assessed and from these data the mean cumulative sprout length per spheroid was calculated.
Scratched wound assay
Migration of HUVECs was detected using a “scratched wound assay.”25 Briefly, HUVECs were grown on 6-cm dishes previously labeled with a traced line. The cell monolayer was scraped with a sterile cell scraper to create a cell-free zone (width approximately 14 mm). Thereafter, cells were washed with medium and stimulated as indicated. Endothelial-cell migration was quantified by measuring the width of the cell-free zone (distance between the edges of the injured monolayer) at the time of injury and after 24 hours of cultivation using a computer-assisted microscope (Zeiss) at 5 distinct positions (every 5 mm).
RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). Afterward, 2 μg RNA from each sample was reverse transcribed into cDNA and subjected to conventional PCR using the oligonucleotide primers summarized in Table 1. Semiquantitative analysis was performed densitometrically using the Scion Image software (version 4.0.2, Scion, Frederick, MD).
Real-time quantitative PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). Afterward, 2 μg RNA from each sample was reverse transcribed into cDNA and subjected to real-time PCR using the LightCycler FastStart DNA Master SYBR Green I Kit (Roche Applied Science, Indianapolis, IN). LightCycler reactions were performed with the LightCycler 2.0 System and LightCycler3 quantification software (Roche Applied Science, Mannheim, Germany).
Statistical analysis
Data are expressed as mean ± SEM. Two treatment groups were compared with the independent samples t test, and 3 or more groups by one-way analysis of variance followed by post-hoc analysis adjusted with a least significant difference correction for multiple comparisons (SPSS, Munich, Germany). Results were considered statistically significant at P values of less than .05.
Results
MLL regulates endothelial-cell migration and sprouting
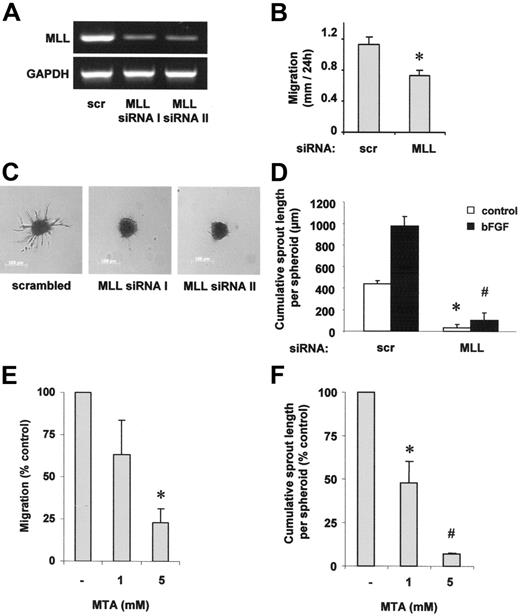
To investigate the role of MLL for angiogenesis, we measured endothelial-cell migration and sprout formation after silencing of MLL expression with siRNA (Figure 1A). The reduction of MLL expression significantly suppressed HUVEC migration in a scratched wound assay (Figure 1B). Moreover, suppression of MLL significantly inhibited basal and bFGF-stimulated endothelial-cell sprout formation when we assessed the angiogenic capacity of endothelial cells by measuring capillary sprouting in a 3-dimensional collagen-embedded spheroid culture assay (Figure 1C-D). As a control for the specificity of MLL silencing, a second MLL siRNA was generated, which reduced capillary sprout formation to a similar extent as compared to MLL siRNA I (Figure 1A,C). The reduction in endothelial-cell migration and sprouting was not secondary to unspecific effects on cell growth or death following knockdown of MLL because MLL siRNA transfection did not significantly alter apoptosis or proliferation (data not shown).
MLL regulates endothelial-cell migration and sprouting. (A,C) HUVECs were transfected with 2 different siRNAs against MLL (I and II) or scrambled oligonucleotides. (A) RT-PCR analysis of MLL mRNA expression. A representative gel is shown. GAPDH serves as loading control. (B,D) HUVECs were transfected with MLL siRNA or scrambled oligonucleotides. (B) Cell migration was measured using a scratched wound assay. Data are shown as mean ± SEM. *P < .05, n = 3. (C) A spheroid assay was performed to analyze basal endothelial sprouting capacity. Representative spheroids are shown. (D) Analysis of endothelial sprouting capacity with or without bFGF (30 ng/mL) stimulation. Endothelial sprouting capacity is given as cumulative sprout length per spheroid. Data are shown as mean ± SEM. *P < .01 versus scrambled and #P < .01 versus scrambled + bFGF, n = 3. (E) HUVECs were prestimulated with different doses of the methyltransferase inhibitor MTA as indicated and stimulation was repeated following scratch procedure. Migration was measured using the scratched wound assay. Data are shown as percent control and mean ± SEM. *P < .05 versus control, n = 3. (F) HUVEC capillary sprouting was analyzed in the presence of different doses of MTA as indicated. Data are shown as percent control and mean ± SEM. *P < .05 versus control; #P < .01 versus control, n = 3.
MLL regulates endothelial-cell migration and sprouting. (A,C) HUVECs were transfected with 2 different siRNAs against MLL (I and II) or scrambled oligonucleotides. (A) RT-PCR analysis of MLL mRNA expression. A representative gel is shown. GAPDH serves as loading control. (B,D) HUVECs were transfected with MLL siRNA or scrambled oligonucleotides. (B) Cell migration was measured using a scratched wound assay. Data are shown as mean ± SEM. *P < .05, n = 3. (C) A spheroid assay was performed to analyze basal endothelial sprouting capacity. Representative spheroids are shown. (D) Analysis of endothelial sprouting capacity with or without bFGF (30 ng/mL) stimulation. Endothelial sprouting capacity is given as cumulative sprout length per spheroid. Data are shown as mean ± SEM. *P < .01 versus scrambled and #P < .01 versus scrambled + bFGF, n = 3. (E) HUVECs were prestimulated with different doses of the methyltransferase inhibitor MTA as indicated and stimulation was repeated following scratch procedure. Migration was measured using the scratched wound assay. Data are shown as percent control and mean ± SEM. *P < .05 versus control, n = 3. (F) HUVEC capillary sprouting was analyzed in the presence of different doses of MTA as indicated. Data are shown as percent control and mean ± SEM. *P < .05 versus control; #P < .01 versus control, n = 3.
In addition to the genetic knockdown of MLL by siRNA transfection, we tested whether methyltransferase activity is required for endothelial-cell migration and sprouting. Because a specific inhibitor of SET-like H3K4-directed methyltransferases is currently not available, we tested the effect of the broad-spectrum methyltransferase inhibitor MTA.26 MTA dose dependently reduced both endothelial-cell migration as well as sprouting (Figure 1E-F).
HoxA9 and HoxD3 expression in endothelial cells depends on MLL
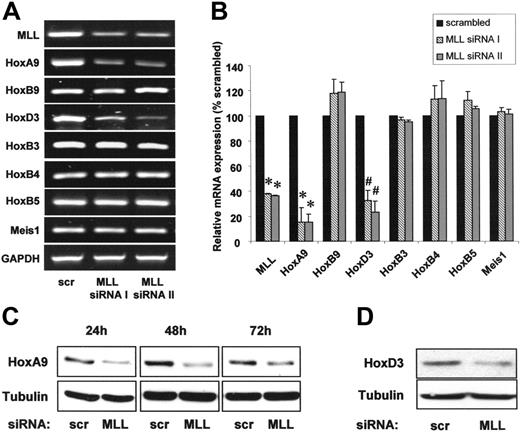
Because the regulation of hematopoiesis by MLL depends on homeobox transcription factors, we measured the expression of several Hox transcription factors by quantitative real-time PCR after transfection of endothelial cells with 2 different MLL-specific siRNA oligonucleotides. Suppression of MLL reduced the transcription factor HoxA9 on mRNA and protein levels (Figure 2A-C). To assess the specificity of the MLL-dependent regulation of HoxA9, we determined whether the expression of homeodomain factors with an established relevance for hematopoietic and endothelial-cell function other than HoxA9 also depends on MLL. Indeed, siRNA-mediated suppression of MLL down-regulated HoxD3 mRNA and protein, whereas the expression of HoxB9, HoxB3, HoxB4, and HoxB5 remained unchanged (Figure 2A-B,D). Likewise, expression of Meis1, which cooperates with HoxA9 to transactivate gene expression, was not affected by siRNA-mediated suppression of MLL in endothelial cells (Figure 2A-B).
The histone methyltransferase MLL specifically regulates HoxA9 and HoxD3 expression. (A-B) HUVECs were transfected with 2 different siRNAs against MLL (I and II) or scrambled oligonucleotides. (A) RT-PCR analysis of MLL and Hox mRNA expression. Representative gels are shown. GAPDH is shown as loading control. (B) Quantitative analysis of MLL and Hox gene expression using real-time PCR. Data are shown as percent scrambled and mean ± SEM. *P < .01 versus scrambled, #P < .05 versus scrambled, n = 3. (C-D) HUVECs were transfected with siRNA against MLL or scrambled oligonucleotides. (C) Western blot analysis of HoxA9 expression. Samples were taken at several time points following siRNA transfection as indicated. Tubulin is shown as loading control. (D) Western blot analysis of HoxD3 expression. Tubulin is shown as loading control.
The histone methyltransferase MLL specifically regulates HoxA9 and HoxD3 expression. (A-B) HUVECs were transfected with 2 different siRNAs against MLL (I and II) or scrambled oligonucleotides. (A) RT-PCR analysis of MLL and Hox mRNA expression. Representative gels are shown. GAPDH is shown as loading control. (B) Quantitative analysis of MLL and Hox gene expression using real-time PCR. Data are shown as percent scrambled and mean ± SEM. *P < .01 versus scrambled, #P < .05 versus scrambled, n = 3. (C-D) HUVECs were transfected with siRNA against MLL or scrambled oligonucleotides. (C) Western blot analysis of HoxA9 expression. Samples were taken at several time points following siRNA transfection as indicated. Tubulin is shown as loading control. (D) Western blot analysis of HoxD3 expression. Tubulin is shown as loading control.
Histone H3 lysine K4 methylation is present at the HoxA9 and HoxD3 genes
To further dissect the molecular mechanisms that link MLL to Hox mRNA expression in endothelial cells, we examined histone methylation within the HoxA9 and HoxD3 genes by ChIP using an antibody specific for K4-methylated histone H3 (Figure 3A-B). As previously reported in HeLa cells,8,27 endothelial cells also displayed histone H3 methylation within the promoter region of the HoxA9 gene (Figure 3C). In addition, ChIP against K4-methylated histone H3 revealed the presence of histone methylation also in the promoter and exon regions of the HoxD3 gene (Figure 3D).
Histone H3 lysine K4 methylation is present at the HoxA9 and HoxD3 genes. (A-D) ChIP was performed in HUVEC lysates using an antibody directed against di-methylated histone H3 lysine K4. (A) Overview of primer pairs (P1-P3 for HoxA9 and P4-P6 for HoxD3) amplifying the indicated promoter or exon regions within the HoxA9 and HoxD3 genes. (B) Western blot analysis was performed to confirm the immunoprecipitation against di-methylated histone H3 lysine K4. (C-D) Representative PCR with primers detecting the indicated promoter or exon regions of the HoxA9 or HoxD3 genes are shown, n = 3. Ab indicates antibody control.
Histone H3 lysine K4 methylation is present at the HoxA9 and HoxD3 genes. (A-D) ChIP was performed in HUVEC lysates using an antibody directed against di-methylated histone H3 lysine K4. (A) Overview of primer pairs (P1-P3 for HoxA9 and P4-P6 for HoxD3) amplifying the indicated promoter or exon regions within the HoxA9 and HoxD3 genes. (B) Western blot analysis was performed to confirm the immunoprecipitation against di-methylated histone H3 lysine K4. (C-D) Representative PCR with primers detecting the indicated promoter or exon regions of the HoxA9 or HoxD3 genes are shown, n = 3. Ab indicates antibody control.
Role of HoxA9 and HoxD3 for MLL-dependent endothelial-cell migration and angiogenesis
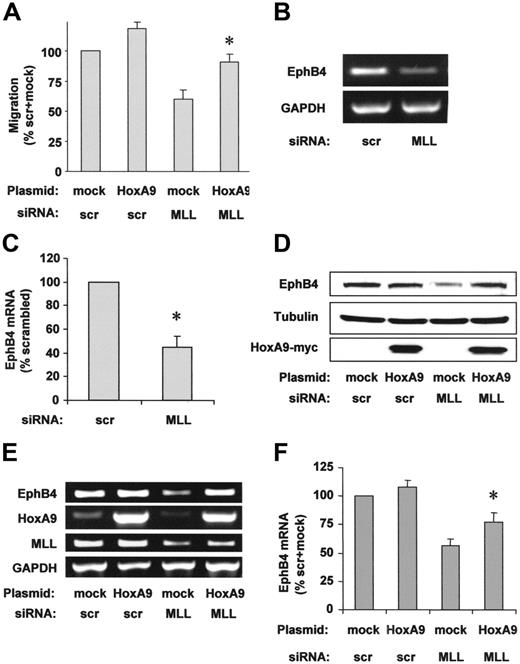
To address whether the reduction of HoxA9 contributes to the reduced endothelial-cell migration after MLL siRNA transfection, we overexpressed HoxA9 wild-type protein. HoxA9 overexpression partially rescued the impaired migration of MLL siRNA-transfected cells in the scratched wound assay (Figure 4A). The axon guidance molecule EphB4, the receptor for the EphrinB2 ligand, is essential for proper vascular morphogenesis during embryonal development28 and for cancer cell migration.29 Because EphB4 was previously identified as a downstream target of HoxA9 in endothelial cells,22 we investigated a potential effect of MLL on EphB4 expression in endothelial cells. Suppression of MLL by siRNA markedly reduced EphB4 mRNA and protein (Figure 4B-D). Simultaneous overexpression of HoxA9 partially compensated for the reduced mRNA and protein expression of EphB4 in MLL siRNA-transfected endothelial cells (Figure 4D-F), suggesting that HoxA9 mediates the regulation of EphB4 in an MLL-dependent signaling pathway.
HoxA9 specifically rescues endothelial-cell migration and EphB4 expression. (A) HUVECs were transfected with siRNA against MLL or scrambled (scr) oligonucleotides and empty vector or HoxA9 wild-type plasmid. HUVEC migration was analyzed using the scratched wound assay. Data are shown as percent scrambled + mock and mean ± SEM. *P < .05 versus mock + MLL siRNA, n = 4. (B) RT-PCR analysis of EphB4 mRNA expression following MLL siRNA or scrambled oligonucleotide transfection. A representative gel is shown. (C) Expression is given as percent scrambled. Data are mean ± SEM. *P < .05, n = 3. (D-F) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxA9 wild-type plasmid. (D) Western blot analysis was performed to investigate EphB4 protein expression. Tubulin is shown as loading control. (E) RT-PCR analysis of EphB4 mRNA expression. A representative gel is shown. GAPDH serves as loading control. (F) Semiquantitative analysis of EphB4 mRNA expression. Data are shown as percent scrambled + mock and mean ± SEM, *P < .05 versus mock + MLL siRNA, n = 6.
HoxA9 specifically rescues endothelial-cell migration and EphB4 expression. (A) HUVECs were transfected with siRNA against MLL or scrambled (scr) oligonucleotides and empty vector or HoxA9 wild-type plasmid. HUVEC migration was analyzed using the scratched wound assay. Data are shown as percent scrambled + mock and mean ± SEM. *P < .05 versus mock + MLL siRNA, n = 4. (B) RT-PCR analysis of EphB4 mRNA expression following MLL siRNA or scrambled oligonucleotide transfection. A representative gel is shown. (C) Expression is given as percent scrambled. Data are mean ± SEM. *P < .05, n = 3. (D-F) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxA9 wild-type plasmid. (D) Western blot analysis was performed to investigate EphB4 protein expression. Tubulin is shown as loading control. (E) RT-PCR analysis of EphB4 mRNA expression. A representative gel is shown. GAPDH serves as loading control. (F) Semiquantitative analysis of EphB4 mRNA expression. Data are shown as percent scrambled + mock and mean ± SEM, *P < .05 versus mock + MLL siRNA, n = 6.
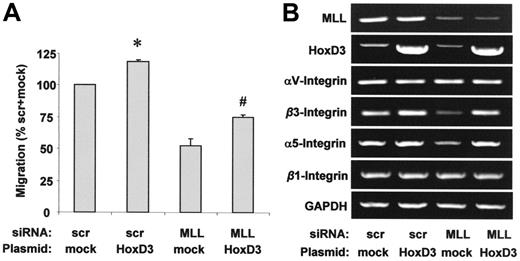
In parallel with HoxA9, HoxD3 is regulated in an MLL-dependent manner in HUVECs (Figure 2A-B) and was also shown to regulate gene expression during angiogenesis in endothelial cells.30,31 Therefore, we investigated whether the regulation of HoxD3 is causally linked to the requirement of MLL for endothelial-cell migration. Simultaneous overexpression of HoxD3 partially compensates for the impaired migration following treatment with MLL siRNA (Figure 5A). Because HoxD3 regulates integrin expression in the endothelium,30,31 we also determined the dependency of integrin αvβ3 and α5β1 expression on MLL. siRNA-mediated suppression of MLL reduced the expression of integrin β3 and α5 mRNA, which was compensated for by simultaneous overexpression of HoxD3, suggesting that the MLL-sensitive expression of HoxD3 causally underlies the observed regulation of integrin α5 and β3 expression by MLL (Figure 5B). In line with previous reports, mRNA levels of the integrin subunits αv and β1 were not affected by MLL siRNA or HoxD3 overexpression (Figure 5B).30,31
Effect of HoxD3 on endothelial-cell migration and integrin expression. (A-B) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxD3 wild-type plasmid. (A) HUVEC migration was analyzed using the scratched wound assay. Data are shown as percent scrambled + mock and mean ± SEM. *P < .05 versus mock + scrambled siRNA, #P < .05 versus mock + MLL siRNA, n = 3. (B) RT-PCR analysis of MLL, HoxD3, and integrin mRNA expression. GAPDH serves as loading control.
Effect of HoxD3 on endothelial-cell migration and integrin expression. (A-B) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxD3 wild-type plasmid. (A) HUVEC migration was analyzed using the scratched wound assay. Data are shown as percent scrambled + mock and mean ± SEM. *P < .05 versus mock + scrambled siRNA, #P < .05 versus mock + MLL siRNA, n = 3. (B) RT-PCR analysis of MLL, HoxD3, and integrin mRNA expression. GAPDH serves as loading control.
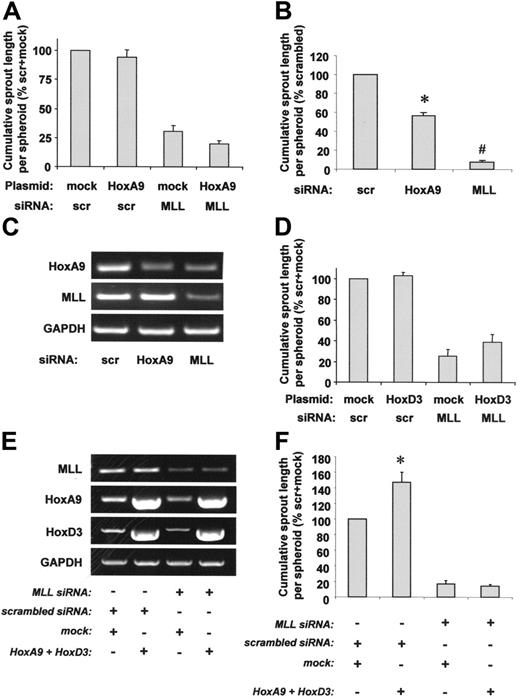
To further dissect the contribution of HoxA9 or HoxD3 for capillary sprout formation, we performed a 3-dimensional spheroid assay. Surprisingly, HoxA9 overexpression did not affect defective capillary sprouting of MLL siRNA-transfected cells (Figure 6A). Similar results were obtained when HoxA9 was overexpressed with an adenovirus in MLL siRNA-treated cells (data not shown), suggesting that insufficient transfection efficiency was not the reason for the failure to rescue sprouting. Thus, HoxA9 may play a more pronounced role as an MLL downstream target for endothelial-cell migration, whereas additional HoxA9-independent MLL targets appear to be involved in complex 3-dimensional capillary sprout formation. In line with these findings, the effect of MLL siRNA on capillary sprouting exceeded the effect of HoxA9 siRNA, which suppressed sprout formation by approximately 50% (Figure 6B), although HoxA9 expression was similarly reduced in HoxA9 siRNA- or MLL siRNA-transfected cells (Figure 6C), suggesting that HoxA9-independent mechanisms contribute to MLL-regulated endothelial-cell sprouting. Because MLL also regulates the expression of HoxD3, we tested whether the effect of HoxD3 down-regulation might be responsible for MLL siRNA-mediated inhibition of sprout formation. However, HoxD3 overexpression was not capable of compensating for abrogated sprout formation in HUVECs lacking MLL (Figure 6D). To explore a potential synergistic role of HoxA9 and HoxD3 for endothelial-cell signaling during sprout formation, we simultaneously overexpressed both genes in MLL siRNA-treated cells (Figure 6E). However, MLL siRNA still reduced capillary sprout formation even in the presence of overexpression levels of HoxA9 and HoxD3, each of which was sufficient to partially rescue defective migration in MLL siRNA-treated cells (Figures 6F, 4A, and 5A). In summary, these data demonstrate that the blockade of endothelial-cell migration by MLL silencing in a scratched wound assay is partially mediated via suppression of HoxA9 and HoxD3 expression, whereas the defect in 3-dimensional sprout formation was neither rescued by HoxA9 nor by HoxD3.
Role of HoxA9 and HoxD3 for MLL-dependent endothelial-cell sprouting. (A) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxA9 wild-type plasmid. Endothelial sprouting was assessed using the spheroid assay. Data are shown as percent scrambled + mock and mean ± SEM. (B-C) HUVECs were transfected with scrambled, HoxA9, or MLL siRNA oligonucleotides. (B) A spheroid assay was performed to investigate endothelial sprouting. Data are shown as percent scrambled and mean ± SEM. *P < .05 versus scrambled; #P < .05 versus scrambled and HoxA9 siRNA, n = 3. (C) RT-PCR analysis of HoxA9 and MLL expression. A representative gel is shown. GAPDH serves as loading control. (D) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxD3 wild-type plasmid. Endothelial sprouting was assessed using the spheroid assay. Data are shown as percent scrambled + mock and mean ± SEM, n = 4. (E-F) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxA9 wild-type + HoxD3 wild-type plasmids. (E) RT-PCR analysis of MLL and Hox expression. GAPDH serves as loading control. (F) A spheroid assay was performed to analyze the effect of simultaneous HoxA9 and HoxD3 overexpression on sprouting capacity. Data are shown as percent scrambled + mock and mean ± SEM. *P < .05 versus scrambled + mock, n = 4.
Role of HoxA9 and HoxD3 for MLL-dependent endothelial-cell sprouting. (A) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxA9 wild-type plasmid. Endothelial sprouting was assessed using the spheroid assay. Data are shown as percent scrambled + mock and mean ± SEM. (B-C) HUVECs were transfected with scrambled, HoxA9, or MLL siRNA oligonucleotides. (B) A spheroid assay was performed to investigate endothelial sprouting. Data are shown as percent scrambled and mean ± SEM. *P < .05 versus scrambled; #P < .05 versus scrambled and HoxA9 siRNA, n = 3. (C) RT-PCR analysis of HoxA9 and MLL expression. A representative gel is shown. GAPDH serves as loading control. (D) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxD3 wild-type plasmid. Endothelial sprouting was assessed using the spheroid assay. Data are shown as percent scrambled + mock and mean ± SEM, n = 4. (E-F) HUVECs were transfected with scrambled or MLL siRNA oligonucleotides and empty vector or HoxA9 wild-type + HoxD3 wild-type plasmids. (E) RT-PCR analysis of MLL and Hox expression. GAPDH serves as loading control. (F) A spheroid assay was performed to analyze the effect of simultaneous HoxA9 and HoxD3 overexpression on sprouting capacity. Data are shown as percent scrambled + mock and mean ± SEM. *P < .05 versus scrambled + mock, n = 4.
Discussion
Here, we investigated the regulation of angiogenic endothelial-cell functions by the histone methyltransferase MLL. Our data show that MLL is essential for both migration and sprout formation of endothelial cells.
By demonstrating an essential angiogenic function of MLL in endothelial cells, the results of our experiments add to the growing recognition of the physiologic role of native (not fusion protein-linked) MLL. In embryonic stem cells derived from MLL-deficient mice that lack HoxA9 expression, Ernst et al reported a reduced capability of hematopoietic progenitor cells to form colonies, which could be restored by ectopic re-expression of HoxA9.9 In accordance, MLL was intrinsically required for the completion of later stages during hematopoiesis.10,11 Although embryonal vasculogenesis was less affected in these animals,10 our data show that MLL exerts essential angiogenic functions in endothelial cells in vitro. One may speculate that the early embryonic lethality of MLL-deficient mice may prevent recognition of angiogenesis defects. Moreover, MLL-dependent endothelial-cell migration and capillary sprouting could well be relevant during later stages of embryonal angiogenesis or for reactive vascular growth in the adult organism. Indeed, postischemic neovascularization is severely impaired in HoxA9-deficient mice,17 although embryonal angiogenesis appears to proceed normally.
The dependency of HoxA9 gene expression on MLL is attributed to the function of the SET-like lysine methyltransferase domain of MLL, which methylates lysine residue K4 of histone H3 within the promoter region of cluster A Hox genes.7,8 Our data indeed demonstrate that histone H3 methylation is present within the HoxA9 promoter in endothelial cells. Besides direct histone methylation, MLL may play a coactivating role for the regulation of the HoxA9 gene by acting as an assembly factor recruiting the histone acetyltransferase MOF into a multiprotein supercomplex at the HoxA9 promoter.27 In accordance, oncogenic MLL fusion proteins lacking the SET-like HMT domain still transactivate HoxA932 through a mechanism that does not involve H3 K4 methylation,7 whereas, however, an MLL construct with a mutated SET domain and, thus, deficient in methyltransferase activity was not capable of inducing expression from the HoxC8 promoter.7 Moreover, in addition to physical interaction with the HoxA9 promoter region, MLL also occupies and regulates promoter regions of miRNAs33 that could affect HoxA9 indirectly. Our data demonstrate that histone H3 methylation is also detectable within the promoter and exon region of the HoxD3 gene in endothelial cells. The presence of histone H3 methylation within the exon of HoxD3 could be explained by recent findings, that genome-wide histone methylation occurs also within coding regions of actively transcribed genes.34
Histone methylation-dependent regulation of gene expression was previously reported to be involved in the regulation of the endothelial nitric oxide synthase (eNOS) gene.35 In this paper, histone methylation at the eNOS promoter was found at increased levels in endothelial cells compared to nonendothelial cells, and the methylation inhibitor MTA reduced eNOS mRNA expression.35 In endothelial progenitor cells, HoxA9 transactivates eNOS transcription during cell maturation ex vivo,17 providing a second clue for the potential involvement of eNOS in MLL-dependent endothelial-cell signaling. However, exogenous NO did not compensate for the suppressed migration in MLL-siRNA-treated cells (data not shown). Therefore, a major causal role for eNOS in this pathway appears unlikely. Rather, our data show that the axon guidance factor EphB4 is located downstream of MLL as an angiogenic effector molecule, which is regulated by MLL in a HoxA9-mediated way. Importantly, EphB4 does not only signal endothelial-cell migration,36 but was recently also identified as an essential regulator of cancer cell migration29 and metastatic potential.37 Thus, MLL-dependent HoxA9- and EphB4-mediated angiogenic signals may provide an attractive therapeutic target for antiangiogenesis and antineoplastic therapeutic strategies.38 In addition to the MLL-HoxA9-EphB4 axis, the MLL-dependent regulation of integrin subunits α5 and β3 via HoxD3 appears to be involved in endothelial-cell migration. These findings are in accordance with the previously described role of HoxD3 for the regulation of gene expression during angiogenesis in endothelial cells.30,31
Strikingly, our data reveal distinct pathways mediating MLL-dependent endothelial-cell migration in contrast to capillary sprouting. Although HoxA9 was previously shown to be sufficient and essential for migration of endothelial cells in a scratched wound assay,22 overexpression of HoxA9 or HoxD3 separately or a combination of both was incapable of rescuing MLL-dependent sprouting in a 3-dimensional assay, suggesting that different mechanisms are required for 3-dimensional sprouting of endothelial cells into a collagen matrix compared to 2-dimensional cell movement in a culture dish. This is conceivable because the 2-dimensional migration assay mainly involves endothelial-cell spreading and proliferation.25,39 In the 3-dimensional spheroid assay, a variety of cellular processes are involved, including the development of intracellular vacuoles, degradation and invasion into the collagen matrix, cell elongation, tube formation, transition from proliferation to differentiation, and maturation of the capillary structures.40 Thus, MLL may regulate additional targets, which are essential for capillary sprout formation during angiogenesis. Therefore, we determined the transcriptional regulation of other homeobox genes as putative MLL-dependent targets. However, neither HoxB4 nor HoxB5 or HoxB9, a paralogous gene of HoxA9, were regulated by MLL. Meis1, another transcription factor, which cooperates with HoxA9 to transactivate gene expression and is required for proper angiogenesis as evidenced by lethal hemorrhages in Meis1-deficient mice,41 is also not regulated by siRNA-mediated suppression of MLL in endothelial cells, although MLL binding to the Meis1 promoter region was shown previously in U937 cells.33 Interestingly, the genetic region encoding for the vascular remodelling factor Notch 4 is characterized by an endothelial cell-specific histone H3 K4 methylation pattern.42 Thus, other homeodomain or nonhomeodomain transcription factors beyond those investigated could mediate MLL-dependent endothelial sprout formation, possibly in a complex regulatory network that might well involve more than one single downstream target of MLL.
In summary, our data reveal a novel physiologic role for MLL in the regulation of endothelial-cell migration via HoxA9 and EphB4 and via HoxD3 and integrins, whereas its essential function for capillary sprout formation requires MLL-dependent targets beyond HoxA9 and HoxD3.
Authorship
Contribution: F.D. performed research and collected and analyzed data; L.R. designed research, collected data, and wrote the paper; A.M.Z. wrote and edited the paper; S.D. designed research and wrote and edited the paper; and C.U. designed research, collected data, and wrote the paper. All authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
F.D. and L.R. contributed equally to this study.
Correspondence: Stefanie Dimmeler, Molecular Cardiology, Department of Internal Medicine III, University of Frankfurt, Theodor-Stern-Kai 7, 60590 Frankfurt, Germany; e-mail: dimmeler@em.uni-frankfurt.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dorit Lüthje, Andrea Knau, and Nicole Konecny for expert technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (C.U.; SFB-TR23, subproject B5) and from the Deutsche Krebshilfe (no. 107154; L.R.).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal