Abstract
Neutrophil granulocytes (Gs) represent highly abundant and short-lived leukocytes that are constantly regenerated from a small pool of myeloid committed progenitors. Nuclear receptor (NR) family members are ligand-activated transcription factors that play key roles in cellular proliferation and differentiation processes including myelopoiesis. Retinoid X receptor alpha (RXRα) represents the predominant NR types I and II homo- and heterodimerization partner in myeloid cells. Here we show that human myeloid progenitors express RXRα protein at sustained high levels during macrophage colony-stimulating factor (M-CSF)–induced monopoiesis. In sharp contrast, RXRα is down-regulated during G-CSF–dependent late-stage neutrophil differentiation from myeloid progenitors. Down-regulation of RXRα is critically required for neutrophil development since ectopic RXRα inhibited granulopoiesis by impairing proliferation and differentiation. Moreover, ectopic RXRα was sufficient to redirect G-CSF–dependent granulocyte differentiation to the monocyte lineage and to promote M-CSF–induced monopoiesis. Functional genetic interference with RXRα signaling in hematopoietic progenitor/stem cells using a dominant-negative RXRα promoted the generation of late-stage granulocytes in human cultures in vitro and in reconstituted mice in vivo. Therefore, our data suggest that RXRα down-regulation is a critical requirement for the generation of neutrophil granulocytes.
Introduction
Polymorphonuclear neutrophils (PMNs) are regenerated from a small proliferative pool of lineage-committed bone marrow (BM) progenitor cells. These cells express myeloid lineage antigens (my+) but lack secondary granules, associated with the postproliferative neutrophil pool. Most (more than 90%) BM granulopoietic cells express the secondary granule marker lactoferrin (LF), and thus show a terminally differentiated phenotype.1,2 The large daily demand on differentiated LF+ PMNs is therefore met by the coordinated proliferation and differentiation of a small pool of immature my+LF− BM progenitors. Induced expression of LF protein has not been demonstrated in differentiation models of human myeloid cell lines. Furthermore, acute myeloid leukemia (AML) cells consistently lack LF.3,4 Therefore, still rather little is known on the molecular mechanisms underlying terminal neutrophil differentiation.
Nuclear hormone receptors (NRs), such as vitamin D3 receptor (VDR) and retinoic acid receptor (RAR), represent key transcriptional regulators of granulomonopoiesis. Addition of vitamin D3 (VD3) to AML cell lines or primary cells in vitro consistently induces monocyte (Mo) characteristics and inhibits alternative differentiation pathways.5 Conversely, addition of retinoids such as all-trans retinoic acid (ATRA) or 9-cis retinoic acid (9cRA) to AML blasts or primary granulocytic/monocytic (G/M) progenitors promotes granulopoiesis.6,7 In addition, transduction of BM cells with a COOH terminal–truncated retinoic acid receptor alpha (RARα) immortalizes myeloid progenitors.8 Similarly, acute promyelocytic leukemia (APL) is induced by aberrant expression of RARα fusion proteins.9,10 APL cells phenotypically resemble normal my+LF− BM cells.4,11 Supraphysiological concentrations of ATRA induce neutrophil differentiation of APL cells in vivo and induce clinical remission in APL.12 However, neutrophil development in vivo seems to take place in the absence of any RAR or VDR.13,14 The observation that vitamin A deficiency (VAD) leads to increased numbers of neutrophils in mice14,15 even indicates a repressive role of retinoids on neutrophil development under steady-state conditions.
RARα or VDR requires heterodimer formation with a second subfamily of NRs known as retinoic X receptors (RXRs).16 RXRα is the most abundant RXR in myeloid cells.17-19 Apart from RXRα heterodimers, RXRα homodimers also exist, and these seem to activate distinctive sets of genes.20,21 RXRα plays a nonredundant essential role in fetal development22,23 and conditional organ-specific knock-out revealed severe alterations in tissue differentiation.24,25 Approaches to investigate the role of RXRα in myeloid development showed that RXR ligands have relatively modest effects on their own, but synergize with RAR ligands in activating gene expression, inhibiting clonal growth, and inducing differentiation of myeloid leukemia cell lines,26-28 suggesting a subordinated role of RXRα in heterodimers. However, other studies showed that RXR agonists can induce terminal neutrophil differentiation of NB4 leukemia cells or myeloid progenitor cells expressing a dominant-negative RARα,29,30 thus supporting the existence of an independent RXR signaling pathway in some myeloid leukemia cell lines.
The regulation of RXRα protein expression and its functional consequences in primary cells undergoing normal neutrophil granulocyte versus monocyte differentiation has not been analyzed. Therefore, we asked 2 consecutive questions. First, is RXRα protein expression up- or down-regulated by primary human myeloid progenitor cells undergoing neutrophil granulocyte or monocyte differentiation in response to lineage specific cytokines? Second, does the level of RXRα expression in progenitor cells determine lineage fate decision and/or differentiation progression along neutrophil or monocyte pathways? We found that RXRα is down-regulated during G–colony-stimulating factor (CSF)–dependent neutrophil but not during M-CSF–dependent monocyte differentiation of human myeloid progenitors. Ectopic expression of RXRα impaired proliferation as well as terminal differentiation in the G but not Mo lineage. Moreover, we found that high RXRα expression in granulopoietic progenitors is sufficient to shift their lineage differentiation toward monocytes. Furthermore, neutrophil generation was promoted by functional inhibition using a dominant-negative RXRα in human primary cells in vitro and in mice in vivo.
Materials and methods
Cytokines and reagents for human in vitro cultures of CD34+ cells
Human stem cell factor (SCF), thrombopoietin (TPO), G-CSF, and macrophage (M)-CSF were purchased from PeproTech (London, United Kingdom); fms-related tyrosine kinase 3 ligand (Flt3L) was obtained from Amgen (Seattle, WA); interleukin (IL)–6 was kindly provided by Novartis Research Institute (Vienna, Austria). The following NR ligands were purchased from Sigma-Aldrich (Vienna, Austria): VD3, 9cRA, and ATRA. RXR agonist LG100268 was kindly provided by Ligand Pharmaceuticals (San Diego, CA).
Isolation of cord blood CD34+ cells and adult blood cells
Cord blood samples from healthy donors were collected during normal full-term deliveries. Approval was obtained from the Medical University of Vienna institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki. Cord blood mononuclear cells (MNCs) were isolated as previously described.31 CD34+ cells were isolated from MNCs by the magnetic-activated cell-sorter (MACS) Direct CD34 Progenitor Cell Isolation Kit (Miltenyi Biotech, Bergisch Gladbach, Germany). CD14+ monocytes were isolated from the MNC fraction of healthy adult blood by the MACS anti-PE kit (Miltenyi Biotech) according to the manufacturer's instructions. Blood granulocytes were obtained after red blood cell (RBC) lysis of the lower fraction after density gradient centrifugation at a purity greater than 98%.
CD34+ cell expansion and monocyte and granulocyte generation
Serum-free X-VIVO15 medium (BioWhittaker, Walkersville, MD) was supplemented with GlutaMAX (2.5 mM; Gibco/Invitrogen, Carlsbad, CA) and penicillin/streptomycin (P/S; 125 U/mL each). Progenitor cell expansion cultures contained Flt3L, SCF, and TPO, each at 50 ng/mL. To generate monocytes, 3-day– to 4-day–expanded CD34+ cells were plated at a density of 2 to 3 × 104/mL in serum-free medium supplemented with M-CSF (100 ng/mL), IL-6 (20 ng/mL), Flt3L (50 ng/mL), and SCF (20 ng/mL) in the absence or presence of VD3 (60 nM), 9cRA, ATRA, or LG100268 (100 nM each), or vehicle control (0.001% DMSO) for 8 days; ligand concentrations were selected according to initial titration experiments to give optimal cellular responses in the absence of cellular toxicity (data not shown). Identical substance concentrations were used in previous in vitro studies of primary cells.21,32 Granulocytes were generated from 2 to 3 × 104/mL expanded CD34+ progenitors in the presence of G-CSF (100 ng/mL), SCF (20 ng/mL), and, where indicated, VD3, 9cRA, ATRA, LG100268, or vehicle over a period of 12 to 14 days. Medium containing cytokines and ligands was renewed every 4 days throughout the culture period. X-VIVO15 was tested by high-performance liquid chromatography (HPLC) for vitamin A and radioimmunoassays (DiaSorin, Stillwater, MN) specific for the vitamin D metabolites 25(OH)VD3 and 1,25(OH)2VD3.
Murine BM transplantation
C57BL/6J mice were purchased from Harlan-Winkelmann (Borchen, Germany) and bred and maintained in the animal facility at the Medical University in Vienna. All animal experiments were done according to protocols approved by the Federal Ministry for Education, Science, and Art. Transduction and transplantation of BM cells was done as previously described.33,34 Briefly, C57BL/6J donor mice were given intraperitoneal injections of 5-fluorouracil (5-FU; Sigma, Aldrich, Vienna, Austria) at 10 mg/mL 4 days prior to BM isolation. After red blood cell lysis, BM cells were transduced with retroviral vectors. After infection (48 hours), BM cells were injected at 1 to 2 × 106 cells/mouse into the tail veins of lethally irradiated recipient mice. γ irradiation was performed as described.34 Thymus, lymph nodes, and spleens were removed from humanely killed animals, and single-cell suspensions were made. BM cells were harvested from reconstituted mice by flushing femurs and tibiae with PBS/2% FCS. After hypotonic lysis of RBCs (0.15 M NH4Cl, 1.0 mM KHCO3, and Na2EDTA [pH 7.2]), cells were analyzed by flow cytometry.
Retroviral vectors and gene transduction
cDNA of mouse RXRα was kindly provided by P. Chambon (Institut Génétique et de Biologie Moléculaire et Cellulaire [IGBMC], Strasbourg, France); cDNA for RXRαΔ (RXRα lacking amino acid [aa] 1-197) was obtained from a functional genetic retroviral cDNA library screen detailed elsewhere (S.T., Mario Kumerz, Florian Göbel, and H.S., manuscript in preparation). Briefly, U937Te cells were transduced with a retroviral cDNA library (human fetal liver library; Stratagen, La Jolla, CA), and U937Te cells that were refractory to VD3-induced up-regulation of CD14/CD11b were sorted. Retroviral cDNA inserts were amplified from single-cell clones and sequenced. RXRα or RXRαΔ cDNAs were subcloned into the MIG-R1 retroviral vector (obtained from H. Singh, Chicago, IL), upstream of an internal ribosome entry site (IRES) followed by green fluorescent protein (GFP). For generating ecotropic retrovirus, vectors were transfected by calcium-phosphate precipitation into the packaging cell line Phoenix-e. For generating amphotropic virus the Phoenix-Gag-Pol cell line (kindly provided by G. P. Nolan, Stanford, CA) was cotransfected with a vector of interest and a plasmid encoding the gibbon ape leukemia virus (GALV) envelope (obtained from D. B. Kohn, Los Angeles, CA). Infection of target cells was done as previously described.31 Briefly, RetroNectin (Takara Bio, Shiga, Japan)–coated non–tissue culture (TC) plates were coated with virus for 3 to 5 hours, followed by the addition of target cells (5 × 104-1 × 105 cells/well). Infections were repeated 2 to 3 times at intervals of 12 to 24 hours. Human CD34+ cells were infected in the presence of SCF, Flt3L, and TPO (50 ng/mL each), and within 60 to 72 hours after the first transduction cycle, cells were harvested and recultured in lineage-specific growth media. Murine BM cells enriched for hematopoietic stem cells (HSCs) were prestimulated for 24 hours in DMEM medium, 10% FCS, P/S, and l-glutamine (complete DMEM) supplemented with recombinant murine SCF (5 U/mL), IL-6 (10 000 U/mL), and IL-3 (6 U/mL) (PeproTech). Infection was done by incubation of 1 to 2 × 107 BM cells with viral supernatant supplemented with murine SCF, IL-3, and IL-6 at the same concentrations indicated for 2 to 3 rounds.
Cell lines
Phoenix cells were maintained in complete DMEM medium. HL60 cells expressing the ecotropic Moloney murine leukemia virus (MMLV) receptor (HL60e) were kindly provided by B. Fletcher (Gainesville, FL). U937T cells35 were obtained from G. Grosveld (Memphis, TN). U937Te cells were generated by transducing U937T cells with pBMNeco-receptorIRESmCD8α, and mCD8αhi cells were isolated by fluoresence-activated cell sorting (FACs) to obtain ecotropic virus–infectable cells. HL60e and U937Te cells were cultured in complete RPMI medium.
Flow cytometry
Flow cytometry staining and analysis were performed as previously described.31 For analyzing human cells, murine monoclonal antibodies (mAbs) of the following specificities were used: FITC-conjugated mAb specific for CD15 (BD Biosciences, Palo Alto, CA) and CD34 (BD Pharmingen, San Diego, CA); phycoerythrin (PE)–conjugated mAbs specific for CD54 (BD Pharmingen), CD11b (BD Biosciences), MPO, CD45-RA (Caltag Laboratories, Hamburg, Germany), and lactoferrin (Caltag, An der Grub, Austria); and biotinylated mAbs specific for CD11b (BD Pharmingen) and M-CSFR (CD115; R&D Systems GmbH, Wiesbaden, Germany); second-step reagents were either streptavidin (SA)–PerCP or SA-APC (BD Pharmingen), and allophycocyanin (APC)–conjugated mAbs specific for CD14 (Caltag Laboratories). Isotype control mAbs were kindly provided by O. Majdic (Vienna, Austria). For FACS sorting we used the BD FACSAria flow cytometer (BD Biosciences). For combined cell surface versus cytoplasmic stainings we used the reagent combination Fix & Perm from Caltag according to the manufacturer's recommendations. For analyzing murine cells, the following mAbs were used: PE-conjugated mAbs specific for CD8 and Gr-1 (Caltag); biotinylated mAbs specific for IgM, CD11b, (Caltag), and Ter119 (BD Pharmingen); Tri-Color–conjugated mAbs specific for CD4 (Caltag); and APC-conjugated mAbs specific for B220 (Caltag). The flow cytometric analysis was performed using a FACSCalibur (BD Biosciences), and data were analyzed with CellQuest Pro software (BD Biosciences). For in vitro cell proliferation studies the PKH26 red fluorescent cell linker-kit (Sigma-Aldrich, St Louis, MO) was used.
Western blot analysis
Whole-cell lysates were prepared as previously described.36 Briefly, U937 cells were lysed in an appropriate volume of lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 2.5 mM EDTA, and 1% Triton X-100) supplemented with 1× protease inhibitor cocktail set III (Calbiochem, San Diego, CA). The protein concentration of the extracts was determined using a Bradford-based protein assay (Bio-Rad, Hercules, CA). Prior to loading, SDS-loading buffer was added and samples were heated for 5 minutes to 95°C. To avoid autodegradation of protein in primary cell lysates of expanded CD34+ cells, G or Mo, cells were directly lysed in 1× SDS-loading dye at 95°C for 5 minutes. For Western blot analysis, 30 to 40 μg protein per lane was loaded on 12% SDS-PAA gels. Resolved proteins were transferred to a polyvinylene-difluoride membrane (Immobilon-P; Millipore, Billerica, MA) and probed with anti-RXRα (sc774; Santa Cruz Biotechnology, San Diego, CA) or antiactin (A-2066; Sigma-Aldrich, Vienna, Austria), followed by horseradish peroxidase (HRP)–conjugated goat anti–rabbit or goat anti–mouse IgG antibodies (Pierce Biotechnology, Rockford, IL). Detection was performed with the chemiluminescent substrate SuperSignal WestPico or WestDura (Pierce Biotechnology).
Statistical analysis
Statistical analysis was performed using a general linear model with the fixed factors NR ligand and vector, and random factor cord blood donor. Where indicated, a paired, 2-tailed Student t test was performed; a P value less than .05 was considered significant. Statistical calculations were carried out using SAS Version 8 (SAS Institute, Cary, NC).
Results
Reciprocal regulation of RXRα protein in granulopoiesis versus monopoiesis
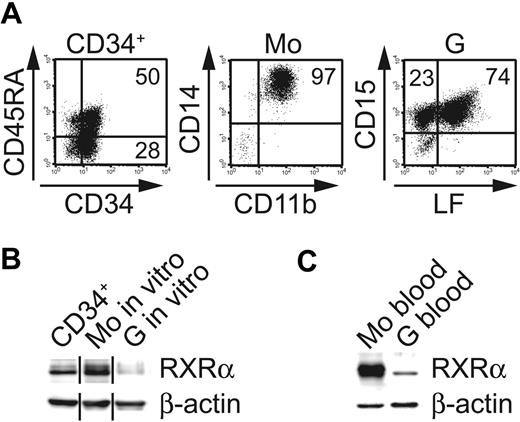
Previous studies demonstrated differentiation stage-dependent alterations in RXRα expression levels in leukemic hematopoiesis.17 However, it is not known whether endogenous RXRα expression is induced or inhibited in normal Mo-versus-G differentiation from progenitor cells. To address this question, we established specific serum-free differentiation cultures for the selective generation of Mo's or Gs from CD34+ human umbilical cord blood progenitor cells. These cultures were devoid of retinoids or VD3. CD34+ cells were expanded in the presence of cytokines promoting myeloid progenitors and subsequently cultured in Mo- or G-selective cytokine conditions. These cultures gave rise to high percentages of Mo's (> 80% CD14+CD11b+) or Gs (> 95% CD15+), respectively. G cultures were further analyzed for LF, a lysosomal protein that serves as a marker for the postmitotic granulocyte pool (from the myelocyte stage onwards). Most cells (70%-80%) in G cultures were LF+ (Figure 1A). These in vitro–generated cell populations (myeloid progenitors, Gs, and Mo's) were analyzed for RXRα expression (Figure 1B). RXRα could be detected in day-3–expanded CD34+ progenitors (Figure 1B). Relative to progenitor cells, in vitro–generated Gs showed substantially lower levels of RXRα (Figure 1B). Conversely, RXRα was found to be strongly expressed in FACS-sorted CD14hiCD11bhi Mo's (Figure 1A-B). Similarly, purified CD14+ Mo's and LF+ neutrophils from normal peripheral blood differed markedly in endogenous RXRα expression levels (ie, high expression in Mo's and low/no expression in Gs; Figure 1C). Therefore, RXRα protein expression is selectively down-regulated during neutrophil granulopoiesis from human myeloid progenitors.
Differential expression of RXRα by monocytes versus neutrophil granulocytes. (A) Representative phenotypic analysis of human CD34+ cells generated in serum-free 72-hour expansion cultures (left panel; FL, SCF, TPO) or after subsequent culture for 10 days in granulocyte conditions (right panel; G-CSF, SCF). Center diagram shows cells from monocyte cultures (M-CSF, IL-6, FL, SCF) after FACS sorting for CD11b and CD14. (B) Western blot analysis (RXRα or β-actin control) of in vitro–generated cells shown in panel A (ie, CD34+ cells after 72-hour expansion; Mo's or Gs). Different lanes from 1 blot were grouped. Data are representative of 5 experiments. (C) Western blot analysis (RXRα vs actin control) of more than 95% pure peripheral blood CD14+ Mo's or CD15+LF+ Gs. (B-C) Protein extracts were prepared using SDS loading dye (“Materials and methods”).
Differential expression of RXRα by monocytes versus neutrophil granulocytes. (A) Representative phenotypic analysis of human CD34+ cells generated in serum-free 72-hour expansion cultures (left panel; FL, SCF, TPO) or after subsequent culture for 10 days in granulocyte conditions (right panel; G-CSF, SCF). Center diagram shows cells from monocyte cultures (M-CSF, IL-6, FL, SCF) after FACS sorting for CD11b and CD14. (B) Western blot analysis (RXRα or β-actin control) of in vitro–generated cells shown in panel A (ie, CD34+ cells after 72-hour expansion; Mo's or Gs). Different lanes from 1 blot were grouped. Data are representative of 5 experiments. (C) Western blot analysis (RXRα vs actin control) of more than 95% pure peripheral blood CD14+ Mo's or CD15+LF+ Gs. (B-C) Protein extracts were prepared using SDS loading dye (“Materials and methods”).
Validation of a novel dominant-negative RXRα in myeloid cells
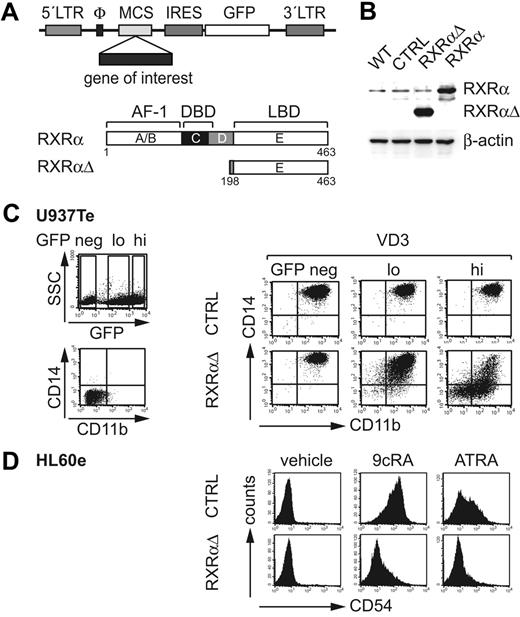
In order to study regulation of myeloid development by RXRα, wild-type RXRα, or an N-terminal–truncated RXRα, which lacks amino acids 1 to 197, including the ligand-independent transactivation domain (AF-1) and the DNA-binding domain (designated RXRαΔ), were inserted upstream of IRES-GFP into retroviral vectors (Figure 2A). Ectopic protein expression was confirmed using Western blot analysis of GFP+ FACS-sorted U937Te cells (Figure 2B). We previously isolated RXRαΔ from a functional genetic cDNA library screen based on its capacity to inhibit VD3-induced up-regulation of CD14 and CD11b in U937Te cells (S.T., Mario Kumerz, Florian Göbel, and H.S., manuscript in preparation). U937Te cell cultures transduced with control vector underwent homogenous differentiation into CD11bhiCD14hi Mo's, regardless of whether GFP−, GFPlo, or GFPhi fractions were analyzed (Figure 2C). Compared with these, GFPhi RXRαΔ-transduced cells showed strongly diminished CD14 and CD11b expression (Figure 2C). GFPlo cells also showed reduced expression of both marker molecules, albeit to a much lower extent (Figure 2C). These data indicate a dosage-dependent inhibitory effect of RXRαΔ on VD3-induced Mo differentiation of U937 cells. We next tested whether RXRαΔ similarly inhibits retinoid-induced myeloid cell differentiation. HL60e cells up-regulated CD54 (ICAM-1) in response to ATRA or 9cRA. GFPhi RXRαΔ-transduced cells effectively inhibited CD54 induction (Figure 2D). Therefore, RXRαΔ inhibits VD3- and retinoid-induced up-regulation of marker molecules in myeloid cell lines in a dosage-dependent manner.
Retroviral expression of RXRα constructs. (A) Schematic representation of full-length wild-type RXRα and truncated RXRα (RXRαΔ) containing the ligand binding domain (LBD) lacking the 5′ AF-1 and DNA-binding domains (DBD). cDNAs were inserted into a retroviral backbone 5′ of an IRES-GFP cassette. (B) Western blot analysis of untransduced (WT) or gene-transduced U937Te cells. GFP+ cells were sorted prior to analysis (CTRL indicates empty control vector; RXRαΔ- or RXRα-encoding vectors). Protein extracts were prepared using cell lysis buffer (“Materials and methods”). (C) U937Te cells were transduced with RXRαΔ or empty control vector. After transduction (48 hours), cells were stimulated with VD3 (60 nM) for 48 hours and were then analyzed for GFP versus CD11b and CD14 surface expressions. GFPhi, GFPlo, and GFP− cells were separately gated and analyzed for CD11b versus CD14. (D) Representative FACS analysis of CD54 of gene-transduced GFPhi HL60e cells. HL60e cells were gene transduced with RXRαΔ or empty control vector (CTRL). After transduction (48 h), cells were stimulated with 9cRA (100 nM), ATRA (100 nM), or vehicle (0.001% DMSO). Data in panel B are representative of 2 experiments. Data in panels C and D are representative of 5 experiments.
Retroviral expression of RXRα constructs. (A) Schematic representation of full-length wild-type RXRα and truncated RXRα (RXRαΔ) containing the ligand binding domain (LBD) lacking the 5′ AF-1 and DNA-binding domains (DBD). cDNAs were inserted into a retroviral backbone 5′ of an IRES-GFP cassette. (B) Western blot analysis of untransduced (WT) or gene-transduced U937Te cells. GFP+ cells were sorted prior to analysis (CTRL indicates empty control vector; RXRαΔ- or RXRα-encoding vectors). Protein extracts were prepared using cell lysis buffer (“Materials and methods”). (C) U937Te cells were transduced with RXRαΔ or empty control vector. After transduction (48 hours), cells were stimulated with VD3 (60 nM) for 48 hours and were then analyzed for GFP versus CD11b and CD14 surface expressions. GFPhi, GFPlo, and GFP− cells were separately gated and analyzed for CD11b versus CD14. (D) Representative FACS analysis of CD54 of gene-transduced GFPhi HL60e cells. HL60e cells were gene transduced with RXRαΔ or empty control vector (CTRL). After transduction (48 h), cells were stimulated with 9cRA (100 nM), ATRA (100 nM), or vehicle (0.001% DMSO). Data in panel B are representative of 2 experiments. Data in panels C and D are representative of 5 experiments.
Granulopoiesis is inhibited by specific activation of ectopic wild-type RXRα in primary cell cultures
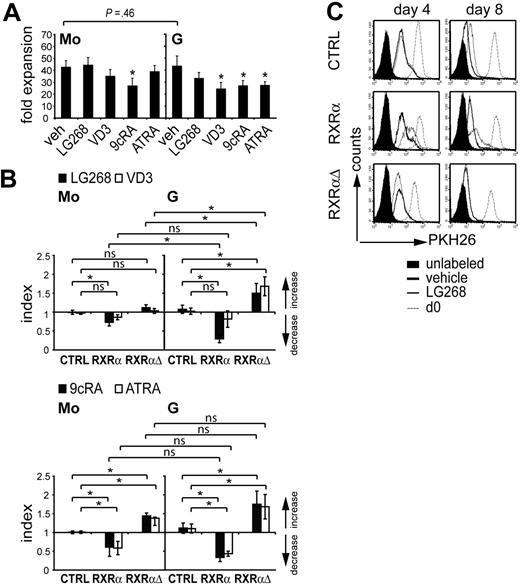
RXRα was down-regulated during granulocyte but not monocyte differentiation of primary myeloid progenitors (Figure 1A-B). Progenitor cells in both culture systems (G or Mo) proliferated vigorously and gave rise to similar cell numbers (Figure 3A). Therefore, we asked whether down-regulation of RXRα in G cultures is required for their proliferative capacity. Thus, CD34+ cells were transduced with RXRα, RXRαΔ, or control vector, and were cultured under Mo- or G-specific defined serum-free conditions (as shown in Figure 1A). To functionally activate RXRα, we added the RXRα-specific agonist LG100268.37 For comparison, VD3, retinoids, or vehicle only (0.001% DMSO) were added to parallel cultures. The addition of LG100268, VD3, or retinoic acid (RA) to untransduced cultures did not impair or only moderately impaired overall cell numbers (Figure 3A). GFP expression levels were monitored during culture to assess relative loss or enrichment of gene-transduced cells (“Index” is a measure for the relative increase or decrease of GFP+ [gene transduced] cells). In the absence of NR ligands there were no detectable changes in the percentage of GFP+ cells for either condition (RXRα, RXRαΔ, or control; data not shown). Addition of the RXRα-selective ligand LG100268 led to a profound loss of RXRα-transduced cells in G but not in Mo cultures (Figure 3B; top right panel vs left panel). Conversely, LG100268 led to a relative increase in the percentage of RXRαΔ-transduced cells, and this effect was more pronounced in G cultures compared with Mo cultures (Figure 3B). VD3 impaired cell proliferation in G cultures (Figure 3A). In line with this, we observed increased percentages of RXRαΔ-transduced cells in G cultures in the presence of VD3 (Figure 3B). Conversely, VD3 had little effect on the frequency of gene-transduced cells in Mo cultures (Figure 3B). Addition of retinoids led to diminished frequencies of RXRα-transduced cells in G and Mo conditions (Figure 3B; bottom right panel versus left panel). This was reverted by ectopic expression of RXRαΔ (Figure 3B). Therefore, ligation of ectopic RXRα by the RXRα-selective agonist LG100268 inhibited cell generation in G cultures. In addition, the frequency of RXRα-transduced granulopoietic cells was consistently diminished by all other ligands.
Ectopic RXRα impairs cell proliferation in granulocyte-specific cultures. (A) Human CD34+ progenitor cells that were expanded for 72 hours were cultured for 10 days in the presence of M-CSF, IL-6, FL, and SCF (Mo cultures), or G-CSF plus SCF (G cultures). NR ligands were added at initiation (day 0) of Mo and G cultures. Fold expansion represents the ratio of total cell numbers at day 10 over total cell number at day 0. Values represent the mean and SD of 6 (Mo) or 4 (G) independent experiments. (B) CD34+ cells were transduced with RXRαΔ or empty control vector shown in Figure 2A. After gene transduction cells were subcultured (48 hours) in Mo- or G-specific serum-free cultures in the presence of the RXR-selective agonist LG100268 (100 nM), VD3 (60 nM), 9cRA (100 nm), ATRA (100 nm), or vehicle (veh). The percentage of GFP+ cells was determined by FACS. Index indicates the ratio of the percentage of GFP+ cells in the presence of NR ligands over the percentage of GFP+ cells in the absence of ligand. Bar diagrams represent the mean and SD calculated from 6 independent experiments (A-B) *Significant differences at P < .05 according to a general linear statistical model; ns indicates not significant. (C) CD34+ cells were transduced as in panel B. After gene transduction (48 hours), cells were harvested and labeled with PKH26. Cells were then cultured in G-specific cultures in the presence or absence of the RXRα-selective agonist LG100268 (100 nM). PKH26 fluorescence was analyzed by FACS at days 0, 4, or 8. Histograms represent gated GFP+ cells analyzed for PKH26 fluorescence intensity.
Ectopic RXRα impairs cell proliferation in granulocyte-specific cultures. (A) Human CD34+ progenitor cells that were expanded for 72 hours were cultured for 10 days in the presence of M-CSF, IL-6, FL, and SCF (Mo cultures), or G-CSF plus SCF (G cultures). NR ligands were added at initiation (day 0) of Mo and G cultures. Fold expansion represents the ratio of total cell numbers at day 10 over total cell number at day 0. Values represent the mean and SD of 6 (Mo) or 4 (G) independent experiments. (B) CD34+ cells were transduced with RXRαΔ or empty control vector shown in Figure 2A. After gene transduction cells were subcultured (48 hours) in Mo- or G-specific serum-free cultures in the presence of the RXR-selective agonist LG100268 (100 nM), VD3 (60 nM), 9cRA (100 nm), ATRA (100 nm), or vehicle (veh). The percentage of GFP+ cells was determined by FACS. Index indicates the ratio of the percentage of GFP+ cells in the presence of NR ligands over the percentage of GFP+ cells in the absence of ligand. Bar diagrams represent the mean and SD calculated from 6 independent experiments (A-B) *Significant differences at P < .05 according to a general linear statistical model; ns indicates not significant. (C) CD34+ cells were transduced as in panel B. After gene transduction (48 hours), cells were harvested and labeled with PKH26. Cells were then cultured in G-specific cultures in the presence or absence of the RXRα-selective agonist LG100268 (100 nM). PKH26 fluorescence was analyzed by FACS at days 0, 4, or 8. Histograms represent gated GFP+ cells analyzed for PKH26 fluorescence intensity.
Ligation of ectopic RXRα confers a proliferative disadvantage to primary granulopoietic cells
We next asked whether the reduction of RXRα-transduced cells in the presence of its selective agonist in G cultures involves an impairment of cell proliferation. PKH26 dye dilution assessed by FACS is a measure for cell proliferation. Gated GFP+ cells were analyzed for PKH26 mean fluorescence intensity (MFI) at days 0, 4, and 8 of culture under G-specific conditions. In the absence of LG100268, cells showed similar proliferation under all conditions (control, RXRα, or RXRαΔ; Figure 3C). Addition of LG100268 reduced proliferation by day 4 for RXRα but not for control or RXRαΔ (Figure 3C). Furthermore, RXRαΔ promoted cell proliferation in the presence of LG100268 (Figure 3C). At day 8, an impairment of cell proliferation by LG100268 was observed even in control-transduced cells and was further enhanced in RXRα-transduced cells (Figure 3C). Together, these experiments show that ectopic RXRα impairs granulocyte generation, and this effect is associated with inhibition of proliferation.
Ectopic RXRα inhibits the generation of late LF+ granulocytes from primary myeloid progenitors
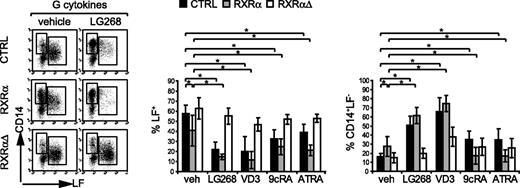
Intracellular LF marks the late (postmitotic) pool of neutrophil granulocytes. LF+ neutrophils express substantially lower levels of RXRα protein than do CD14+ monocytes (Figure 1A-B). Furthermore, ectopic expression of RXRα inhibited granulopoietic cell proliferation (Figure 3). We next asked whether, in addition to a negative effect on proliferation, ectopic RXRα also influences G differentiation. GFP+ cells from G cultures were analyzed for LF versus CD14 expression (Figure 4; FACS and bar diagrams). Control-transduced cells were subdivided into a major portion of LF+ cells and a small portion of CD14+LF− cells. LG100268 substantially reduced the frequency of LF+ cells (Figure 4; for percentages, see bar diagrams). Unliganded ectopic RXRα slightly decreased the percentage of LF+ cells and increased the percentage of CD14+LF− cells, an effect further enhanced by addition of LG100268 (Figure 4). Qualitatively similar reductions in the percentage of LF+ cells were observed for VD3 or the retinoids 9cRA or ATRA (Figure 4; top bar diagram). However, differences were observed for the frequency of CD14+LF− cells for LG100268 and VD3 versus retinoids. When added to RXRα-transduced cultures, ATRA or 9cRA decreased rather than increased the percentage of CD14+LF− cells (Figure 4; bar diagrams). As a result, in the presence of retinoids ectopic RXRα favored the generation of LF−CD14− cells (representing most of all GFP+ gated gene-transduced cells). Most of these cells expressed myeloperoxidase (MPO) and represented early myeloid cells (data not shown). Dominant-negative RXRαΔ consistently reverted phenotypic changes induced by RXRα, in that RXRαΔ restored terminal granulopoiesis in LG100268-treated cells as well as VD3- and RA-treated cells (Figure 4; bar diagrams). In aggregate, forced expression of RXRα inhibits the generation of LF+ late granulocytes in serum-free G-specific cultures. In addition, elevated RXRα alone is sufficient to increase the frequency of CD14+ monocytes in G cultures, and this effect is enhanced by specific RXRα ligation.
Ectopic RXRα impairs the induction of LF+ granulocytes. Human CD34+ cells were transduced with vectors encoding RXRα, RXRαΔ, or empty control vector (CTRL). After transduction (48 hours), cells were subcultured in serum-free G-specific cultures in the absence (vehicle) or presence of NR ligands (100 nM) as indicated. Generated cells were analyzed at day 12 by FACS for GFP versus intracellular LF and cell-surface CD14. FACS diagrams represent gated GFP+ cells analyzed for LF versus CD14. Data are representative of 6 independent experiments. Bars represent the mean and SD of the percentage of LF+ or the percentage of LF−CD14+ cells among gated GFP+ cells of 6 independent experiments. *Significant differences at P < .05 according to a general linear statistical model; ns indicates not significant.
Ectopic RXRα impairs the induction of LF+ granulocytes. Human CD34+ cells were transduced with vectors encoding RXRα, RXRαΔ, or empty control vector (CTRL). After transduction (48 hours), cells were subcultured in serum-free G-specific cultures in the absence (vehicle) or presence of NR ligands (100 nM) as indicated. Generated cells were analyzed at day 12 by FACS for GFP versus intracellular LF and cell-surface CD14. FACS diagrams represent gated GFP+ cells analyzed for LF versus CD14. Data are representative of 6 independent experiments. Bars represent the mean and SD of the percentage of LF+ or the percentage of LF−CD14+ cells among gated GFP+ cells of 6 independent experiments. *Significant differences at P < .05 according to a general linear statistical model; ns indicates not significant.
Ectopic RXRα promotes Mo differentiation
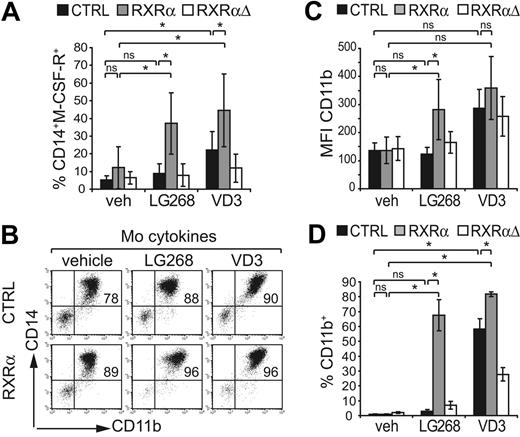
M-CSFR (CD115) expression marks monopoietic cells. In line with an increased frequency of CD14+LF− cells in RXRα-transduced G cultures (Figure 4; bottom bar diagram), we also observed elevated percentages of CD14+CD115+ cells in the presence of LG100268 (Figure 5A). Since CD14+ Mo's show increased endogenous RXRα expression, RXRα up-regulation might enhance Mo differentiation. Therefore, we next analyzed whether LG100268 also induces Mo features under Mo-promoting conditions. Serum-free M-CSF–dependent cultures gave rise to high percentages of CD14hiCD11bhi cells (Figure 5B; Table 1). Ectopic RXRα in the presence of LG100268 or VD3 further increased the percentage of CD14+CD11b+ cells (Figure 5B; Table 1). Furthermore, ectopic RXRα led to a significant increase in CD11b expression densities (MFI CD11b) in the presence of LG100268 (Figure 5C; Table 2). In comparison, stimulation of control or RXRα-transduced cells with VD3 consistently enhanced MFI CD11b values (Table 2). HL60e is a leukemia cell line arrested at the promyelocyte stage and showed moderate endogenous RXRα expression levels (data not shown). Ectopic RXRα markedly induced CD11b and CD14 by HL60e cells in the presence of its specific agonist LG100268 (Figure 5D; data not shown). Together, these results show that increased RXRα promotes monopoiesis in primary human progenitor cells.
Ectopic RXRα augments monocyte features. (A) Human CD34+ progenitor cells were transduced with vectors encoding RXRα, RXRαΔ, or empty control (CTRL) under progenitor expansion conditions. After transduction (48 hours), cells were subcultured in serum-free G-specific cultures in the absence (veh) or presence of LG100268 (100 nM) or VD3 (60 nM) as indicated. Generated cells were analyzed by FACS for GFP versus CD14 and CD115 (M-CSFR). Bars represent the mean percentage and SD of the percentage of CD14+M-CSFR+ cells among gated GFP+ cells observed in 4 independent experiments. (B-C) CD34+ cells were transduced with empty control (CTRL) RXRα- or RXRαΔ-encoding vectors under progenitor expansion conditions. After transduction (48 hours), cells were replated in serum-free Mo-specific cultures in the absence (vehicle) or presence of LG100268 or VD3 as indicated. GFP+ cells were analyzed by FACS for CD11b versus CD14. (B) One representative of 5 independent experiments is shown (P < .05 for the percentage of CD11b+CD14+). Bars in panel C represent the average CD11b mean fluorescence intensity and SD calculated from 5 independent experiments. (D) HL60e cells transduced with empty control vector, RXRα, or RXRαΔ. Cells were cultured for 48 hours in the absence or presence of LG100268 or VD3 as indicated. GFP+ cells were gated and analyzed for CD11b. Bars represent the mean and SD calculated from 3 independent experiments. (A-D) *Significant differences at P < .05 according to a general linear statistical model; ns indicates not significant.
Ectopic RXRα augments monocyte features. (A) Human CD34+ progenitor cells were transduced with vectors encoding RXRα, RXRαΔ, or empty control (CTRL) under progenitor expansion conditions. After transduction (48 hours), cells were subcultured in serum-free G-specific cultures in the absence (veh) or presence of LG100268 (100 nM) or VD3 (60 nM) as indicated. Generated cells were analyzed by FACS for GFP versus CD14 and CD115 (M-CSFR). Bars represent the mean percentage and SD of the percentage of CD14+M-CSFR+ cells among gated GFP+ cells observed in 4 independent experiments. (B-C) CD34+ cells were transduced with empty control (CTRL) RXRα- or RXRαΔ-encoding vectors under progenitor expansion conditions. After transduction (48 hours), cells were replated in serum-free Mo-specific cultures in the absence (vehicle) or presence of LG100268 or VD3 as indicated. GFP+ cells were analyzed by FACS for CD11b versus CD14. (B) One representative of 5 independent experiments is shown (P < .05 for the percentage of CD11b+CD14+). Bars in panel C represent the average CD11b mean fluorescence intensity and SD calculated from 5 independent experiments. (D) HL60e cells transduced with empty control vector, RXRα, or RXRαΔ. Cells were cultured for 48 hours in the absence or presence of LG100268 or VD3 as indicated. GFP+ cells were gated and analyzed for CD11b. Bars represent the mean and SD calculated from 3 independent experiments. (A-D) *Significant differences at P < .05 according to a general linear statistical model; ns indicates not significant.
Ectopic RXRα and LG100268 cooperatively augment CD14+CD11b+ Mo's
| Experiment no. . | CD11b+CD14+, %* . | |||||
|---|---|---|---|---|---|---|
| Control . | RXRα . | |||||
| Vehicle . | LG100268 . | VD3 . | Vehicle . | LG100268 . | VD3 . | |
| 1† | 77 | 77 | 88 | 93 | 94 | 95 |
| 2 | 87 | 90 | 93 | 90 | 90 | 96 |
| 3 | 64 | 70 | 77 | 79 | 89 | 93 |
| 4 | 84 | 85 | ND | 93 | 96 | 98 |
| 5 | 79 | 88 | 90 | 89 | 95 | 95 |
| 6 | 75 | 78 | 89 | 76 | 93 | 93 |
| Experiment no. . | CD11b+CD14+, %* . | |||||
|---|---|---|---|---|---|---|
| Control . | RXRα . | |||||
| Vehicle . | LG100268 . | VD3 . | Vehicle . | LG100268 . | VD3 . | |
| 1† | 77 | 77 | 88 | 93 | 94 | 95 |
| 2 | 87 | 90 | 93 | 90 | 90 | 96 |
| 3 | 64 | 70 | 77 | 79 | 89 | 93 |
| 4 | 84 | 85 | ND | 93 | 96 | 98 |
| 5 | 79 | 88 | 90 | 89 | 95 | 95 |
| 6 | 75 | 78 | 89 | 76 | 93 | 93 |
CD34+ cells were transduced with empty control or RXRα-encoding vectors under progenitor expansion conditions. After transduction (48 hours), cells were replated in serum-free Mo-specific cultures in the absence (Vehicle) or presence of LG100268 or VD3 as indicated. ND indicates not determined.
GFP+ cells were analyzed by FACS for CD11b and CD14.
Six independent cord blood donor experiments are shown.
Ligation of ectopic RXRα increases the MFI of CD11b
| Experiment no. . | MFI CD11b* . | |||||
|---|---|---|---|---|---|---|
| Control . | RXRα . | |||||
| Vehicle . | LG100268 . | VD3 . | Vehicle . | LG100268 . | VD3 . | |
| 1† | 166 | 123 | 303 | 132 | 332 | 364 |
| 2 | 130 | 137 | 241 | 112 | 232 | 291 |
| 3 | 124 | 99 | 202 | 114 | 179 | 212 |
| 4 | 160 | 157 | 305 | 220 | 447 | 435 |
| 5 | 97 | 102 | 380 | 105 | 220 | 493 |
| Experiment no. . | MFI CD11b* . | |||||
|---|---|---|---|---|---|---|
| Control . | RXRα . | |||||
| Vehicle . | LG100268 . | VD3 . | Vehicle . | LG100268 . | VD3 . | |
| 1† | 166 | 123 | 303 | 132 | 332 | 364 |
| 2 | 130 | 137 | 241 | 112 | 232 | 291 |
| 3 | 124 | 99 | 202 | 114 | 179 | 212 |
| 4 | 160 | 157 | 305 | 220 | 447 | 435 |
| 5 | 97 | 102 | 380 | 105 | 220 | 493 |
CD34+ cells were transduced with empty control or RXRα-encoding vectors under progenitor expansion conditions. After transduction (48 hours), cells were replated in serum-free Mo specific cultures in the absence (Vehicle) or presence of LG100268 or VD3 as indicated.
GFP+ cells were analyzed by FACS for the MFI of CD11b.
Five independent cord blood donor experiments.
RXRαΔ promotes granulocyte generation from hematopoietic stem cells in mice in vivo
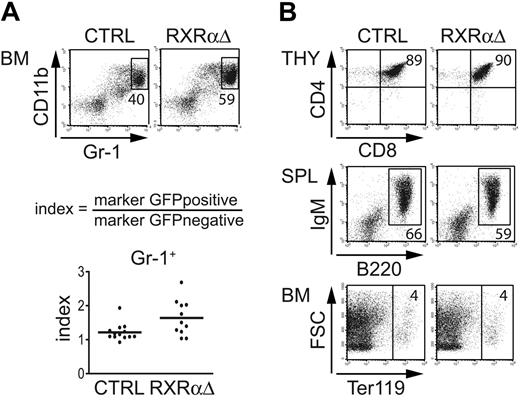
Our data indicate that down-regulation of RXRα is required for neutrophil generation from primary human progenitor cells. To analyze whether down-regulation of RXRα is required for terminal neutrophil development in vivo, we transduced murine BM cells with RXRαΔ-IRES-GFP or empty control vector followed by BM reconstitution experiments. Engrafted cells were analyzed for GFP and informative lineage marker molecules. To normalize for interindividual variations we calculated ratios of gene-transduced (GFP+) over nontransduced (GFP−) fractions observed in individual mice. We found that RXRαΔ-transduced splenic and BM fractions contained elevated percentages of Gr-1hiCD11b+ granulocytes compared with those of control samples (Figure 6A; data not shown). Thus, inhibition of NR signaling by ectopic expression of a truncated RXRα molecule increased the frequency of Gs in vivo. We further analyzed whether RXRαΔ expression might interfere with other lineages. Similar populations of thymocytes (CD8+CD4+), B cells (B220+IgM+), erythroid cells (Ter119+), and splenic dendritic cells were observed among GFP+ cells for RXRαΔ- and control-transduced reconstituted mice (Figure 6B; data not shown). In addition, we analyzed GFP+ BM fractions from RXRαΔ-transduced mice for the percentage of c-kit+lin− progenitor/stem cells. Similar percentages of c-kit+lin− cells were observed in control-transduced or RXRαΔ-transduced recipient BM samples (data not shown). In summary, down-regulation of RXRα function enhances terminal neutrophil development in mice in vivo, but does not alter progenitor cell frequency or alternative lymphoid or erythroid development.
RXRαΔ promotes granulopoiesis in vivo. (A) Murine BM cells transduced with RXRαΔ-IRES-GFP or empty control vector were injected into irradiated recipients. BM cells were analyzed between days 38 and 66 after BM transplantation. FACS diagrams represent gated GFP+ cells analyzed for Gr-1 versus CD11b. Values depict the percentages of total Gr-1hiCD11b+ BM cells (top panel). Values in index charts were calculated according to the formula shown. Each dot represents data from 1 mouse (bottom diagram). Horizontal bars represent the mean values. P < .05 according to a paired 2-tailed Student t test. (B) Analysis of gated GFP+ cells (CD4+CD8+ thymocytes, B220+IgM+ splenocytes, and Ter119+ BM erythroid cells) from mice reconstituted with CTRL- or RXRαΔ-transduced BM. Numbers depict percentages of cells in each quadrant or region. Data are representative of 3 independent BMT experiments (n = 11 mice).
RXRαΔ promotes granulopoiesis in vivo. (A) Murine BM cells transduced with RXRαΔ-IRES-GFP or empty control vector were injected into irradiated recipients. BM cells were analyzed between days 38 and 66 after BM transplantation. FACS diagrams represent gated GFP+ cells analyzed for Gr-1 versus CD11b. Values depict the percentages of total Gr-1hiCD11b+ BM cells (top panel). Values in index charts were calculated according to the formula shown. Each dot represents data from 1 mouse (bottom diagram). Horizontal bars represent the mean values. P < .05 according to a paired 2-tailed Student t test. (B) Analysis of gated GFP+ cells (CD4+CD8+ thymocytes, B220+IgM+ splenocytes, and Ter119+ BM erythroid cells) from mice reconstituted with CTRL- or RXRαΔ-transduced BM. Numbers depict percentages of cells in each quadrant or region. Data are representative of 3 independent BMT experiments (n = 11 mice).
Discussion
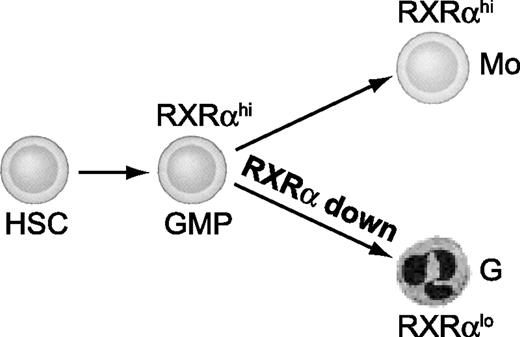
Here we demonstrate that normal granulomonopoiesis is regulated by the expression level of RXRα in human and mouse. We found that RXRα protein is down-regulated during neutrophil development. Conversely, RXRα levels remain high in monocyte differentiation of primary human myeloid progenitors. Sustained high expression of RXRα in cells undergoing neutrophil differentiation impaired proliferation as well as terminal differentiation. In addition, we found that high RXRα expression in G-CSF–dependent cultures is sufficient to shift their lineage differentiation to monocytes. In line with this, ectopic RXRα augmented M-CSF–dependent monocyte generation. In contrast, neutrophil generation was promoted by functional inhibition of RXRα signaling in vitro and in vivo. Since M-CSF– versus G-CSF–dependent signals differentially regulate RXRα protein expression, and RXRα signaling is sufficient to redirect G-to-Mo differentiation, our data strongly suggest that RXRα down-regulation is a key requirement for normal neutrophil generation (Figure 7).
RXRα is down-regulated during neutrophil granulocyte differentiation. Myeloid progenitor cells show sustained high RXRα expression as they differentiate to monocytes. Conversely, RXRα is down-regulated concomitant with granulocyte differentiation. We here show that RXRα down-regulation is critically required for the development of neutrophils.
RXRα is down-regulated during neutrophil granulocyte differentiation. Myeloid progenitor cells show sustained high RXRα expression as they differentiate to monocytes. Conversely, RXRα is down-regulated concomitant with granulocyte differentiation. We here show that RXRα down-regulation is critically required for the development of neutrophils.
We show that down-regulation of RXRα protein is required for neutrophil differentiation. Previous studies were hampered by the fact that the RXRα-null mutation in mice is lethal at the embryonic stage.22 Most current knowledge on NR signaling in granulomonopoiesis is based on differentiation studies of myeloid cell lines.8,38,39 However, human cell lines unequivocally fail to express secondary granule markers in response to exogenous differentiation stimuli. In addition, many cell line models aberrantly express truncated RARα molecules, thus showing perturbed NR signaling.6,8
RXRα redirected G-to-Mo differentiation in primary CD34+-derived cultures. In contrast to G differentiation, we observed sustained high RXRα expression levels in Mo's relative to their progenitors. Also, in vivo, human peripheral blood Mo's showed much higher RXRα protein levels than did neutrophils. In line with this, forced expression of RXRα in G cultures was sufficient to redirect these cells to become Mo's. A similar shift from G-to-Mo differentiation was observed for “master” transcriptional regulators of monocytes/macrophages.40,41 How RXRα interacts with these factors remains to be shown. It is interesting to speculate that subcellular localization of RXRα (cytoplasmic versus nuclear compartment) might differ during myeloid development.42,43 This may contribute to myeloid lineage fate decisions. Future studies should address this possibility.
We observed marked proliferation inhibition of G-CSF–dependent cultures concomitant with G-to-Mo redirection by liganded ectopic RXRα. Conversely, proliferation of M-CSF–induced Mo's was not affected by liganded ectopic RXRα. These data suggest that overexpression of RXRα is incompatible with G proliferation, but is not generally antiproliferative. In vivo, G precursors have to undergo high rates of multiplication to replenish neutrophils on a daily basis. Our data suggest that RXRα down-regulation is not only required for transition of myeloid progenitors to late-stage neutrophils (LF+), but also for ordered proliferation of neutrophil precursors.
The serum-free culture models used by us to generate Gs or Mo's from primary human myeloid progenitors were devoid of exogenous NR ligands. This facilitated studying a potential interplay of cytokine receptor signals and RXRα protein regulation in normal granulomonopoiesis. We found that G-CSF cosignaling diminished RXRα. Conversely, M-CSF increased RXRα expression. Thus, G or Mo lineage-inducing cytokine combinations differentially regulate RXRα expression, and RXRα protein abundance might then solidify G-versus-Mo lineage differentiation downstream of cytokine signals. While virtually nothing was known on RXRα protein levels during normal granulomonopoiesis, our data are in line with previous observations demonstrating that retinoids and cytokines cooperate in the regulation of target genes.44,45 Furthermore, it was recently shown that RXRα is “desubordinated” in RAR/RXR heterodimers mediated by cyclic AMP/protein kinase A activation, a stimulus inducing Mo differentiation in primary cells and cell lines.46-49 Therefore, RXRα may modulate cytokine-induced signaling and may itself be regulated by cytokines.
We found that expression of a “dominant-negative” RXRα promoted neutrophil generation and re-established late Gs in the presence of retinoids in vitro and in vivo, but did not alter progenitor cell frequency or alternative lymphoid or erythroid development. Consistent with our results, vitamin A deficiency as well as administration of an RARα antagonist promotes neutrophil generation in vivo.14,15 Therefore, RXRα/RARα heterodimers seem to constitute a negative regulatory pathway for neutrophil generation in vivo. Down-regulation of RXRα protein will desensitize developing neutrophils to retinoids. Thus, our data suggest that developing neutrophils have adopted RXRα down-regulation to escape repression by retinoids in vivo. This is supported by the inhibition of proliferation and terminal differentiation of Gs observed in RXRα-expressing primary cell cultures.
We observed that the RXRα-specific ligand LG100268 displayed effects similar to those of VD3 in vitro, suggesting activation of RXR/VDR heterodimers. Although we cannot exclude up-regulation of VDR by RXR ligation in G cultures, the activation of VDR was ruled out by using a serum-free culture medium devoid of any VD3 and retinoid derivatives (S. Brecht [Cambrex Bio Science, Verviers, France], written personal communication, April 7, 2006; and independent confirmation at our institution by HPLC and radioimmunoassay [RIA] analysis for RA or VD3, respectively; data not shown). Therefore, the effects on G and Mo differentiation observed upon RXRα ligation were mediated via RXRα homodimers or alternatively, via RXRα heterodimers with “permissive” partners. In support for this assumption, activation of “nonpermissive” RXRα heterodimers, such as RXR/VDR by LG100268, was not reported to our knowledge.
RXRα transduction will preferentially result in RXRα homodimer formation. We showed that specific ligation of RXRα induces a monocyte differentiation program. Since we found that monocytes show high endogenous RXRα protein expression, RXRα homodimer signaling might play an important physiologic role in monopoiesis. Interestingly, RXRα homodimers can activate proliferator-activated receptor (PPAR) γ target genes even in the presence of endogenous PPARγ,50 suggesting that homodimers can be activated even in the presence of a “permissive” partner, such as PPARγ. In contrast to an RXRα-selective ligand, which redirected G-to-Mo differentiation, retinoids known to signal via “nonpermissive” RXRα/RARα heterodimers failed to do so. Instead, retinoid addition to RXRα-transduced G cultures induced early MPO+LF−CD14− myeloid progenitors phenotypically resembling promyelocytes, supporting previous observations.51 Thus, high RXRα expression in myeloid progenitors might prime these cells to become monocytes or stay immature depending on whether RXRα homodimers or RXRα/RARα heterodimers are activated.
In conclusion, our study demonstrates that down-regulation of RXRα is critically required for normal neutrophil differentiation in human in vitro cultures as well as in mouse in vivo, whereas sustained high expression of RXRα redirects myeloid progenitors toward the Mo lineage. In APL, similar to normal hematopoiesis, RXRα is required, but is sequestered by aberrantly expressed PML/RARα fusion proteins.52 Furthermore, RXRα is expressed at elevated levels in patients with AML who exhibit monocyte features, suggesting a role for RXRα as a therapeutic target.17 Indeed, RXR-selective ligands hold promise for the treatment of AML. New concepts propose enhancer drugs to improve this therapy by targeting pathways that functionally activate subordinated RXRα in leukemia cells.53 As suggested by our study, signaling pathways that modulate RXRα protein expression levels in myeloblasts might be exploited additionally to further improve existing therapeutic concepts.
Authorship
Author contributions: S.T. designed and performed research, analyzed data, and wrote the paper; C.K. performed research; B.P. and A.J. contributed analytical tools; W.E. designed research; T.B. analyzed data; and H.S. designed research and wrote the paper.
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: Herbert Strobl, Institute of Immunology, Medical University Vienna, Lazarettgasse 19, A-1090 Vienna, Austria; e-mail: herbert.strobl@meduniwien.ac.at.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Acknowledgments
We thank the collaborating nurses and doctors from the obstetric departments at Kaiser Franz Josef Hospital and the General Hospital, Vienna. We thank M. Leibowitz (Ligand Pharmaceuticals) for providing LG100268, P. Chambon for the RXRα cDNA, and G. Grosveld for providing U937T cells. Furthermore, we thank C. Sillaber and H. Beug for critically reading the manuscript.
Supported by START grants Y-156 (to H.S.) and Y-163 (to W.E.), SFB grants F2304 (to H.S.) and F2305-B13 (to W.E.) from the Austrian Science Fund, and grant 10294 (to H.S.) from the Austrian National Bank.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal