Abstract
Diamond-Blackfan anemia (DBA) typically presents with red blood cell aplasia that usually manifests in the first year of life. The only gene currently known to be mutated in DBA encodes ribosomal protein S19 (RPS19). Previous studies have shown that the yeast RPS19 protein is required for a specific step in the maturation of 40S ribosomal subunits. Our objective here was to determine whether the human RPS19 protein functions at a similar step in 40S subunit maturation. Studies where RPS19 expression is reduced by siRNA in the hematopoietic cell line, TF-1, show that human RPS19 is also required for a specific step in the maturation of 40S ribosomal subunits. This maturation defect can be monitored by studying rRNA-processing intermediates along the ribosome synthesis pathway. Analysis of these intermediates in CD34− cells from the bone marrow of patients with DBA harboring mutations in RPS19 revealed a pre-rRNA–processing defect similar to that observed in TF-1 cells where RPS19 expression was reduced. This defect was observed to a lesser extent in CD34+ cells from patients with DBA who have mutations in RPS19.
Introduction
Diamond-Blackfan anemia (DBA) typically presents as a red blood cell aplasia that affects children in their first year of life. In addition to anemia, patients with DBA present with a heterogeneous mixture of congenital abnormalities.1 Craniofacial abnormalities are observed in approximately 50% of patients with DBA, while other defects, including growth failure, thumb malformation, and cardiac and urogenital defects, are observed less frequently.
Approximately 25% of patients with DBA have mutations in the gene encoding ribosomal protein S19, 1 of 33 ribosomal proteins that together with 18S rRNA constitutes the 40S ribosomal subunit.2-4 The etiology of the remaining cases of DBA is unknown. DBA is the first and only human disease known to be caused by mutations in a gene encoding a ribosomal protein. Interestingly, several other bone marrow (BM) failure syndromes have been linked to factors involved in ribosome synthesis.5 These syndromes include dyskeratosis congenita (DC), cartilage hair hypoplasia (CHH), and Shwachman Diamond syndrome (SDS). The proteins and RNAs affected in these diseases include the DKC1 gene in X-linked DC, which encodes a pseudouracil synthase,6 dyskerin involved in rRNA modification, the gene RMRP involved in CHH, which participates in rRNA processing,7 and SBDS, the gene affected in SDS which encodes a protein thought to function in RNA metabolism.8-11 The exact role of a defect in ribosome synthesis in each of these marrow failure syndromes is obscured by the fact that some of these proteins and RNAs are part of complexes that have multiple functions within cells. Dyskerin is a component of a number of ribonucleoprotein complexes, including telomerase,12-14 whereas RMRP is a component of an endoribonuclease involved in mRNA decay in addition to rRNA processing.15
The only other known function for ribosomal protein S19 (RPS19) is as a monocyte attractant, leaving open the possibility that the loss of a nonribosomal function for RPS19 is responsible for DBA.16 However, the recent identification of reduced ribosomal protein gene expression in DBA patients with normal RPS19 strongly favors a ribosome synthesis defect as the underlying cause of DBA.17 Previous studies have shown that the yeast homologs of RPS19 are required for the maturation of the 3′ end of 18S rRNA and the formation of active 40S ribosomal subunits. 40S subunit precursors that accumulate in cells depleted of the yeast RPS19 proteins are retained in the nucleus and fail to recruit factors required for late steps in the maturation of 40S subunits.18
To investigate the role of the human RPS19 protein in rRNA processing and the maturation of 40S ribosomal subunits, we turned to the TF-1 erythroleukemia cell line in which expression of RPS19 was reduced using siRNAs directed against RPS19 mRNA.19 Reduced expression of RPS19 in TF-1 cells preferentially affects erythroid differentiation and leads to increased apoptosis. Here we show that like the yeast RPS19 protein, human RPS19 is involved in the maturation of 40S ribosomal subunits and is required for specific steps in the maturation of the 3′ end of 18S rRNA. In light of the processing defect observed in TF-1 cells expressing siRNA against RPS19 mRNA, we examined pre-rRNA processing in CD34+ and CD34− cells from patients with DBA. Our data indicate that patient cells exhibit an rRNA-processing defect similar to that observed in TF-1 cells. These data are the first to show a pre-rRNA–processing defect in cells from patients with DBA who have mutations in RPS19, providing further support for the view that defects in ribosome synthesis may contribute to DBA.
Materials and methods
Cell lines and culture conditions
Construction of TF-1 cell lines expressing inducible siRNAs targeting RPS19 mRNA (TF-1 A and TF-1 B) and a scrambled control siRNA (TF-1 Sc) was described previously.19 The TF-1 cell lines were maintained in RPMI media supplemented with 10% fetal bovine serum and antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin). Granulocyte-macrophage colony-stimulating factor (GM-CSF; 5 ng/mL) was added to the media to support growth of the cytokine dependent TF-1 cell lines. Doxycycline (DOX) was included in the culture medium at a concentration of 0.5 μg/mL to induce siRNA expression.
Mononuclear BM cell samples
BM samples were collected after informed consent from healthy donors and patients with DBA. Mononuclear BM cells were isolated using a Lymphoprep density gradient (Nycomed, Oslo, Norway). Midi MACS LS separation columns and the CD34 MicroBead Kit (Miltenyi Biotec, Auburn, CA) were used to separate CD34+ and CD34− mononuclear BM cells. Cells were frozen in DMEM supplemented with 20% fetal calf serum and 10% dimethylsulfoxide and stored at −80°C. DBA patients 1 through 6 do not have mutations in the RPS19 gene. We previously described patients DBA-7, DBA-8, and DBA-9 as patients 2, 1, and 4, respectively.20 Patient DBA-7 has a chromosomal break in intron 3 on the RPS19 gene, patient DBA-8 has a total deletion of the RPS19 gene, and patient DBA-9 has a (TT157-158AA, 160 insertion CT) mutation encoding a truncated form of RPS19. Patients DBA-7 and DBA-8 were transfusion dependent and patient DBA-9 was in spontaneous remission at the time of the study. Patients DBA-7, DBA-8, and DBA-9 display impaired erythroid development in vitro, which can be improved by RPS19 gene transfer, proving that the erythroid defect is a result of RPS19 deficiency.21
RNA analysis
Total RNA was isolated from TF-1 cells or patient samples using an RNaqueous small-scale RNA isolation kit from Ambion (Austin, TX). Total RNA was recovered from 0.5 to 1 × 106 cells following the manufacturer's instructions for isolating RNA from suspension cultures. 5 to 10 μg total RNA was fractionated on 1.5% formaldehyde agarose gels and transferred to Zetaprobe membrane (Biorad Inc, Hercules, CA). Membranes were washed overnight at 55°C with 2 × SSC (0.3M NaCl and 0.03M Na citrate [pH 7.0]) and 1% sodium dodecyl sulfate and prehybridized for a minimum of 4 hours with ULTRAhyb oligonucleotide hybridization buffer (Ambion). The oligonucleotides used were: α, 5′-ACCGGTCACGACTCGGCA-3′ (complementary to sequences 1786-1804 in ETS1 of the rRNA transcription unit); β, 5′-GCATGGCTTAATCTTTGAGACAAGCATAT-3′ (complementary to sequences 3681-2709 in 18S rRNA); γ, 5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′ (complementary to sequences 5520-5547 spanning the boundary between 18S rRNA and internal transcribed sequence 1 [ITS1]); δ, 5′-TCTCCCTCCCGAGTTCTCGGCTCT-3′ (complementary to sequences 5687-5710 in the 5′ portion of ITS1); and ϵ, 5′-CTAAGAGTCGTACGAGGTCG-3′ (complementary to sequences 6613-6632 spanning the boundary between ITS1 and 5.8S rRNA). The probes (30 pmol) were labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Beverly MA). Membranes were hybridized overnight at 37°C in ULTRAhyb oligonucleotide hybridization buffer and washed the following morning 3 times with 6 × SSC at 37°C. Washed membranes were subjected to phosphorimage analysis (Phosphorimager SF; Molecular Dynamics, Sunnyvale, CA).
TF-1 cells transduced with lentiviral vectors expressing either a scrambled siRNA or RPS19 siRNA B were used for pulse-chase analysis. Cells were grown in RPMI media containing GM-CSF (5 ng/mL) in the presence or absence of DOX (5 μg/mL) for 4 days. Approximately 1 × 106 cells were harvested and washed with RPMI media lacking methionine (RPMI-Met). GM-CSF was included in the RPMI-Met media and DOX when appropriate. Cells were suspended in 3 mL RPMI-Met media and incubated for 2 hours at 37°C. Each cell suspension was treated with 150 μL (0.037 MBq/mL [1 μCi/μL]) [methyl-3H]-Met and pulse-labeled for 30 minutes. Cells were pelleted by centrifugation, media were removed, and cells resuspended in 3.2 mL RPMI media with methionine. Aliquots (1 mL) were chased for 0, 45, and 90 minutes in Met-containing media, after which cells were harvested and total RNA was isolated using RNaqueous kits. Total RNA was fractionated on 1.5% formaldehyde agarose gels and transferred to zetaprobe membranes. Membranes were baked for 2 hours at 80°C and exposed to BioMax MS film at −80°C using a BioMax LE intensifying screen (Eastman Kodak Co, Rochester NY).
Polysome analysis
TF-1 A cells were grown for 4 days in RPMI media containing GM-CSF with or without DOX. Extracts for polysome analysis were prepared as described by Tang et al.22 Extracts were layered on 16 mL 15% to 55% sucrose gradients and centrifuged in a SW28.1 rotor (Beckman Instruments, Fullerton CA) for 5 hours at 67 000g. Gradients were fractionated and absorbance at 254nm monitored on an ISCO model 185 gradient fractionator and a UA-6 absorbance detector (Lincoln, NE). Chart records were digitized using Adobe Photoshop (San Jose, CA).
Results
RPS19-deficient TF-1 cells exhibit a defect in pre-rRNA processing
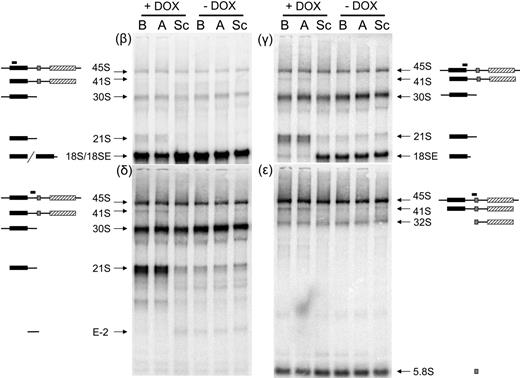
To assess the effect of reductions in RPS19 expression on ribosome synthesis in human hematopoietic cells we used a system in which RPS19 expression is controlled by a DOX-induced siRNA against RPS19 mRNA.19 The cell line used for these studies is the hematopoietic progenitor cell line TF-1, which can be induced to differentiate along the erythroid and myeloid lineages. Cells containing lentiviruses encoding 1 of 2 siRNAs against the RPS19 message or a scrambled siRNA control were either untreated or induced to express siRNAs by the addition of DOX to the culture media. Previous studies using these TF-1 cells have shown that by day 5 after DOX induction the steady-state level of the RPS19 protein is decreased by 40% to 60%.19 The growth rate of cells expressing the siRNAs targeted to RPS19 begins to decline relative to scrambled controls by day 4, suggesting that the effects of depleting RPS19 may appear at this time point (data not shown). We therefore harvested cells after 4 days of DOX induction and total RNA was isolated, fractionated on formaldehyde-agarose gels, and blotted with a series of oligonucleotide probes complementary to regions within the human rRNA repeat unit. The human rRNA-processing pathway and the probes used for Northern blot analysis are shown in Figure 1. Oligonucleotide β hybridizes to sequences within the coding region for 18S rRNA. Cell lines expressing siRNAs targeted to RPS19 (siRNAs A and B plus DOX) show a reduction in 18S rRNA compared with the same cell lines in the absence of DOX or cell lines containing scrambled siRNAs (Figure 2; β). In addition to a reduction in 18S rRNA, cell lines expressing siRNA to RPS19 have a novel species migrating just above 18S rRNA (labeled 21S).
Pre-rRNA processing in human cells. The major rRNA-processing pathways in human cells as initially derived from Hadjiolova et al23 and modified by Rouquette et al.24 Mature rRNA species are shown as filled boxes: 18S, ▪; 5.8S, ⊡; and 28S, ▨. External and internal transcribed sequences are shown as lines and are labeled above the primary transcript. Cleavage sites are designated with numbered and lettered arrows. Oligonucleotide probes used in Northern blot analysis are shown as lines below the primary transcript and are labeled with Greek letters. Two alternative pathways observed in human cells are shown below the 45S′ pre-rRNA that differ in the order of cleavages 1 and 2.
Pre-rRNA processing in human cells. The major rRNA-processing pathways in human cells as initially derived from Hadjiolova et al23 and modified by Rouquette et al.24 Mature rRNA species are shown as filled boxes: 18S, ▪; 5.8S, ⊡; and 28S, ▨. External and internal transcribed sequences are shown as lines and are labeled above the primary transcript. Cleavage sites are designated with numbered and lettered arrows. Oligonucleotide probes used in Northern blot analysis are shown as lines below the primary transcript and are labeled with Greek letters. Two alternative pathways observed in human cells are shown below the 45S′ pre-rRNA that differ in the order of cleavages 1 and 2.
Northern blot analysis demonstrates abnormal pre-rRNA processing in TF-1 cells depleted of RPS19. Total RNA was isolated from TF-1 cells, fractionated on 1.5% formaldehyde-agarose gels, transferred to zetaprobe, and hybridized with oligonucleotides complementary to different regions of the rRNA primary transcript. The siRNAs present in each cell line are listed above each lane. Cell lines in lanes labeled A and B express 2 different siRNAs targeting RPS19. Sc indicates scrambled siRNA. Cell lines were grown for 4 days in the presence (+DOX) or absence (−DOX) of 0.5 μg/mL DOX. Panels are designated according to the oligonucleotide used for hybridization. Pre-rRNAs hybridizing with the different oligonucleotide probes are designated with arrows to the right or left of the panels. Illustrations of rRNA species hybridizing with different probes are included to the sides of each image. Filled boxes represent mature rRNAs: 18S, ▪; 5.8S, ⊡; 28S, ▨.
Northern blot analysis demonstrates abnormal pre-rRNA processing in TF-1 cells depleted of RPS19. Total RNA was isolated from TF-1 cells, fractionated on 1.5% formaldehyde-agarose gels, transferred to zetaprobe, and hybridized with oligonucleotides complementary to different regions of the rRNA primary transcript. The siRNAs present in each cell line are listed above each lane. Cell lines in lanes labeled A and B express 2 different siRNAs targeting RPS19. Sc indicates scrambled siRNA. Cell lines were grown for 4 days in the presence (+DOX) or absence (−DOX) of 0.5 μg/mL DOX. Panels are designated according to the oligonucleotide used for hybridization. Pre-rRNAs hybridizing with the different oligonucleotide probes are designated with arrows to the right or left of the panels. Illustrations of rRNA species hybridizing with different probes are included to the sides of each image. Filled boxes represent mature rRNAs: 18S, ▪; 5.8S, ⊡; 28S, ▨.
In yeast cells depleted of RPS19, a 21S pre-rRNA accumulates at high levels relative to wild-type strains.18 The 21S pre-RNA extends through the 3′ end of mature 18S rRNA into the ITS1. To determine whether the novel 18S-related species observed in cells expressing siRNA to RPS19 extended into ITS1, the membrane was probed with oligonucleotide γ, which spans cleavage site 3 at the 3′ end of 18S rRNA. In Figure 2, γ shows that in control cell lines oligonucleotide γ recognizes a species referred to as 18SE, which extends 56 nucleotides downstream of the 3′ end of 18S rRNA to a newly identified cleavage site E in ITS1.24 In contrast, cell lines expressing siRNA targeted to RPS19 show a dramatic reduction in18SE and a corresponding increase in 21S rRNA. These data indicate that the 21S rRNA extends beyond the 3′ end of 18S rRNA into ITS1 and that cleavage at site E is affected in cells depleted of RPS19. We have used probes internal to ITS1 (oligonucleotide δ) and at the 3′ end of ITS1 overlapping with the 5′ end of 5.8S rRNA (oligonucleotides ϵ) to determine how far 21S rRNA extends into ITS1. In Figure 2, δ and ϵ indicate that 21S rRNA hybridizes with the internal probe but not with the 3′ probe, indicating that 21S rRNA likely terminates at site 2 within ITS1.
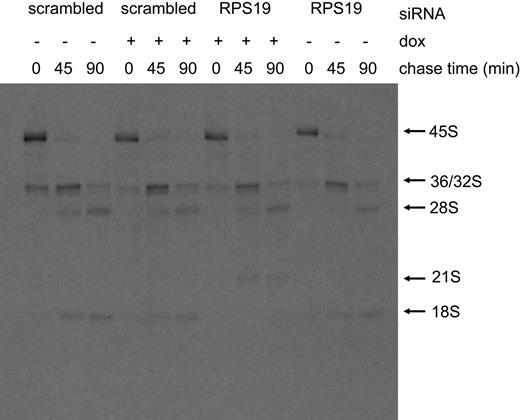
To further examine the rRNA-processing defect in cells expressing siRNA against RPS19 we turned to pulse-chase studies. In these experiments, cells expressing RPS19 siRNA B were compared with cells expressing a scrambled siRNA. Cells were grown in the presence or absence of DOX, and approximately 1 × 106 cells were harvested 4 days after DOX induction. Each cell line was grown for 2 hours in 3 mL RPMI media lacking methionine followed by the addition of 5.55 MBq (150 μCi) of [methyl-3H]-methionine and further incubation at 37°C for 30 minutes. During the pulse-labeling period the added methionine rapidly equilibrates with the S-adenosyl-methionine pool, which is subsequently used to methylate rRNA precursors. After the pulse period, the radiolabel was removed from the culture media and 3 mL RPMI media containing unlabeled methionine was added. Aliquots (1 mL) from each cell line were withdrawn, and cells were harvested after 0, 45, or 90 minutes of chase. Figure 3 shows that cells expressing siRNA against RPS19 produced very little mature 18S rRNA over the course of the 90-minute chase. In contrast, each of the other cell lines had detectable levels of 18S rRNA present in the 45-minute chase period. The decrease in 18S rRNA production in cell lines expressing siRNA to RPS19 was accompanied by an increase in the 21S rRNA species. These data are consistent with the Northern blot analysis data, providing further evidence that cells expressing siRNA targeting RPS19 failed to efficiently cleave 21S pre-rRNAs at the E site within ITS1 forming the mature 3′ end of 18S rRNA.
Pulse-chase analysis demonstrates abnormal pre-rRNA processing in TF-1 cells depleted of RPS19. Pulse-chase was carried out as described in “Materials and methods.” TF-1 cells infected with lentiviruses containing either siRNA B targeted to RPS19 (RPS19) or a scrambled siRNA (scrambled) were grown for 4 days in the presence (+) or absence (−) of DOX. Chase periods are shown above each lane.
Pulse-chase analysis demonstrates abnormal pre-rRNA processing in TF-1 cells depleted of RPS19. Pulse-chase was carried out as described in “Materials and methods.” TF-1 cells infected with lentiviruses containing either siRNA B targeted to RPS19 (RPS19) or a scrambled siRNA (scrambled) were grown for 4 days in the presence (+) or absence (−) of DOX. Chase periods are shown above each lane.
Cleavage site E in human ITS1 appears to correspond to cleavage site A2 in yeast ITS1, which is the major site affected in yeast cells depleted of RPS19.18 In yeast, when cleavage at site A2 is inhibited, processing still occurs at site A3 in ITS1.25 The 27S A3 pre-RNA resulting from A3 cleavage can proceed down the large subunit pathway, giving rise to mature 60S ribosomal subunits. On the other hand, 21S pre-RNA that extends from the mature 5′ end of 18S RNA through the A2 site to the A3 site is retained in the nucleus and is not efficiently processed to mature 18S rRNA.18 Consequently, a failure to efficiently cleave at the A2 site within ITS1 preferentially affects the production of 18S RNA and 40S ribosomal subunits. If cleavage sites E and 2 within ITS1 of human cells were comparable to sites A2 and A3 in yeast, respectively, inhibition of cleavage at site E would not be expected to have a dramatic affect on the production of 60S ribosomal subunits.
RPS19 is required for the maturation of 40S ribosomal subunits
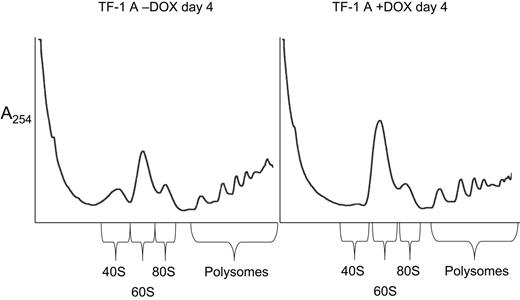
To address whether TF-1 cells expressing siRNA against RPS19 have a selective deficiency of 40S ribosomal subunits, cell extracts were prepared and polysome profiles examined after sucrose gradient centrifugation. The TF-1 cells used in Figure 4 (RPS19 siRNA A) were grown for 4 days in the presence and absence of DOX. Cells grown in the presence of DOX showed a reduction in free 40S subunits, an increase in free 60S subunits, and a shift toward smaller polysomes compared with cells grown in the absence of DOX. This profile is expected for cells with a deficiency of 40S ribosomal subunits.
Altered polysome profiles in TF-1 cells depleted of RPS19. Cells extracts were prepared for polysome analysis as described in “Materials and methods.” TF-1 cells infected with a lentivirus containing siRNA A targeted to RPS19 were grown for 4 days in the presence (+DOX) or absence (−DOX) of 0.5 μg/mL DOX. Extracts were layered on 15% to 55% sucrose gradients, and centrifugation was carried out for 5 hours at 67 000g. Gradients were fractionated using an ISCO-type 185 gradient fractionator, and absorbance at 254nm was monitored with a UA-6 absorbance detector.
Altered polysome profiles in TF-1 cells depleted of RPS19. Cells extracts were prepared for polysome analysis as described in “Materials and methods.” TF-1 cells infected with a lentivirus containing siRNA A targeted to RPS19 were grown for 4 days in the presence (+DOX) or absence (−DOX) of 0.5 μg/mL DOX. Extracts were layered on 15% to 55% sucrose gradients, and centrifugation was carried out for 5 hours at 67 000g. Gradients were fractionated using an ISCO-type 185 gradient fractionator, and absorbance at 254nm was monitored with a UA-6 absorbance detector.
BM cells from DBA patients with mutations in RPS19 exhibit abnormal processing of pre-rRNA
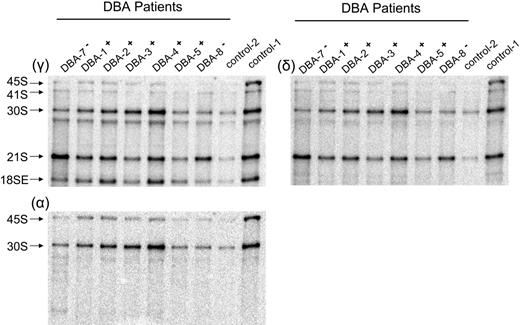
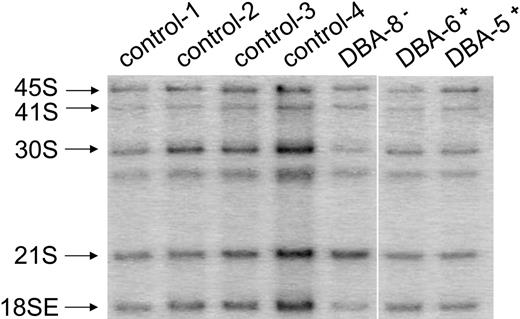
The data derived from TF-1 cells indicate that cells expressing suboptimal levels of RPS19 have defects in the maturation of 40S ribosomal subunits. Failure to efficiently mature 40S ribosomal subunits could therefore play an important role in the pathophysiology of DBA. Because of the high degree of sensitivity of the rRNA-processing assay, we used Northern blot analysis to examine the maturation of 40S subunits in cells derived from patients with DBA and healthy controls. Some of the cells from patients with DBA used in these studies contained mutations in RPS19; however, most did not (Tables 1–2).Both CD34− and CD34+ mononuclear BM cells were studied. We began our analysis with the more abundant CD34− cell populations. Total RNA isolated from CD34− cells was isolated, fractionated on 1.5% formaldehyde agarose gels, transferred to zeta-probe membrane, and blotted with oligonucleotide probes to different regions of the rRNA primary transcript. In Figure 5, the γ panel shows an increase in the ratio of 21S to 18SE pre-rRNA in patients with DBA who have mutations in RPS19 relative to healthy controls. Specific pre-RNA assignments in these primary cell populations were confirmed in blots with other oligonucleotide probes (δ and α panels). The δ panel, where hybridization was carried out with oligonucleotide δ internal to ITS1, shows that 21S pre-rRNA is detected, whereas the α panel, using an oligonucleotide complementary to sequences within ETS1, as expected, shows no evidence of hybridization with 21S pre-rRNA. The ratio of 21S to 18SE pre-rRNA in RPS19− patient samples was 3- to 4-fold higher than in healthy individuals and patients with DBA lacking RPS19 mutations (Table 1). These data indicate that like TF-1 cells, CD34− cells from patients with DBA who have mutations in RPS19 fail to efficiently cleave rRNA precursors at the E site within ITS1.
Abnormal pre-rRNA processing in CD34− cells from patients with DBA who have mutations in RPS19
| Clinical status* . | RPS19† . | No. sample runs‡ . | 21S/18SE ratio§ . |
|---|---|---|---|
| Control-1 | — | 2 | 1.3 |
| Control-2 | — | 5 | 1 |
| DBA-1 | RPS19+ | 2 | 1.2 |
| DBA-2 | RPS19+ | 2 | 1.2 |
| DBA-3 | RPS19+ | 2 | 1.2 |
| DBA-4 | RPS19+ | 2 | 1.2 |
| DBA-5 | RPS19+ | 2 | 0.8 |
| DBA-6 | RPS19+ | 2 | 0.9 |
| DBA-7 | RPS19−/breakpoint intron 3 | 2 | 3.1 |
| DBA-8 | RPS19−/complete deletion | 4 | 3.3 |
| DBA-9 | RPS19−/frameshift | 2 | 3.8 |
| Clinical status* . | RPS19† . | No. sample runs‡ . | 21S/18SE ratio§ . |
|---|---|---|---|
| Control-1 | — | 2 | 1.3 |
| Control-2 | — | 5 | 1 |
| DBA-1 | RPS19+ | 2 | 1.2 |
| DBA-2 | RPS19+ | 2 | 1.2 |
| DBA-3 | RPS19+ | 2 | 1.2 |
| DBA-4 | RPS19+ | 2 | 1.2 |
| DBA-5 | RPS19+ | 2 | 0.8 |
| DBA-6 | RPS19+ | 2 | 0.9 |
| DBA-7 | RPS19−/breakpoint intron 3 | 2 | 3.1 |
| DBA-8 | RPS19−/complete deletion | 4 | 3.3 |
| DBA-9 | RPS19−/frameshift | 2 | 3.8 |
Comparison of the DBA RPS19+ patient group data sets (rows 3-7) with the control group (rows 1-2) data sets: P = .5; DBA RPS19− patient group data sets (rows 8-11) with the control group: P < .001.
— indicates not sequenced.
Patients diagnosed with DBA are listed as DBA-1 to DBA-9.
The RPS19 gene was sequenced in each DBA patient. RPS19+ indicates no mutations were found. RPS19− indicates mutations were found and the nature of the mutation.
The number of times each sample was run on a different agarose gel.
Average ratio of 21S to 18SE pre-rRNA after phosphorimage analysis. The 21S/18SE ratio for each sample was normalized against the control-2 ratio in the same gel.
Pre-rRNA–processing defect in CD34+ cells from patients with DBA who have mutated RPS19
| Clinical status* . | RPS19 status† . | No. sample runs‡ . | 21S/18SE ratio§ . |
|---|---|---|---|
| Control-1 | — | 3 | 1.4 |
| Control-2 | — | 3 | 1 |
| Control-3 | — | 2 | 1.2 |
| Control-4 | — | 3 | 1.2 |
| DBA-5 | RPS19+ | 3 | 1 |
| DBA-6 | RPS19+ | 3 | 0.9 |
| DBA-8 | RPS19−/complete deletion | 3 | 1.7∥ |
| Clinical status* . | RPS19 status† . | No. sample runs‡ . | 21S/18SE ratio§ . |
|---|---|---|---|
| Control-1 | — | 3 | 1.4 |
| Control-2 | — | 3 | 1 |
| Control-3 | — | 2 | 1.2 |
| Control-4 | — | 3 | 1.2 |
| DBA-5 | RPS19+ | 3 | 1 |
| DBA-6 | RPS19+ | 3 | 0.9 |
| DBA-8 | RPS19−/complete deletion | 3 | 1.7∥ |
— indicates not sequenced.
Patients diagnosed with DBA are listed as DBA-5, DBA-6, and DBA-8.
The RPS19 gene was sequenced in each patient with DBA. RPS19+ indicates no mutations were found. RPS19− indicates mutations were found.
Number of times each sample was run on a different agarose gel.
Average ratio of 21S to 18SE pre-rRNA after phosphorimage analysis. The 21S/18SE ratio for each sample was normalized against the control-2 ratio in the same gel.
P < .003. The P value reported is for a comparison of the RPS19− data set with the combined control data sets using the Student t test.
Northern blot analysis of CD34− BM cells reveals abnormal pre-rRNA processing in patients with DBA who have mutations in RPS19. Total RNA was isolated from CD34− cells and prepared for Northern blot analysis as described in Figure 2. Panels are designated according to oligonucleotides used for hybridization. Patients with DBA who have mutations in RPS19 are designated DBA-7− and DBA-8−, while patients with normal RPS19 are designated DBA-1+ to DBA-5+. Samples from DBA patients labeled DBA-7− and DBA-8− have a chromosome breakpoint mutation in RPS19 and a complete deletion of RPS19, respectively. Ratios listed in Table 1 are derived from phosphorimage analysis of signals for the RNA species listed. Not all samples listed in Table 1 are shown in here.
Northern blot analysis of CD34− BM cells reveals abnormal pre-rRNA processing in patients with DBA who have mutations in RPS19. Total RNA was isolated from CD34− cells and prepared for Northern blot analysis as described in Figure 2. Panels are designated according to oligonucleotides used for hybridization. Patients with DBA who have mutations in RPS19 are designated DBA-7− and DBA-8−, while patients with normal RPS19 are designated DBA-1+ to DBA-5+. Samples from DBA patients labeled DBA-7− and DBA-8− have a chromosome breakpoint mutation in RPS19 and a complete deletion of RPS19, respectively. Ratios listed in Table 1 are derived from phosphorimage analysis of signals for the RNA species listed. Not all samples listed in Table 1 are shown in here.
It is possible that CD34− from patients with DBA who have normal RPS19 could have a defect in steps along the rRNA-processing pathway that is different from patients with RPS19 mutations. Several studies in yeast have shown that mutations in genes encoding different ribosomal proteins can have distinct effects on rRNA processing.18,26 Examination of the patterns in Figure 5 (γ and δ panels) show a modest increase in ratio of 30S to 18SE, 1.5- and 1.2-fold, in patients DBA-3 and DBA-4, respectively, relative to control samples. Further studies will be necessary to determine if these increases are significant and contribute to the pathophysiology in these patients. Other DBA patient samples with normal RPS19 showed no obvious differences among each other and with control rRNA-processing patterns.
The primary hematopoietic defect in patients with DBA is thought to reside in the differentiation and amplification of early progenitor cells in the erythroid lineage. We therefore also examined rRNA processing in CD34+ cells from patients with DBA and healthy controls. Figure 6 shows a blot of CD34+ cells hybridized with oligonucleotide γ, the probe which revealed the rRNA-processing defect linked to a reduction in functional RPS19 in TF-1 cells and CD34− cells. Surprisingly, the CD34+ cells from the patient sample with mutated RPS19 (DBA-8) showed only a modest but statistically significant increase in the 21S-to-18SE ratio relative to other samples (Table 2). Thus, the rRNA-processing defect observed in CD34− cells from a patient with DBA who has an RPS19 mutation appears to be reduced in magnitude in CD34+ cells from the same patient.
Northern blot analysis displays defective pre-rRNA processing in CD34+ cells from patients with DBA who have mutations in RPS19. The figure shows a representative Northern blot using total RNA isolated from CD34+ cells and prepared for Northern blot analysis as described in Figure 2. Pre-rRNAs were hybridized with oligonucleotide γ (Figure 1) to examine the ratio of 21S to 18SE pre-rRNA.
Northern blot analysis displays defective pre-rRNA processing in CD34+ cells from patients with DBA who have mutations in RPS19. The figure shows a representative Northern blot using total RNA isolated from CD34+ cells and prepared for Northern blot analysis as described in Figure 2. Pre-rRNAs were hybridized with oligonucleotide γ (Figure 1) to examine the ratio of 21S to 18SE pre-rRNA.
Discussion
Previous studies have shown that the yeast RPS19 protein is required for a specific step in the maturation of 40S ribosomal subunits.18 In yeast cells depleted of RPS19, pre-40S particles accumulate in the nucleus with a corresponding decrease in the amount of mature 40S subunits in the cytoplasm. The pre-40S subunits that accumulate in RPS19-depleted cells contain a 21S precursor to mature 18S rRNA. This precursor begins at the mature 5′ end of 18S rRNA and extends past the mature 3′ end of 18S rRNA to the A3 cleavage site within ITS1 of the rRNA transcription unit. Thus, yeast cells depleted of RPS19 fail to efficiently cleave pre-RNAs at the A2 cleavage site within ITS1, resulting in immature subunits that have failed to mature the 3′ end of 18S rRNA.
Our goal here was to monitor rRNA processing and 40S subunit maturation in human cells depleted of RPS19. Cells used for these studies were human TF-1 cells, a hematopoietic progenitor cell line expressing siRNA against the RPS19 mRNA, and cells from patients with DBA heterozygous for mutations in the RPS19 gene. Results from TF-1 cells indicate that like yeast, human cells depleted of RPS19 accumulate a 21S pre-rRNA extended through the mature 3′ end of 18S rRNA into ITS1. We have not specifically defined the 3′ end of the 21S pre-rRNA, but show that it extends through the recently identified E site within ITS1 of the human transcription unit.24 Since this precursor does not extend to the extreme 3′ end of ITS1 it most likely terminates at cleavage site 2 within human ITS1. In this respect, cleavage sites E and 2 within the human transcription unit would be comparable to sites A2 and A3 of yeast, respectively. Pulse-chase analysis showed a precursor product relationship between the 21S pre-rRNA and mature 18S rRNA, and polysome profiles revealed that TF-1 cells depleted of RPS19 had a deficiency of 40S ribosomal subunits. Thus, like its yeast ortholog, the human RPS19 protein is required for the maturation of 40S subunits and specifically affects a cleavage step within ITS1 needed for the formation of the mature 3′ end of 18S rRNA.
Maturation of the 3′ end of 18S rRNA in humans cells occurs through a stepwise pathway involving cleavage first at site 2 in ITS1, followed by cleavage at site E, and finally formation of the mature 3′ end of 18S rRNA by cleavage at site 3.24 Northern blot analysis of TF-1 cells depleted of RPS19 show a dramatic decrease in 18SE pre-rRNA that parallels the increase in 21S pre-rRNA. Thus, the human RPS19 protein is required for efficient E site cleavage. We therefore used the ratio of 21S to 18SE as a signature of RPS19 function in cells from patients with DBA and healthy controls.
Analysis of CD34− cells from patients with DBA who have mutations in RPS19 showed a 3- to 4-fold increase in the ratio of 21S to 18SE pre-RNAs relative to healthy controls and patients with DBA lacking RPS19 mutations. This increase is less pronounced than the 20- to 40-fold difference observed in TF-1 cells. However, BM-derived CD34− cells represent a complex mixture of different cell types, with approximately 1 in 4 being of the erythroid lineage, mainly in the form of erythroblasts.27 Moreover, previous studies have shown that the BM of patients with DBA is relatively devoid of erythroid precursors.28 As such, the rRNA-processing defect measured in the CD34− cells from patients with DBA who have mutations in RPS19 may be derived from only a small number of cells within the total population.
The finding of a functional defect in 40S subunit maturation linked to suboptimal levels of RPS19 supports the notion that defective ribosome synthesis may be the underlying molecular basis for DBA. However, only 25% of DBA cases have been linked to mutations in RPS19. We were therefore interested in determining whether we could measure changes in rRNA processing in CD34− cells from patients with DBA who have normal RPS19. While there is a modest increase in the ratios of 30S to 21S pre-rRNA in DBA-3 and DBA-4 CD34− cells relative to controls, suggesting a processing defect elsewhere in the pathway, the significance of this small change is unclear at the present time. The ability to identify specific cell populations manifesting the rRNA-processing defect linked to mutations in RPS19 may allow us to enrich for these populations in other DBA patient samples, thereby increasing the sensitivity of the rRNA-processing assay which, in turn, may unmask other rRNA-processing defects.
We also examined CD34+ cells for an rRNA-processing defect similar to that observed in CD34− cells. The 21S-to-18SE ratio in CD34+ cells from patient DBA-8 was less than that observed for CD34− cells from the same patient. The decreased magnitude of the rRNA-processing defect in CD34+ relative to CD34− cells from this patient with a complete deletion of RPS19 could be a reflection of relative percentage of cells expressing the rRNA-processing defect in CD34− and CD34+ cell populations derived from this individual. Alternatively, these data may suggest that the defect in ribosome synthesis manifests itself to a greater extent at later steps in erythroid differentiation after the CD34 antigen is lost. Da Costa et al29 have shown that RPS19 expression decreases during terminal erythroid differentiation. Expression of RPS19 has also been monitored in normal human hematopoietic BM cells by Northern blot and quantitative reverse transcription–polymerase chain reaction (Q-RT-PCR). RPS19 mRNA levels are relatively high in populations enriched for multipotent progenitors and gradually decrease in more differentiated cell populations.20,21 Interestingly, the population containing erythroblasts, a cell that has unusually high levels of free ribosomes causing the basophilic cytoplasm, express relatively low levels of RPS19 mRNA.30 It is therefore possible that RPS19 haploinsufficiency may not become limiting for 40S subunit maturation until the latter stages of erythroid differentiation, when the level of RPS19 is naturally reduced.
In conclusion, we have shown that a human progenitor cell line expressing siRNAs to RPS19 exhibits a specific defect in rRNA processing and the maturation of 40S ribosomal subunits. The rRNA-processing defect can be measured in patients with DBA patients who have mutant alleles of RPS19. The defect appears in CD34− cells and, to a somewhat lesser extent, in CD34+ cells derived from the BM of the patients with DBA. Identification of the specific cell types manifesting this defect in 40S subunit maturation should contribute to our understanding of the molecular mechanisms underlying the pathophysiology of DBA.
Authorship
Author contributions: J.F. and A.A. performed research and analyzed data; J.C.B. and K.M. contributed reagents; J.M.C. contributed to early phases of the study; and S.K. and S.R.E. have responsibility for the entire manuscript.
Conflict-of-interest statement: The authors declare no competing financial interests.
J.F. and A.A. contributed equally to the present work.
Correspondence: Steven R. Ellis, Department of Biochemistry and Molecular Biology, University of Louisville School of Medicine, Louisville, KY 40292; e-mail: srellis@louisville.edu; or Stefan Karlsson, Department of Molecular Medicine and Gene Therapy, Lund Stem Cell Center, Lund University Hospital, BMC A12, 221 84, Lund, Sweden; e-mail: stefan.karlsson@molmed.lu.se.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We wish to thank Dr Isao Hamaguchi, Dr Thomas Kiefer, and Karin Olsson for scientific discussions and help with collecting patient bone marrow cells. We also thank Drs Niklas Dahl and Johan Richter for help in recruiting patients for this study.
Supported by National Institutes of Heath grant RO1 HL079583 (S.R.E.), the Kentucky Lung Cancer Research Program (S.R.E.), the Ronald McDonald Foundation (J.F.), the Royal Physiographic Society in Lund (J.F.), the Swedish Cancer Society (S.K.), the European Commission (CONCERT) (S.K.), the Swedish Medical Council (S.K.), Crafoordska Stiftelsen (S.K.), the Swedish Children Cancer Foundation (S.K.), a clinical research award from Lund University Hospital (S.K.), and grants from the Diamond Blackfan Anemia Foundation (S.R.E. and S.K.). The Joint Program on Stem Cell Research at Lund University Hospital is supported by the Juvenile Diabetes Research Foundation and the Swedish Medical Research Council. The Lund Stem Cell Center is supported by a Center of Excellence grant in Life Sciences from the Swedish Foundation for Strategic Research.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal