To the editor:
We read with interest the online version of the paper by Gonzalez-Paz et al.1 Having recently examined the relationship of methylation and expression of p16INK4a (CDKN2A) in multiple myeloma (MM) tumors and cell lines, we were aware that many commercial assays for human p16 RNA—including all current Affymetrix arrays—do not distinguish the overlapping p16INK4A and p14ARF genes, both of which have unique exon 1 but common exon 2 and 3 sequences. Indeed, the real-time reverse-transcription–polymerase chain reaction (RT-PCR) kit (CDKN2A catalog no., Hs00233365_m1; Applied Biosystems, Foster City, CA) used in the online version of this paper actually measures both p16 and p14. We notified the authors of this error and suggested alternative p16-specific primers. Their corrected real-time RT-PCR data of p16 expression is shown in Figure 1 of a revised article. They conclude that p16 expression was similar in all MM tumors, regardless of whether p16 is methylated.
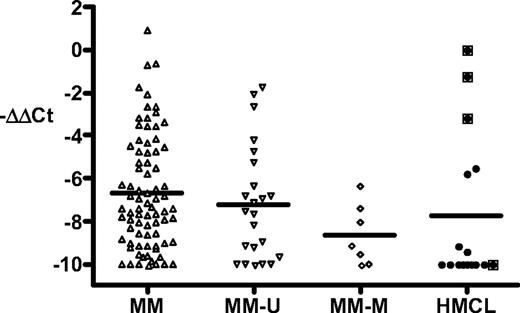
We have additional data for MM tumors and cell lines (Figure 1).Seven (23%) of 30 MM tumors had methylated p16, whereas 12 (75%) of 16 MM cell lines had methylated p16, consistent with results published by Gonzalez-Paz et al1 and others.5-7 We determined the expression of p16 RNA in 79 tumors (including 7 with methylated p16 and 23 with unmethylated p16) and in 16 cell lines. Three cell lines (EJM, OPM-2, PE-1) with unmethylated p16 express substantial levels of p16, with the highest level of expression in EJM being comparable with small-cell lung cancer lines that express high levels of p16 (data not shown).8 The expression of p16 in the other cell lines is low or not detectable (including SKMM-2 with unmethylated p16). Although most MM tumors express only low levels of p16, a significant fraction expresses p16 at a level comparable with that of the 3 cell lines just mentioned. All 7 tumors with methylated p16 express only low levels of p16, distinct from the expression pattern seen for all 79 MM tumors (P = .04). We conclude that most MM tumors express little or no p16, regardless of whether there is methylation of p16. However, in contrast to the results of Gonzalez-Paz et al,1 our results on a larger data set show that a significant fraction of MM tumors and MM cell lines express moderately high levels of p16 RNA.
Methylation and expression of p16INK4A in multiple myeloma tumors and cell lines. Purification of MM tumor cells, growth of cell lines (EJM, OPM-2, PE-1, Delta-47, ANBL6, KMS12-BM, JIM-3, H1112, KHM-11, KMS-11, H929, 8226, Karpas-620, LP1, SKMM-2, JJN-3), and preparation of RNA and cDNA have been described previously.2 The methylation status of p16 was determined as described elsewhere.3 Quantitative real-time PCR was done using USF2 as an internal control.4 The p16 primers (reference sequence L27211) were from exon 1 (nt 163-183) and exon 2 (nt 218-205), and the p16 probe was 5′VIC-CATGACCTGGATCGGCCTCGGA-TAMRA-3′. The data are calculated as ΔCt = Ct.p16-Ct.USF2, and then normalized as ΔΔCt = sample.ΔCt − EJM.ΔCt (∼ 0-1 in most experiments). The results are shown as −ΔΔCt and cover a more than 1000-fold range, with all values less than −10 shown as −10. MM indicates all MM tumors; MM-U and MM-M, MM tumors with unmethylated and methylated p16, respectively; and HMCL, MM cell lines. The boxed symbols indicate the 4 unmethylated HMCLs. The mean is shown as a horizontal line. The results for the MM and MM-M groups differ by a t test (P = .04).
Methylation and expression of p16INK4A in multiple myeloma tumors and cell lines. Purification of MM tumor cells, growth of cell lines (EJM, OPM-2, PE-1, Delta-47, ANBL6, KMS12-BM, JIM-3, H1112, KHM-11, KMS-11, H929, 8226, Karpas-620, LP1, SKMM-2, JJN-3), and preparation of RNA and cDNA have been described previously.2 The methylation status of p16 was determined as described elsewhere.3 Quantitative real-time PCR was done using USF2 as an internal control.4 The p16 primers (reference sequence L27211) were from exon 1 (nt 163-183) and exon 2 (nt 218-205), and the p16 probe was 5′VIC-CATGACCTGGATCGGCCTCGGA-TAMRA-3′. The data are calculated as ΔCt = Ct.p16-Ct.USF2, and then normalized as ΔΔCt = sample.ΔCt − EJM.ΔCt (∼ 0-1 in most experiments). The results are shown as −ΔΔCt and cover a more than 1000-fold range, with all values less than −10 shown as −10. MM indicates all MM tumors; MM-U and MM-M, MM tumors with unmethylated and methylated p16, respectively; and HMCL, MM cell lines. The boxed symbols indicate the 4 unmethylated HMCLs. The mean is shown as a horizontal line. The results for the MM and MM-M groups differ by a t test (P = .04).
Mutant or absent p16 is a critical oncogenic factor in murine plasmacytomas, but the situation is less clear in MM.9 One individual from a melanoma-prone family with a germ-line mutant p16 gene developed MM, and his tumor cells showed selective loss of the normal p16 allele.10 Yet there are no other reports of p16 mutations in MM. We agree with Gonzalez-Paz et al1 that methylation of p16 may be an epiphenomenon—often occurring as a late progression event—that is not responsible for the inactivation of p16. In addition, our results indicate that inactivation of p16 is not an essential oncogenic event in at least some, if not all, MM tumors.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Kuehl, National Cancer Institute, Naval Hospital, Bldg 8, Rm 5101, Bethesda, MD 20889-5105; e-mail: wmk@helix.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal