Abstract
Erythropoiesis occurs in erythroblastic islands, where developing erythroblasts closely interact with macrophages. The adhesion molecules that govern macrophage-erythroblast contact have only been partially defined. Our previous work has implicated the rat ED2 antigen, which is highly expressed on the surface of macrophages in erythroblastic islands, in erythroblast binding. In particular, the monoclonal antibody ED2 was found to inhibit erythroblast binding to bone marrow macrophages. Here, we identify the ED2 antigen as the rat CD163 surface glycoprotein, a member of the group B scavenger receptor cysteine-rich (SRCR) family that has previously been shown to function as a receptor for hemoglobin-haptoglobin (Hb-Hp) complexes and is believed to contribute to the clearance of free hemoglobin. CD163 transfectants and recombinant protein containing the extracellular domain of CD163 supported the adhesion of erythroblastic cells. Furthermore, we identified a 13–amino acid motif (CD163p2) corresponding to a putative interaction site within the second scavenger receptor domain of CD163 that could mediate erythroblast binding. Finally, CD163p2 promoted erythroid expansion in vitro, suggesting that it enhanced erythroid proliferation and/or survival, but did not affect differentiation. These findings identify CD163 on macrophages as an adhesion receptor for erythroblasts in erythroblastic islands, and suggest a regulatory role for CD163 during erythropoiesis.
Introduction
The functional unit for definitive erythropoiesis in the bone marrow is the erythroblastic island, a multicellular structure composed of a central macrophage surrounded by erythroblasts at various stages of differentiation.1–3 The contact between erythroblasts and macrophages supports the growth, survival, and differentiation of erythroblasts, and allows for phagocytosis of the extruded erythroid nucleus. Thus far, the molecular interaction(s) that mediate the formation of erythroblastic islands have only been partly defined.2,4 First, interactions between vascular cell adhesion molecule-1 (VCAM-1) on macrophages and α4 integrins on erythroblasts have been implicated in the contact between these cells.5 Furthermore, a molecule called erythroblast macrophage protein (Emp) has been identified, which is expressed on macrophages, erythroblasts, and other cells.6–8 Emp is believed to mediate erythroblast adhesion to macrophages, probably by homophilic interaction. Of relevance, Emp-deficient fetuses, which die perinatally, have significantly reduced numbers of erythroblastic islands and defective erythropoiesis. This phenotype appears to result from a deficiency in both macrophage development as well as disturbed erythroblast nuclear extrusion. Finally, the erythroid intercellular adhesion molecule-4 (ICAM-4) has been demonstrated to bind to the αv integrin expressed by macrophages in erythroblastic islands, and this has also been shown to contribute to erythroblastic island formation in vivo.9,10
We have previously identified the rat macrophage ED2 antigen, which is expressed on resident bone marrow macrophages (RBMMφ) as well as other subsets of mature tissue macrophages, as a candidate receptor for erythroblasts.11 In particular, we have shown that binding of rat fetal liver erythroblasts to RBMMφ can be blocked by the ED2 monoclonal antibody (mAb). In order to provide further insight into the molecular interactions between erythroblasts and macrophages and the contributions of these interactions to erythropoiesis, we further characterized the role of the ED2 antigen in erythroblast-macrophage adhesion. Our results identify the ED2 antigen as the macrophage hemoglobin scavenger receptor CD163. Furthermore, we show that CD163 can directly support interactions with erythroblasts and identify a motif in the second scavenger domain of CD163 that, by binding to a putative counterreceptor on erythroblasts, promotes their growth and/or survival. This implicates CD163 in erythroblast adhesion to macrophages and suggests a possible regulatory role for this molecule in erythropoiesis.
Materials and methods
The use of human cord blood was approved by the Medical Ethical Committee of the Utrecht Medical Center. The use of animals was approved by the Experimental Animal Committee of the Vrije University (VU) Medical Center, Amsterdam, The Netherlands. Approval was granted in accordance with Dutch legislations. Informed consent was obtained in accordance with the Declaration of Helsinki.
FACS staining
Immunofluorescence staining of stably transfected CHO cells12 was performed on 100 000 cells using anti-human CD163 monoclonal antibody (mAb; EDHu113 ). Cells were stained with EDHu1 (10 μg/mL) for 60 minutes at 4°C in phosphate buffered saline (PBS)–0.1% bovine serum albumin (BSA; Boehringer-Mannheim, Mannheim, Germany) and washed. Next, the cells were stained with FITC-conjugated rabbit anti-mouse (1:300; DAKO, Copenhagen, Denmark) for 45 minutes. After washing, cells were resuspended in PBS–0.1% BSA, and fluorescence intensity was determined using a FACScalibur flow cytometer (Becton Dickinson, San Jose, CA).
Immunohistochemistry
Fresh spleen cryosections of 7-day-old normal male DA rats and adult Lewis rats that had been infected with Plasmodium berghei 6 days earlier were double-immunostained for red pulp macrophages (ED2; peroxidase-black) and rat transferrin receptor (OX26; alkaline phosphatase–red) by the indirect immunoenzyme method as described previously.14
Affinity purification and peptide sequencing of the ED2 antigen
The mouse anti-rat mAb ED2 was described previously11,15,16 and is available from Serotec (Oxford, United Kingdom). The ED2 antigen was affinity isolated from a rat spleen lysate (ie, 60 spleens from Wistar; Fisher F1 and Fisher F334 rats (Harlan-CPB, Zeist, NC) homogenized in 1% NP40; 0.01 M triethanolamine/HCl [pH 7.8]; and 0.15 M NaCl containing the protease inhibitors aprotinine, pepstatine, leupeptine, and PMSF) on an ED2–CNBr–sepharose column (4.3 mg IgG/mL) after preclearance on a bovine IgG (10 mg IgG/mL) column. The ED2 antigen was eluted using 50 mM glycine [pH 2.5], 0.1% Triton X-100, and 0.15 M NaCl, yielding a total of approximately 100 to 120 μg protein. After further purification of the 175-kDa band (Figure 2A) on tricine sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and semidry blotting to PVDF membrane, N-terminal sequencing was performed by Edman degradation on a protein sequencer (Applied Biosystems 473A; Warrington, United Kingdom). Internal peptide sequences were obtained by 2 methods. First, peptides from an endolysine C digest were separated by high-performance liquid chromatography (HPLC) and sequenced. Second, ED2 antigen purified from a silver-stained acrylamide gel was subjected to quadrupole time-of-flight (Q-TOF) analysis (Micromass, Waters, Etten-Leur, The Netherlands). Where indicated, fractions were treated with peptide: N-Glycosidase F (PNGase F; New England BioLabs, Ipswich, MA) according to the manufacturer's instructions.
Isolation of erythroblasts
Rat fetal liver erythroblasts were isolated as described previously.11 Human erythroblasts were generated in vitro from CD34+ hematopoietic stem cells (HSCs) obtained from healthy-donor umbilical cord blood as previously described,17 with minor modifications. Briefly, mononuclear cells were isolated from umbilical cord blood by density centrifugation over a ficoll-paque solution. Magnetic-activated cell sorting (MACS; Miltenyi Biotec, Auburn, CA) using a hapten-conjugated antibody against CD34 coupled to beads was used to isolate CD34+ cells. CD34+ cells were cultured at 104 cells/mL for 8 days in serum-free medium containing the indicated sources or iron, transferrin, and other supplements as well as stem cell factor, IL-3, erythropoietin (EPO), and hydrocortisone. After culture, the purity (> 95%) and maturity of erythroblasts was checked with May-Grunwald-Giemsa stainings of cytospin preparations. Both rat and human erythroblast suspensions were less than 1.5% positive for CD163 as evaluated by flow cytometry.
Transfectants and adhesion assays
K562 cells were labeled with 0.5 μM 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein, acetoxymethyl ester (BCECF-am) for 15 minutes in HEPES-buffered RPMI-1640 at 37°C, washed, and 105 cells in 100 μL medium were allowed to adhere for 60 minutes at 37°C to CHO cells stably expressing full-length human CD16312 or to parental cells cultured to confluency in flat-bottomed 96-well plates. After washing, cells were lysed in 0.1 M NaOH, and fluorescence was determined in a microplate reader (Fluostar Galaxy; BMG Laboratories, Offenburg, Germany) at an excitation of 485 nm and an emission of 535 nm. The percentage of adherence was calculated using a K562 cell calibration curve.
Solid phase binding of K562 cells or freshly isolated rat fetal liver erythroblasts,11 using 105 cells/well (100 μL), to synthetic peptides was performed as described previously,18,19 omitting Tween-20 from the buffers. Adhesion of nucleated cells was quantified by SYTO-13 staining (Invitrogen, Carlsbad, CA) and measured in the microplate reader at 488 nm excitation and 509 nm emission.
In vitro erythroid maturation was performed by culturing rat erythroblasts in serum-free medium (HyQ CCM5; Hyclone, Logan, UT) and 2 IU/mL recombinant human erythropoietin (Roche, Welwyn Garden City, United Kingdom) in triplicate. Cultured erythroblasts were detached and then counted using a hemocytometer. Maturation was determined by May-Grunwald-Giemsa staining of cytospin preparations.
Fluorescent bead adhesion assay
CHO cells were stably transfected with a plasmid to express the extracellular domain of CD163 (amino acids [aa] 1-993) as a fusion protein with human IgG1 Fc (generously provided by Dr R. J. J. van Neerven, Macrozyme, Amsterdam, The Netherlands). Supernatants from these cultures were collected and CD163-Fc derived from stably transfected CD163 CHO cells was purified before coating onto carboxylate-modified TransFluoSpheres (488/645 nm, 1.0 μm; Molecular Probes, Eugene, OR) as described previously.20 Briefly, streptavidin was covalently coupled onto TransFluoSpheres as described by manufacturer. To enable coupling of CD163-Fc, streptavidin-coated beads were allowed to bind to biotinylated goat anti-human anti-Fc Fab2 fragments (6 μg/mL; Jackson ImmunoResearch Laboratories, Bar Harbor, ME) in 0.5 mL TBS containing 2 mM Ca2+ for 2 hours at 37°C. The beads were washed once with TBS-Ca2+ and incubated with human CD163-Fc (1 μg) for 2 days at 4°C. The ligand-coated beads were washed, resuspended in 100 μL TBS-Ca2+, and stored at 4°C. For bead adhesion to K562 cells and rat erythroblasts, cells were resuspended in TBS-Ca2+. The ligand-coated beads (20 beads/cell) were added to 105 cells, and the suspension was incubated for 60 minutes at 37°C. The percentage of K562 cells or rat erythroblasts binding CD163-coated beads was quantified by flow cytometry using the FACScalibur (Becton Dickinson, Oxnard, CA) and expressed as normalized values for comparison of individual experiments. As a control, beads coated with human ICAM-3-Fc21 were used.
Results
Expression of the ED2 antigen in erythroblastic islands
Our previous studies have provided preliminary evidence that the rat ED2 antigen is involved in the adhesion of erythroblasts to RBMMφ.11 In particular, the mAb ED2 was shown to inhibit erythroblast binding to RBMMφ in bone marrow frozen sections (approximately 80% inhibition), or freshly isolated RBMMφ (approximately 60% inhibition), respectively. Also, the ED2 antigen was found to be highly expressed on these RBMMφ that form the central macrophages in erythroblastic islands in the bone marrow. Here, we further characterize the role of the ED2 antigen in erythroblast adhesion.
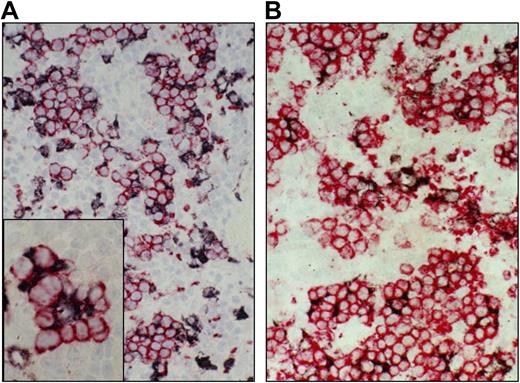
First, we investigated whether the ED2 antigen is expressed by macrophages in erythroblastic islands at extramedullary sites, such as the spleen. In particular we investigated the erythropoietic splenic tissue of both juvenile (1-week-old) rats and adult animals that had been infected 6 days before with the malaria parasite P brucei. Typical erythroblastic islands, identified by staining with transferrin receptor/CD71 (using mAb OX26), that contain macrophages expressing high levels of ED2 antigen were identified in the splenic red pulp area in both conditions (Figure 1). In fact, most if not all erythroblastic islands contained one or more ED2+ macrophages. Conversely, the ED2−ED3+ macrophages, which are located in the marginal zone,22 did not form erythroblastic islands (not shown). In normal adult spleens, which have considerably less erythropoietic activity, the ED2 antigen was still highly expressed on the red pulp macrophages (data not shown; Dijkstra et al16 ), suggesting that the presence of the ED2 antigen on these macrophages per se is not sufficient for the formation of erythroblastic islands. Taken together, these findings demonstrate that the ED2 antigen is expressed on macrophages in erythroblastic islands at both medullary and extramedullary sites.
Expression of the rat ED2 antigen by macrophages in splenic erythroblastic islands. Microphotographs from the red pulp areas from the spleens of a juvenile 7-day-old rat (A), or an adult animal 6 days after P berghei infection (B), double-stained with mAb ED2 for rat CD163 (black) and mAb OX26 for rat transferrin receptor–positive erythroblasts (red).
Expression of the rat ED2 antigen by macrophages in splenic erythroblastic islands. Microphotographs from the red pulp areas from the spleens of a juvenile 7-day-old rat (A), or an adult animal 6 days after P berghei infection (B), double-stained with mAb ED2 for rat CD163 (black) and mAb OX26 for rat transferrin receptor–positive erythroblasts (red).
The ED2 antigen is identical to the scavenger receptor CD163
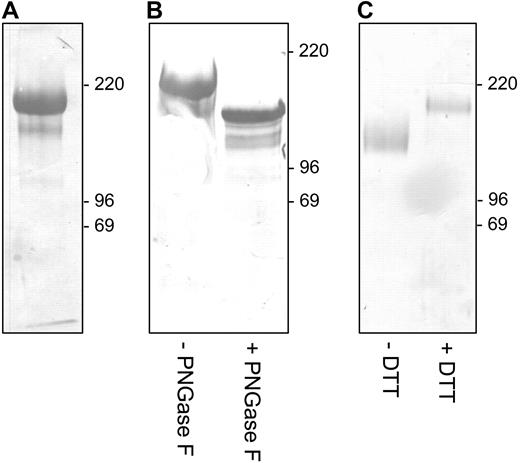
In order to establish the molecular identity of the rat macrophage ED2 glycoprotein antigen, we subjected a rat spleen lysate to affinity chromatography using a column containing the ED2 mAb coupled to sepharose. Previous work had shown that ED2 immunoprecipitates contained 3 (glyco)proteins (175, 160, and 95 kDa).15 However, our subsequent preclearing experiments with specific antibodies showed that the 160- and 95-kDa polypeptides represented contaminating CD11b/CD18 integrin (Mac-1), which was not directly associating with the 175-kDa ED2 antigen (data not shown). Thus, after preclearing the lysate over a column with irrelevant IgG, affinity purification on the ED2 column yielded the major approximately 175-kDa protein (Figure 2A). Amino acid sequencing of the N-terminus and internal peptides (Table 1) identified the ED2 molecule as the rat ortholog of CD163. The minor band from Figure 2A, which was confirmed by peptide sequencing to be rat CD163 too (not shown), probably represents a degradation product of CD163 or a splice- or O-glycosylation variant. The rat CD163 polypeptide sequence is very similar to its human23 and mouse24 counterparts (for an alignment of the complete predicted rat, mouse and human CD163 aa sequences, see Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). Rat spleen CD163 is a glycoprotein carrying a significant amount of N-linked glycans, as indicated by the reduction in molecular weight (Mr) upon PNGase F treatment to approximately 140 kDa (Figure 2B). The latter roughly corresponds to the calculated Mr 128.3 kDa of the predicted rat CD163 1184–aa polypeptide, although clearly there may be some O-glycosylation as well. Furthermore, rat CD163 migrated at a lower Mr (approximately 145 kDa) under nonreducing conditions, suggesting internal disulfide bridges (Figure 2C), which is in line with the presence of scavenger domains that are predicted to have a number of intradomain disulfide bonds. CD163 is a member of the group B scavenger receptor cysteine-rich domain (SRCR-B) family and has been shown to act as a receptor for the binding and uptake of hemoglobin-haptoglobin (Hb-Hp) complexes.12,25,26 Collectively, these results identify the ED2 antigen as the rat CD163 glycoprotein.
Purification and identification of the ED2 antigen as rat CD163. (A) The approximately 175-kDa ED2 antigen was immunoaffinity purified from rat spleen, electrophoresed, blotted, and stained with Coomassie Brilliant Blue. (B) Analysis of N-linked glycosylation of the CD163/ED2 antigen by endoglycosidase F treatment. (C) SDS-PAGE of CD163/ED2 antigen under reducing (+DTT, 175 kDa) or nonreducing (−DTT, 145 kDa) conditions. All samples were run on 10% SDS-PAGE under reducing conditions (+DTT) unless indicated otherwise.
Purification and identification of the ED2 antigen as rat CD163. (A) The approximately 175-kDa ED2 antigen was immunoaffinity purified from rat spleen, electrophoresed, blotted, and stained with Coomassie Brilliant Blue. (B) Analysis of N-linked glycosylation of the CD163/ED2 antigen by endoglycosidase F treatment. (C) SDS-PAGE of CD163/ED2 antigen under reducing (+DTT, 175 kDa) or nonreducing (−DTT, 145 kDa) conditions. All samples were run on 10% SDS-PAGE under reducing conditions (+DTT) unless indicated otherwise.
Identification of the ED2 antigen as rat CD163
| Peptide . | ED2 antigen . | Rat CD163 . | Position . | Identity, % . |
|---|---|---|---|---|
| 1* | VTQAPEGRKKELLLAG | VTQAPEGRKKELRLAG | 30-45 | 94 |
| 2 | WGTVCDDNFSK | WGTVCDDNFSK | 170-180 | 100 |
| 3 | QLGCGSALSFSGSAK | QLGCGSALSFSGSAK | 189-203 | 100 |
| 4 | QLGCPTAITAIGRVNASK | QLGCPTAITAIGRVNASE | 300-318 | 94 |
| 5 | EDAGVTCSDGADLELR | EDAGVTCSDGADLELR | 349-364 | 100 |
| 6 | LVGGEIPCSGR | LVGGEIPCSGR | 475-485 | 100 |
| 7 | GAGQVWRHK | GAGQVWRHK | 633-641 | 100 |
| 8 | VDTLWQCPSSPWK | VDTLWQCPSSPWK | 894-906 | 100 |
| 9 | EAAFGPGTGPIWLNEMK | EAAFGPGTGPIWLNEMK | 976-992 | 100 |
| Peptide . | ED2 antigen . | Rat CD163 . | Position . | Identity, % . |
|---|---|---|---|---|
| 1* | VTQAPEGRKKELLLAG | VTQAPEGRKKELRLAG | 30-45 | 94 |
| 2 | WGTVCDDNFSK | WGTVCDDNFSK | 170-180 | 100 |
| 3 | QLGCGSALSFSGSAK | QLGCGSALSFSGSAK | 189-203 | 100 |
| 4 | QLGCPTAITAIGRVNASK | QLGCPTAITAIGRVNASE | 300-318 | 94 |
| 5 | EDAGVTCSDGADLELR | EDAGVTCSDGADLELR | 349-364 | 100 |
| 6 | LVGGEIPCSGR | LVGGEIPCSGR | 475-485 | 100 |
| 7 | GAGQVWRHK | GAGQVWRHK | 633-641 | 100 |
| 8 | VDTLWQCPSSPWK | VDTLWQCPSSPWK | 894-906 | 100 |
| 9 | EAAFGPGTGPIWLNEMK | EAAFGPGTGPIWLNEMK | 976-992 | 100 |
Peptide sequences of the rat ED2 antigen and comparison (ie, position, percent amino acid identity) to corresponding sequence from the predicted rat CD163 (National Center for Biotechnology Information Sequence Viewer; http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&=62648134). Differences between the determined and predicted sequence are underlined. (An alignment of rat, mouse, and human CD163 is provided in Figure S1.)
N = terminal.
CD163 mediates erythroblast adhesion
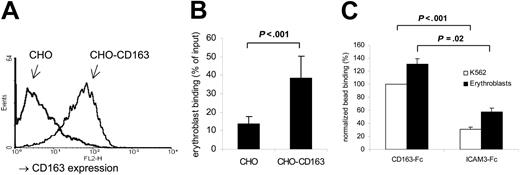
Our previous studies have shown that the binding of fetal liver erythroblasts to CD163-expressing (ie, ED2+) RBMMφ can be strongly inhibited using the ED2 mAb.11 Clearly, it was important to confirm that cellular CD163 expression could confer erythroblast binding. Therefore, we initially tested binding of the human erythroblast leukemic cell line K562 to CHO cells ectopically expressing the full-length human CD163 (Figure 3A). As can be seen in Figure 3B, CD163 expression consistently and significantly supported K562 cell binding. To provide further evidence for a direct interaction between CD163 and erythroblasts, we tested binding to a recombinant protein composed of the extracellular region of CD163, including all 9 scavenger receptor domains, and the Fc portion of the human IgG1 molecule (CD163-Fc). When CD163-Fc was coated onto fluorescent beads, it resulted in enhanced binding compared with control beads coated with an irrelevant control protein (ICAM-3-Fc; Figure 3C). The binding of erythroblasts to either CD163 transfectants or beads was not significantly affected by using Ca2+-free buffer and/or EDTA (results not shown). Although interactions of SRCR family members are often found to be Ca2+ dependent, divalent cation-independent cell-cell interactions by members of this family have also been reported, for instance for the SRCR-B member CD6.27 These findings, in conjunction with our previously reported data,11 suggests that CD163 can directly mediate erythroblast adhesion.
CD163 functions as an adhesion receptor for erythroblasts. (A) Surface expression of CD163, as identified by fluorescence-activated cell sorter (FACS) staining using mAb EDhu1, on CHO cells stably expressing human CD163 (CHO-CD163) or empty vector (CHO). (B) K562 erythroblastic leukemic cell binding to CHO cells expressing CD163. Data shown are the means ± standard deviation (SD) from 4 independent experiments. (C) Binding of K562 cells or freshly isolated rat erythroblasts to fluorescent beads coated with either CD163-Fc or ICAM3-Fc protein. Data shown are the means ± SD from 3 independent experiments.
CD163 functions as an adhesion receptor for erythroblasts. (A) Surface expression of CD163, as identified by fluorescence-activated cell sorter (FACS) staining using mAb EDhu1, on CHO cells stably expressing human CD163 (CHO-CD163) or empty vector (CHO). (B) K562 erythroblastic leukemic cell binding to CHO cells expressing CD163. Data shown are the means ± standard deviation (SD) from 4 independent experiments. (C) Binding of K562 cells or freshly isolated rat erythroblasts to fluorescent beads coated with either CD163-Fc or ICAM3-Fc protein. Data shown are the means ± SD from 3 independent experiments.
An erythroblast adhesion motif in the second CD163 scavenger receptor domain
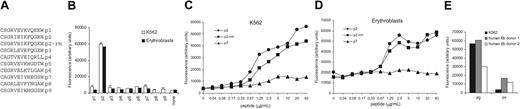
CD163 is composed of 9 extracellular SRCR domains. Previous studies on a closely related glycoprotein gp340/SAG/DMBT1 have mapped its ligand-binding site to a short amino acid peptide motif in the SRCR domain.18,19 The corresponding peptide in the SRCR domain of MARCO, a class A family member, has also been shown to mediate ligand binding.28 Interestingly, molecular modeling18 using the structure of the Mac-2–binding protein as a template29 has localized this 11-aa motif to a putative ligand binding pocket in the scavenger receptor domain. We hypothesized that the corresponding region in one or more of the CD163 scavenger domains may be responsible for erythroblast binding. Therefore, 13-mer peptides were generated encoding these motifs from each of the 9 scavenger domains of human CD163 (Figures 4A; S1) and tested for binding to K562 cells and freshly isolated rat erythroblasts. Among these CD163 peptides, only the one corresponding to the 11-aa motif of the second SRCR domain (CD163p2) of human CD163 displayed specific erythroblast binding (Figure 4B). Binding was dependent on coating concentration, and was also observed when the corresponding peptide of the rat and mouse CD163 domain 2 (p2-rm) was used (Figure 4C-D). Finally, human erythroblasts (95%-98% pure), generated in vitro from human CD34+ HSCs,17 also displayed binding to the CD163p2 peptide (Figure 4E). Taken together, these data suggested that the CD163p2 motif was, at least in part, responsible for mediating the CD163 binding to erythroblasts.
Identification of an erythroblast-binding motif in the second scavenger receptor domain of CD163. (A) Generated 13-mer peptides corresponding to sequences from each of the 9 extracellular scavenger domains of human CD163 (called p1-p9, respectively) and of the second domain of rat/mouse CD163 (called p2-rm). (B) Binding of K562 cells or freshly isolated rat erythroblasts to CD163 peptides. Peptides were used at 40 μg/mL coating concentrations, and cell binding was quantified by staining with the DNA-binding dye SYTO-13 followed by measurement of the fluorescence. Data shown are the means ± SD from at least 5 independent experiments. (C-D) Concentration dependence of K562 (C) and rat erythroblast (D) adhesion to the human (p2) or rodent (p2-rm) CD163-binding motif. Data shown are from 1 representative experiment of 3. (E) Binding of human erythroblasts, generated in vitro from CD34+ HSCs from 2 separate donors, to CD163p2 peptide. CD163p7 peptide is used as a negative control. K562 cell binding is shown for comparison. The 2 cell populations had comparable numbers (> 80%) of erythroblastic cells, with a similar subset distribution.
Identification of an erythroblast-binding motif in the second scavenger receptor domain of CD163. (A) Generated 13-mer peptides corresponding to sequences from each of the 9 extracellular scavenger domains of human CD163 (called p1-p9, respectively) and of the second domain of rat/mouse CD163 (called p2-rm). (B) Binding of K562 cells or freshly isolated rat erythroblasts to CD163 peptides. Peptides were used at 40 μg/mL coating concentrations, and cell binding was quantified by staining with the DNA-binding dye SYTO-13 followed by measurement of the fluorescence. Data shown are the means ± SD from at least 5 independent experiments. (C-D) Concentration dependence of K562 (C) and rat erythroblast (D) adhesion to the human (p2) or rodent (p2-rm) CD163-binding motif. Data shown are from 1 representative experiment of 3. (E) Binding of human erythroblasts, generated in vitro from CD34+ HSCs from 2 separate donors, to CD163p2 peptide. CD163p7 peptide is used as a negative control. K562 cell binding is shown for comparison. The 2 cell populations had comparable numbers (> 80%) of erythroblastic cells, with a similar subset distribution.
Interaction of erythroblasts with the CD163p2 motif promotes erythroid expansion
Upon demonstrating that the CD163p2 motif could contribute to erythroblast adhesion, it was of interest to investigate whether this interaction could also directly affect erythropoiesis. To investigate this possibility, rat erythroblasts were cultured in plates coated with CD163p2 or control peptides. These experiments were performed under conditions (ie, in the presence of EPO but in absence of other factors) that support a limited degree of erythroid differentiation and survival, but only allow minimal growth. By directly counting viable cells, the CD163p2 peptide was shown to specifically stimulate erythroid expansion as peptides corresponding to the other nonbinding CD163 SRCR domain motifs did not (Figure 5A). In contrast, there were no significant effects on erythroid maturation as evaluated by May-Grunwald-Giemsa stainings (Figure 5B). This suggested that binding of erythroblasts to the CD163p2 motif selectively promoted the growth and/or survival of these cells. It should be mentioned that we have not observed the growth promoting effect of CD163p2 peptide under the conditions that support the strong growth of the human CD34+ HSC-derived erythroblast cultures. Whether this is directly related to the high level of proliferation already occurring in these cells is not known.
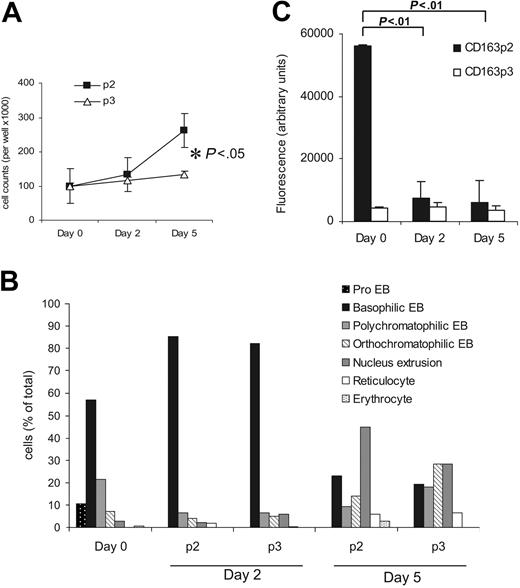
The CD163p2 motif promotes erythroid expansion. (A) Numbers of viable rat erythroblasts after culture for different times on CD163 peptide p2 and control peptide p3. (B) Maturation of rat erythroblasts after culture for different times on CD163 peptide p2 and control peptide p3. Results in panels A and B are from one representative experiment of 3. (C) Binding of rat erythroblasts at different stages after in vitro maturation in the presence of 2 IU/mL EPO to CD163p2 and control p3 peptides. Data shown are the means ± SD from 3 independent experiments. Statistics: 2-tailed t test.
The CD163p2 motif promotes erythroid expansion. (A) Numbers of viable rat erythroblasts after culture for different times on CD163 peptide p2 and control peptide p3. (B) Maturation of rat erythroblasts after culture for different times on CD163 peptide p2 and control peptide p3. Results in panels A and B are from one representative experiment of 3. (C) Binding of rat erythroblasts at different stages after in vitro maturation in the presence of 2 IU/mL EPO to CD163p2 and control p3 peptides. Data shown are the means ± SD from 3 independent experiments. Statistics: 2-tailed t test.
Finally, we investigated whether the adhesion of erythroblasts to CD163p2 is related to their maturation stage. Indeed, in vitro maturation of erythroblasts in the presence of EPO (Figure 5C) resulted in a virtually complete loss of CD163p2 peptide–binding capacity (Figure 5A). The loss of binding during differentiation coincided particularly well with the disappearance of proerythroblasts from the populations, indirectly suggesting that proerythroblasts may be the main erythroblast subset responsible for binding to CD163p2.
Discussion
In the present report we provide evidence that the surface receptor CD163 can function as an adhesion receptor for erythroblasts. CD163 is highly expressed on macrophages in erythroblastic islands both in the bone marrow as well as in extramedullary sites of erythropoiesis. We demonstrate that the rat ED2 antigen, which has previously been implicated in macrophage-erythroblast interactions,11 is the rat CD163 ortholog. Furthermore, we show that CD163, either expressed on cells or as a recombinant protein, can interact directly with erythroblastic cells. We also identify a motif in the second SRCR domain of CD163 that mediates erythroblast binding. Finally, we provide evidence that the interaction of this CD163 motif with erythroblasts promotes the growth and/or survival of these cells. Collectively, this identifies CD163 as a novel adhesion receptor that, together with previously identified molecules and perhaps others,2 mediates the interaction between macrophages and erythroblasts in erythroblastic islands.
Our findings suggest a potential role for CD163 in erythropoiesis, clearly adding a new dimension to the relationship between CD163 and red blood cells (RBCs). CD163 has previously been identified as a receptor for Hb-Hp complexes and, as such, is believed to contribute to the clearance of free hemoglobin, a potent oxidant, from the circulation.12 This appears to be of particular importance during conditions of intravascular hemolysis, such as autoimmune hemolytic anemia, sickle cell disease, or malaria. Consistent with this notion, CD163 is highly expressed on splenic red pulp and liver macrophages,13,16,30 which are major scavengers not only of Hb-Hp but also of senescent RBCs in vivo. Interestingly, our current results (Figure 1) support the idea that these strongly CD163+ macrophage subpopulations can simultaneously support extramedullary erythropoiesis. This occurs both during the indicated pathologic conditions as described above, in this paragraph, which are accompanied by anemia, but also during normal development. Therefore, our findings suggest that CD163 on liver and spleen macrophages could have a dual function (ie, simultaneously mediating the clearance of Hb and promoting erythropoiesis). Such a possible physical link between Hb clearance and erythropoiesis would provide a very efficient mechanism for recycling iron to developing erythroblasts. Of potential interest in this context, the CD163 Hb-Hp binding site appears to be distinct from the erythroblast-binding site, suggesting that binding of Hb-Hp complexes and erythroid precursors is coordinated by CD163 in erythroblastic islands. In particular, the third scavenger receptor domain of CD163 is most likely involved in Hb-Hp binding,12,25 while our current data implicate a motif in the second scavenger domain in erythroblast binding. Consistent with this hypothesis, we have not been able to demonstrate significant inhibition of erythroblast binding to CD163 with antibodies (ie, EDhu1) directed against the third scavenger receptor domains (data not shown), which have previously been shown to efficiently block Hb-Hp binding to CD163.25 As our current peptide analysis was limited to sequences corresponding to the previously identified ligand binding site in the related SRCR-B family member gp340/SAG,18,19 we cannot exclude that CD163 domains other than the CD163p2 motif contribute to erythroblast binding.
A recent study31 has suggested that CD163 is expressed on the surface of a small subset (approximately 2%) of CD34+ hematopoietic stem cells. Furthermore, evidence was reported that antibodies against the third scavenger receptor domain of CD163 (ie, EDhu1 and Mac2-158) can promote erythroid expansion from CD34+ cells in the presence of EPO and IL-3, suggesting that CD163 signaling may somehow promote erythropoiesis. This effect was postulated to mimic previously reported stimulatory effects of Hb on erythropoiesis.31 Our current findings may suggest an alternative explanation for these results. One possibility to consider, and re-evaluate, is that the small fraction of surface CD163+ cells within the CD34+ population may actually be macrophages. If so, then the ligation of CD163, by either Hb-Hp and/or erythroblasts, could induce the generation of erythroid differentiation-promoting factors by macrophages. In line with this idea, other ligands such as EDhu1 and Hb-Hp have previously been shown to trigger hematopoietic and inflammatory cytokine (eg, granulocyte-macrophage colony-stimulating factor [GM-CSF], IL-1β, IL-6, and IL-10) production in CD163-expressing macrophages.13,32 In addition to this, our current data suggest that erythroblasts have a putative ligand for CD163 that specifically recognizes the CD163p2 motif and provides signals that stimulate erythroblast growth and/or survival. The interactions of CD163 on macrophages with this putative ligand on erythroblasts could therefore regulate erythropoiesis by mediating bidirectional signaling in erythroblastic islands.
Our identification of CD163 as an erythroblast adhesion receptor also constitutes the first evidence for a role of CD163 in cell-cell interactions in general. This corresponds to the proposed functions of several other members of the SRCR-B family, including the lymphocyte surface molecules CD627,33,34 and WC1,35 and the soluble macrophage molecule Spα,36 which have also been shown to mediate interactions with cell-surface ligands. In particular, the third SRCR domain of CD6 binds to the activated leukocyte cell adhesion receptor (CD166) on endothelial cells,37 and WC1 interacts with an unknown ligand via its SRCR domains 9 and 11.35 In this context, it will be of interest to evaluate the role of CD163 in interactions between macrophages and other hematopoietic or nonhematopoietic cells as well.
Collectively, these findings show that the macrophage scavenger receptor CD163 can function as an erythroblast adhesion receptor in erythroblastic islands and suggest that this interaction may be instrumental in the regulation of erythropoiesis. Major future challenges will be to identify the natural ligand(s) for CD163 on erythroblasts and to establish the exact role of this interaction in erythropoiesis in vivo.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Connie Jiménez for her contribution to the ED2 peptide sequencing. We are also grateful to Ed Döpp and Priscilla Heijnen for hybridoma culture and antibody production. Kamran Nazmi is acknowledged for his contribution to peptide systhesis. Floris Bikker and Georg Kraal are acknowledged for useful discussions.
Authorship
Contribution: B.O.F., M.M.J.P., R.P.M.V., R.C.v.d.S., A.J.M.L., C.G., and K.M. performed research and analyzed data; L.K.W. contributed vital new reagents; S.K.M. and C.D.D. designed research and analyzed data; T.K.v.d.B. designed research, analyzed data, and wrote the paper; and B.O.F. and M.M.J.P. contributed equally to this work.
Conflict-of-interest disclosure: One author (T.V.D.B.) holds a patent related to the work described in this study. All other authors declare no competing financial interests.
Correspondence: Timo K. van den Berg, Department of Blood Cell Research, Sanquin Research, and Landsteiner Laboratory, Academic Medical Center, University of Amsterdam, PO Box 9190, 1006 AD Amsterdam, the Netherlands; e-mail: t.k.vandenberg@sanquin.nl.
References
Supplemental data
Links: AAK16065 and CAA80542Alignments were performed with the clustalW algorithm version 1.82 and adjusted manually. Amino acid identities for the full-length proteins were rat-mouse 89%, rat-human 74%, and human-mouse 73%. The positions of the peptides identified by amino acid sequencing (Fig. 1D) are indicated in red; the first of these (ie, VTQAPEGRKKELRLAG) also indicates the N-terminus of the mature polypeptide. The peptides that were synthesized and analyzed for erythroblast binding are highlighted in blue. Dotted lines indicate respective leader and transmembrane regions; the scavenger domain boundaries are also indicated.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal