Abstract
The GATA-1–interacting protein Friend Of GATA-1 (FOG-1) is essential for the proper transcriptional activation and repression of numerous GATA-1 target genes. Although FOG-1–independent activation by GATA-1 has been described, all known examples of GATA-1–mediated repression are FOG-1 dependent. In the GATA-1–null G1E cell line, estrogen receptor ligand binding domain (ER) chimeras of either wild-type GATA-1 or a FOG-1–binding defective mutant of GATA-1 repressed several genes similarly upon activation with β-estradiol. Repression also occurred in a FOG-1–null cell line expressing ER–GATA-1 and during ex vivo erythropoiesis. At the Lyl1 and Rgs18 loci, we found highly restricted occupancy by GATA-1 and GATA-2, indicating that these genes are direct targets of GATA factor regulation. The identification of genes repressed by GATA-1 independent of FOG-1 defines a novel mode of GATA-1–mediated transcriptional regulation.
Introduction
FOG-1 is an essential coregulator of GATA-1 during hematopoiesis, mediating both transcriptional activation and repression.1–3 The importance of FOG-1 is underscored by the finding that FOG-1–deficient mice die from anemia with defects resembling loss of GATA-1.3 A substitution mutation at valine 205 within the amino-terminal zinc finger of GATA-1 that impairs the association with FOG-11 occurs in patients with anemia.4 In the GATA-1–null erythroid precursor cell line G1E,5 valine 205 mutations prevent GATA-1 from rescuing the maturation block.2 A compensatory FOG-1 mutation, which restores the interaction with GATA-1 (V205) mutants, rescues the G1E differentiation defects, demonstrating the specificity of the GATA-1 (V205) mutation for FOG-1. Repression by GATA-1 can involve FOG-1–dependent recruitment of the nucleosome remodeling and histone deacetylase (NuRD) complex to GATA-1 target genes.6 However, complementation studies in FOG-1−/− cells indicate that the NuRD binding domain of FOG-1 is required for megakaryocytic but not erythroid differentiation.7
Despite the importance of FOG-1 as a GATA-1 coregulator, examples of FOG-1–independent transcriptional activation by GATA-1 have been reported. Expression of ER–GATA-1(V205G) in G1E cells induced transcripts for erythroid Kruppel-like factor (Eklf), heme-regulated eIF-α-kinase (HRI), and Fog1.2 We described FOG-1–independent activation of Tac2, which encodes a neurokinin-B precursor.8,9 ER–GATA-1–mediated Tac2 expression was biphasic, with FOG-1–independent activation followed by FOG-1–dependent repression. Abrogating the GATA-1 interaction with FOG-1 enhanced the ability of GATA-1 to occupy a Tac2 regulatory region and to activate Tac2 transcription and prevented the late repression phase. Whereas the aforementioned genes are induced by GATA-1 independently of FOG-1, herein we describe a novel mode of GATA-1 repression, operational at target genes in mouse and human systems, in which FOG-1 is absolutely not required.
Materials and methods
Cell isolation and culture
G1E cells were cultured as described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Human primary erythroid progenitors were derived by in vitro culture of CD34+ cells isolated from growth factor–mobilized peripheral blood10 as described in Document S1.
Quantitative real-time RT-PCR
cDNAs were prepared from RNA purified with Trizol (GIBCO/BRL, Carlsbad, CA) as described.11 Primers are described in Document S1.
Quantitative chromatin immunoprecipitation (ChIP) assay
ChIP analysis was conducted as described.9 Primers and antibodies are described in Document S1.
Results and discussion
Transcriptional regulation by GATA-1 has been extensively studied in G1E cells using a ligand-activated estrogen receptor hormone binding domain fusion to GATA-1 (ER–GATA-1).9,11,12 As a discovery tool to identify GATA-1 target genes regulated independently of FOG-1, we conducted expression profiling in a G1E clone expressing the ER–GATA-1 (V205G) mutant. Genes identified on the array as being repressed by ER–GATA-1(V205G) upon β-estradiol treatment (Figure S1) were validated by real-time RT-PCR. Wild-type ER–GATA-1 and ER–GATA-1(V205G) repressed several genes similarly (Figure 1A). Of the targets identified, only lymphoblastic leukemia (Lyl1) has been described as a GATA-1–repressed gene.13 Lyl1 is expressed in hematopoietic stem/progenitor cells and in all hematopoietic lineages except T cells.14,15 Lyl1 overexpression is associated with T-cell acute lymphoblastic leukemia16 and acute myeloblastic leukemia.17 Regulator of G-protein signaling 18 (Rgs18) is expressed in hematopoietic stem cells (HSCs) and to a lesser extent in more committed hematopoietic progenitors18 and is abundant in megakaryocytes.19 Rgs13 is expressed in germinal center B lymphocytes and is implicated in regulating chemokine response.20 Although not identified on the array, Rgs1, positioned between Rgs13 and Rgs18 on chromosome 1, was also down-regulated in G1E cells by ER–GATA-1(V205G). Rgs1 is expressed in B cells21,22 and monocytes.23 Treml2, a member of the triggering receptor expressed on myeloid cells gene cluster, is expressed in B cells, neutrophils, and macrophages.24 Clec4d (Mpcl/mcl/mMCL/Clecsf8) is a C-type lectin with restricted expression in the monocyte/macrophage lineage.25 Adamts5, a disintegrin and metalloprotease with thrombospondin type 1 motif 5, is an aggrecanase present in cartilage.26 Tmem44, a putative transmembrane protein, has not been studied. Whereas the genes identified herein were efficiently repressed by ER–GATA-1(V205G), Gata2, a known FOG-1–dependent GATA-1 target,2,11 was not repressed in these clones. Furthermore, none of the genes were repressed in β-estradiol–treated G1E cells that lacked an ER–GATA-1 chimera, demonstrating that repression is not due to β-estradiol treatment (Figure S2).
Novel mode of GATA-1 function: FOG-1–independent repression. Real-time RT-PCR analysis of mRNA levels in (A) G1E cells expressing either wild-type ER–GATA-1(WT) or ER–GATA-1(V205G) and (B) FOG-1−/− cells expressing wild-type ER–GATA-1. Transcript levels from cells treated with 1 μM β-estradiol for 24 hours were compared with untreated controls. Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript levels and, for each clone, the mean value for untreated samples was set to 1 (mean ± standard error [SE] from 3 independent experiments). (C) Real-time RT-PCR analysis of mRNA levels during ex vivo human erythropoiesis. CD34+ cells isolated from peripheral blood were cultured for up to 14 days. RNA was isolated at days 3, 7, 10, and 14 from 2 independent samples. Transcript levels were normalized to 18S RNA levels and the relative level of transcript from day 3 for each sample was set to 1.
Novel mode of GATA-1 function: FOG-1–independent repression. Real-time RT-PCR analysis of mRNA levels in (A) G1E cells expressing either wild-type ER–GATA-1(WT) or ER–GATA-1(V205G) and (B) FOG-1−/− cells expressing wild-type ER–GATA-1. Transcript levels from cells treated with 1 μM β-estradiol for 24 hours were compared with untreated controls. Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript levels and, for each clone, the mean value for untreated samples was set to 1 (mean ± standard error [SE] from 3 independent experiments). (C) Real-time RT-PCR analysis of mRNA levels during ex vivo human erythropoiesis. CD34+ cells isolated from peripheral blood were cultured for up to 14 days. RNA was isolated at days 3, 7, 10, and 14 from 2 independent samples. Transcript levels were normalized to 18S RNA levels and the relative level of transcript from day 3 for each sample was set to 1.
As the V205G mutation may not completely abrogate the GATA-1:FOG-1 interaction4 in the G1E cell system, we used clones of a FOG-1−/− hematopoietic precursor cell line7 stably expressing ER–GATA-1. Upon β-estradiol treatment, Lyl1, Rgs1, Rgs18, Treml2, and Clec4d were repressed (Figure 1B). Repression of Rgs13, Adamts5, and Tmem44 was not measured, as the respective mRNA levels were extremely low in FOG-1−/− cells, perhaps due to repression by endogenous GATA-1. Whereas ER–GATA-1 repressed Gata2 in G1E cells 15- and 30-fold, Gata2 mRNA in FOG-1−/− clones decreased only 2- to 4-fold, which represents an 86% reduction in the magnitude of repression, despite high-level ER–GATA-1 expression. Although it is unclear why low-level Gata2 repression occurs in FOG-1−/− cells but not G1E cells, many GATA-1 targets are not absolutely FOG-1 dependent but can be regulated by GATA-1 to a certain degree independently of FOG-1 (data not shown).
To assess whether the novel FOG-1–independent GATA-1 targets are down-regulated during erythropoiesis, we measured their expression during ex vivo differentiation of human peripheral blood CD34+ erythroid progenitors. Gata1 and Gata2 are up-regulated and down-regulated, respectively, in this system.8 In 2 independent samples, LYL1, RGS1, RGS18, TREML2, CLEC4D, and TMEM44 were down-regulated, whereas the mRNA levels of the Lyl1 paralog TAL1 were largely unchanged until day 14, in which small increases were evident.
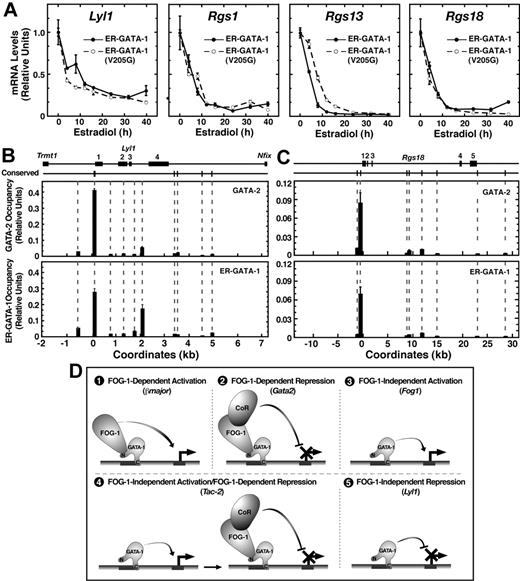
In G1E and FOG-1−/− cells, repression was measured 24 hours after induction of ER–GATA-1 activity by β-estradiol. As such, the ER–GATA-1(V205G) repression kinetics may differ from that of ER–GATA-1. However, mRNA loss rates were nearly identical for Lyl1, Rgs1, and Rgs18 in G1E clones (Figure 2A). Only Rgs13 repression was delayed by the V205G mutation.
Highly restricted GATA factor occupancy at endogenous Lyl1 and Rgs18 domains. (A) Comparison of transcript levels in G1E cells expressing wild-type ER–GATA-1 or ER–GATA-1(V205G) following β-estradiol treatment (1 μM) for various times up to 40 hours. Transcript levels were normalized to GAPDH transcript levels and, for each clone, the mean value for untreated samples was set to 1 (mean ± SE from 3 independent experiments). GATA factor occupancy was measured at (B) the Lyl1 locus and (C) the Rgs18 locus by quantitative real-time ChIP analysis in G1E–ER–GATA-1 cells. For GATA-2 and GATA-1, ChIPs were conducted using untreated or β-estradiol–treated (1 μM, 24 h) cells, respectively (mean ± SD, 2 to 4 independent ChIP experiments). Mean preimmune control signals did not exceed 0.0026 for Lyl1 or 0.0015 for Rgs18 and are not shown. The positions of conserved (mouse to man) WGATAR, NGATAR, and WGATAN sites in each locus are shown above the graph. All nonconserved WGATAR motifs in the Lyl1 locus were also analyzed by ChIP. Coordinates are based upon distance from the conserved WGATAR motif (TTATCA) in the Lyl1 promoter and the transcriptional start site of the Rgs18 locus. (D) Multiple modes of GATA-1–mediated transcriptional regulation. An example of a gene regulated via each mode is indicated in parentheses.
Highly restricted GATA factor occupancy at endogenous Lyl1 and Rgs18 domains. (A) Comparison of transcript levels in G1E cells expressing wild-type ER–GATA-1 or ER–GATA-1(V205G) following β-estradiol treatment (1 μM) for various times up to 40 hours. Transcript levels were normalized to GAPDH transcript levels and, for each clone, the mean value for untreated samples was set to 1 (mean ± SE from 3 independent experiments). GATA factor occupancy was measured at (B) the Lyl1 locus and (C) the Rgs18 locus by quantitative real-time ChIP analysis in G1E–ER–GATA-1 cells. For GATA-2 and GATA-1, ChIPs were conducted using untreated or β-estradiol–treated (1 μM, 24 h) cells, respectively (mean ± SD, 2 to 4 independent ChIP experiments). Mean preimmune control signals did not exceed 0.0026 for Lyl1 or 0.0015 for Rgs18 and are not shown. The positions of conserved (mouse to man) WGATAR, NGATAR, and WGATAN sites in each locus are shown above the graph. All nonconserved WGATAR motifs in the Lyl1 locus were also analyzed by ChIP. Coordinates are based upon distance from the conserved WGATAR motif (TTATCA) in the Lyl1 promoter and the transcriptional start site of the Rgs18 locus. (D) Multiple modes of GATA-1–mediated transcriptional regulation. An example of a gene regulated via each mode is indicated in parentheses.
GATA-2 occupies conserved GATA motifs within the Lyl1 promoter,27 and a 464-bp region of the promoter that includes the GATA motifs is sufficient to drive Lyl1 expression in endothelial and hematopoietic cells.28 GATA-1 occupancy of the Lyl1 locus had not been studied previously. In G1E cells, ChIP analysis revealed GATA-2 occupancy at the Lyl1 promoter and reduced levels within intron 3 containing a nonconserved WGATAR motif. Little to no occupancy was detected at other WGATAR motifs between Trmt1 and Nfix that flank Lyl1 or at conserved (mouse to human) NGATAR and WGATAN motifs within the same region (Figure 2B). Similar to GATA-2, ER–GATA-1 occupied both the promoter and intron 3 upon ER–GATA-1 activation. These sites were also occupied by GATA-1 and -2 in day-4 embryoid bodies (Figure S3). We also tested whether GATA-1 and GATA-2 occupy the Rgs18 locus. Maximal ER–GATA-1 and GATA-2 occupancy occurred at conserved GATA motifs within the promoter (Figure 2C). Little to no occupancy was detected at other conserved GATA motifs within Rgs18. These results provide strong evidence that Lyl1 and Rgs18 repression occurs via direct ER–GATA-1 occupancy at these loci.
FOG-1–independent repression represents an entirely new mode of GATA-1 activity, expanding the repertoire of mechanisms by which GATA-1 regulates transcription (Figure 2D). The segregation of GATA-1 target genes into distinct mechanistic categories lays the groundwork for the identification of new mediators of GATA-1 function and hematopoiesis. It will be particularly instructive to determine whether target genes residing within a single category function in a common biologic pathway or if the mechanistic diversity reflects the need to contend with a spectrum of chromosomal environments.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants DK55700 (E.H.B.), DK68634 (E.H.B.), and DK069488 (K.D.J.).
National Institutes of Health
Authorship
Contribution: K.D.J. designed and performed research, analyzed data, and wrote the manuscript; M.E.B., J.-A.K., and A.W. performed research; A.B.C. provided a vital reagent; and E.H.B. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Emery H. Bresnick, 385 Medical Sciences Center, 1300 University Avenue, Madison, WI 53706; e-mail, ehbresni@wisc.edu.

![Figure 1. Novel mode of GATA-1 function: FOG-1–independent repression. Real-time RT-PCR analysis of mRNA levels in (A) G1E cells expressing either wild-type ER–GATA-1(WT) or ER–GATA-1(V205G) and (B) FOG-1−/− cells expressing wild-type ER–GATA-1. Transcript levels from cells treated with 1 μM β-estradiol for 24 hours were compared with untreated controls. Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript levels and, for each clone, the mean value for untreated samples was set to 1 (mean ± standard error [SE] from 3 independent experiments). (C) Real-time RT-PCR analysis of mRNA levels during ex vivo human erythropoiesis. CD34+ cells isolated from peripheral blood were cultured for up to 14 days. RNA was isolated at days 3, 7, 10, and 14 from 2 independent samples. Transcript levels were normalized to 18S RNA levels and the relative level of transcript from day 3 for each sample was set to 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/12/10.1182_blood-2007-02-072983/4/m_zh80120701980001.jpeg?Expires=1769130204&Signature=Chg60-4Pae5X9MaAg2FE9HS-mF9ODiD1OvNjT1~201hBERhfRUqQ2GjXwIx3aFgo~-okoIASMLWwC~P4TTjTSzQKKhzC1blya-Akcye6zfGqMpcQGWcUvOAN2-~BDP9r-l5QDlnUcw-c1XJKqC60fUPa5fCubKvUIF6icnHPzBYXway-6SbWw6bRGJCVAAD04xgTY98RWl02NWRIL5WPaZHs-SjN2ibl9XgZDf70FA-eNSll0ib2M4lLwWvl~DRswRIgug37mN3~foZ8v7dWeT1Q6QDIgUnEYfQLRT-AmKp2HfZnVSN4eCQE4Xt9~M55Yfr787PpncuToQDtPhLIfg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal