Abstract
The human germinal-center–associated lymphoma (HGAL) gene and its cognate protein are expressed in a germinal center (GC)–specific manner. Its expression in classic Hodgkin lymphoma (cHL) prompted us to address whether HGAL expression could distinguish biologically distinct subgroups of cHL. Tissue microarrays from 145 patients treated with curative intent showed HGAL staining in 75% and was closely correlated with MUM1/IRF4 (92%) expression. BCL6 (26%), CD10 (0%), BCL2 (31%), Blimp1 (0.02%), and Epstein-Barr virus (EBV) (20%) showed no specific correlation; neither did phospho-STAT6, a key mediator of IL-4 and IL-13 signaling that induces HGAL and is implicated in cHL pathogenesis. In our study cohort, the 5-year overall survival (OS) correlated with young age (less than 45 years, P < .001), low stage (stage I and II, P = .04), and low International Prognostic Score (P = .002). In univariate analysis, HGAL expression was associated with improved OS (P = .01) and failure-free survival (FFS) (P = .05) but was not independent of other factors in multivariate analysis of OS or FFS. The expression of the GC-specific marker HGAL in a subset of cHL suggests that these cHLs retain characteristics of GC-derived lymphomas. The association with improved OS in univariate but not multivariate analysis suggests that HGAL expression is related to known clinical parameters of improved survival.
Introduction
Classic Hodgkin lymphoma (cHL) is characterized by scattered large atypical cells in a mixed inflammatory milieu and an immunophenotype unlike that of any normal cell of the hematopoietic system. Studies on single cells have established that cHL cells are derived from clonal B cells and rarely from T cells.1 Cases of B-cell derivation typically harbor rearranged immunoglobulin (Ig) genes containing somatic mutations similar to those of normal B cells that have passed through the germinal center (GC). But cHL cells lack a functional surface B-cell receptor and therefore differ from normal B cells and other B-cell lymphomas. A subset of cHL possesses deleterious (“crippling”) somatic mutations in its Ig genes,2 and recent evidence points to a role for the Epstein-Barr virus (EBV) in enabling the cells' escape from apoptosis that is normally observed in GC B cells with nonfunctional Ig genes.3,4,5 In some cases of cHL the Ig genes have preserved coding capacity but still lack Ig mRNA, apparently due to a defect in Ig transcription.6 Gene-expression profiling studies of cHL cell lines confirm these observations and, in addition, show that cHL cells display down-regulation of B-lineage–specific genes and also of genes for key signaling pathways and transcription factors active in normal B cells.7
At least 80% of patients with cHL are cured with currently available therapies.8 However, biologic markers capable of accurately separating risk groups to better predict treatment failure and to reduce unnecessary treatment have not been identified. The International Prognostic Score (IPS) incorporates 7 clinical and laboratory parameters and is currently considered the gold standard for stratifying patients with advanced-stage cHL.9 The IPS has been reported to be applicable for patients with early-stage Hodgkin lymphoma10 ; however, these patients are usually risk-stratified based on the number of involved regions on the same side of the diaphragm (up to 3 or more than 3), evidence of bulky disease, presence of B symptoms, and erythrocyte sedimentation rate (ESR).11 Recently, members of our group showed that BCL2 protein expression is an independent predictor of poor outcome in cHL patients.12
The human germinal center–associated lymphoma (HGAL) gene was initially identified from an expressed sequence tag that was associated with improved survival in patients with diffuse large B-cell lymphoma (DLBCL).13,14 Molecular profiling using cDNA microarrays had previously established that DLBCL patients with expression of genes similar to germinal center B cells (GCB-like DLBCL) demonstrate a better overall outcome compared with those that express genes found in activated peripheral-blood B cells (ABC-like DLBCL).14 We cloned and characterized HGAL and found that it is induced by interleukin-4 (IL-4).13,14 In addition, we generated a monoclonal antibody to the HGAL protein and showed that it is expressed in normal GC and in GC-derived lymphomas but not in other B-, T- or natural killer (NK)–cell lymphomas.15 We subsequently found that HGAL protein is expressed in 70% of nodular lymphocyte-predominant Hodgkin lymphoma, a tumor that is considered to be derived from GC B cells but, surprisingly, we also found it expressed in 73% of classic Hodgkin lymphoma.15
In the current study, we aimed to confirm our initial observation of HGAL expression in cHL in an independent cohort of patients. We investigated whether those cHLs that express the HGAL protein also retain expression of other GC-specific markers. To this end, we compared the protein-expression pattern of HGAL with GC markers, BCL6 and CD10, and non-GC markers, BCL2, MUM1/IRF4, and Blimp1. Correlation between EBV and HGAL expression was also explored to determine whether EBV is associated with HGAL-positive cHL. Given that HGAL is an IL-4–inducible gene and cHL is characterized by an abundance of inflammatory cells secreting large amounts of cytokines implicated in cHL pathogenesis, we examined whether cytokines deregulated in cHL and components of their specific intracellular signaling pathways contribute to HGAL expression. Lastly, we addressed whether HGAL protein expression could serve to distinguish biologically distinct subgroups of cHL associated with different clinical outcome.
Patients, materials, and methods
Patient selection
A total of 145 informative cases of cHL, 72 from the Cleveland Clinic Foundation and 73 from the Nebraska Lymphoma Study Group, constitute this study cohort. Institutional review board (IRB) approval was obtained from all participating institutions. All patients were staged at initial diagnosis according to the modified Ann Arbor staging system. The following clinical and laboratory data from the time of diagnosis were collected: age, sex, stage, presence or absence of B symptoms or bulky disease (defined as mediastinal widening of at least one third of the thoracic diameter or a mass of at least 10 cm in maximum dimension), number of involved sites, ESR, and subtype of histology. The IPS was computed based on the following 7 designated parameters for high-risk disease: age at least 45 years, male sex, stage IV disease, white blood count (WBC) at least 15 × 109/L (15 000/μL), lymphocyte count less than 0.6 × 109/L (600/μL), serum albumin less than 40 g/L (4 g/dL), and hemoglobin less than 105 g/L (10.5 g/dL). Patients were categorized into a low-risk group if up to 2 adverse factors were present and a high-risk group if 3 or more adverse factors were present. In addition, patients with early stage (I and II) were stratified into favorable- and unfavorable-risk subgroups based on the number of involved regions on the same side of the diaphragm (1 to 3 or more than 3), evidence of bulky disease, and presence of B symptoms, as previously suggested.11 Data on ESR at presentation were available only for a limited number of early-stage patients and thus were not used for risk stratification of patients. None of the patients had a known history of HIV infection or other forms of immunosuppression.
All patients were treated with curative intent. In general, patients with early-stage disease received radiation therapy alone or combination chemotherapy with or without radiation therapy. Patients with advanced-stage disease received combination chemotherapy with or without radiation therapy. Patients with limited-stage disease who either demonstrated resistance or relapsed following initial response to radiation therapy were retreated with combination chemotherapy. Patients who failed initial chemotherapy received salvage chemotherapy or high-dose chemotherapy with autologous stem cell transplantation. Of 120 patients treated with chemotherapy, 30% initially received ChlVPP (chlorambucil, vinblastine, procarbazine, and prednisone), 23% received ABVD (adriamycin, bleomycin, vinblastine, and dacarbazine), 23% received MOPP (mechlorethamine, vincristine, procarbazine, and prednisone [3%] or MOPP/ABV or ABVD hybrid [20%]), and 17% received ChlVPP/ABV. Nine patients (7%) received other therapies. Follow-up information was obtained from the medical records of the Cleveland Clinic Foundation and the Nebraska Lymphoma Study Group. Availability of tissue, clinical follow-up information, and having undergone treatment with curative intent were considered inclusion criteria for this study. The studies were approved by the IRB at Cleveland Clinic and the University of Nebraska. All of the patients signed informed consent, in accordance with the Declaration of Helsinki and IRB instructions.
Tissue microarray construction
Tissue microarrays (TMAs) were constructed using a tissue arrayer (Beecher Instruments, Silver Spring, MD) by a previously described method.16 Standardized methods for tissue fixation (10% buffered formalin) and processing for paraffin-embedded sections were used at all participating centers. Tissue cores were selected for TMA by characteristic morphology based on examination of hematoxylin and eosin (H & E)–stained sections without prior knowledge of immunohistologic stains of individual cases. Two cores of 1.5 mm were taken from all paraffin blocks to maximize inclusion of Hodgkin cells on selected cores. Sections of 4 to 5 μm thickness were cut from the TMAS and placed on glass slides, which were then baked for 1 hour at 60°C before immunohistochemistry was performed.
Immunohistochemistry
Primary antibodies directed against proteins HGAL, BCL6, CD10, MUM1/IRF4, BCL2, Blimp1, LMP2A, and pSTAT6 were used in this study. The conditions used for immunohistologic staining are summarized in Table 1. Serial 4 to 5 μm–thick sections from paraffin-embedded lymphoid tissue or TMAs were deparaffinized in xylene and hydrated in a series of graded alcohols. Heat-induced antigen retrieval was carried out by microwave pretreatment in the appropriate buffer as indicated for each antibody. Detection was carried out using the DAKO Envision system (DAKO, Carpinteria, CA). Staining for all antibodies was optimized on normal paraffin-embedded tonsil sections. For the HGAL antibody, a tyramide amplification system was used (DAKO Catalyzed Signal Amplification System; DAKO). Scoring criteria as a percent of Hodgkin cells for each antibody are listed in Table 1. These cutoffs were chosen, and the scoring of the cases was performed before examining correlation with clinical end points. Cases from both institutions were scored independently by 2 observers (Y.N. and E.D.H.).
All hematoxylin-and-eosin and immunohistochemistry images were analyzed using a Nikon Eclipse E1000M microscope (Nikon, Burlingame, CA) equipped with a 40×/0.75 NA objective lens. Images were photographed using a SPOT RT color camera (Diagnostic Instruments, Sterling Heights, MI) and Ektachrome 100 Plus EPP 135-136 film (Eastman Kodak, Rochester, NY). The resulting photographs were acquired using a Nikon Superscan 4000ED scanner, and digitized images were processed using Adobe Photoshop CS (Adobe Systems, San Jose, CA).
In situ hybridization for Epstein-Barr viral RNA
In situ hybridization was performed on 4 to 5 μm–thick formalin-fixed, paraffin-embedded sections using the INFORM EBER probe (Ventana, Tucson, AZ). Slides were stained on an automated immunostainer (Ventana Benchmark) using the Ventana ISH iVIEW detection kit. Specific nuclear signal was considered positive. An oligo(dT) control to verify intact RNA within the nuclei of the cells composing the tissue and a known positive control were performed.
Induction of HGAL expression by cytokine stimulation
Peripheral-blood mononuclear cells from healthy donors were isolated by Ficoll-Hypaque density centrifugation (Amersham Pharmacia Biotech, Piscataway, NJ). B cells were enriched to more than 90% by human B-cell enrichment cocktail (StemCell Technologies, Vancouver, BC, Canada), as determined by fluorescence-activated cell sorter (FACS) analysis. Enriched B cells were plated at 5× 106 cells per well in 6-well plates (Corning Glassworks, Corning, NY) in IMDM (Gibco BRL, Carlsbad, CA) supplemented with 2% fetal calf serum (FCS), 0.5% bovine serum albumin (Sigma, St Louis, MO), 50 μg/mL human transferrin (Sigma), 5 μg/mL bovine insulin (Sigma), and 15 μg/mL gentamicin (Cellgro-Mediatech, Herndon, VA) at 37°C in 5% CO2. The cells were stimulated with IL-4, IL-10, IL-2, IL-13 (R&D Systems, Minneapolis, MN), or a combination of IL-4 and IL-13 for 6 hours. Concentrations of cytokines used for stimulation are shown in Figure 3.
Isolation of RNA, its quantification, and the reverse transcription (RT) reactions were performed as previously reported.17 HGAL mRNA expression was measured by real-time polymerase chain reaction (PCR) using the Applied Biosystems Assays-on-Demand Gene Expression Product on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA), as we previously reported.17
Statistical analysis
Overall survival was defined as the time interval between the date of diagnosis to the date of death or last follow-up. Failure-free survival was defined as the time interval between the date of initial diagnosis and date of disease progression or death from any cause, whichever came first. The Fisher exact test was used to compare patient groups with respect to HGAL staining, and the generalized Wilcoxon rank sum test was used for univariate comparisons of overall and failure-free survival. The Cox proportional hazards model, stratified by center, was used to simultaneously assess the impact of multiple factors on these outcomes. Hierarchic clustering was used to assess the associations between HGAL, CD10, BCL6, BCL2, MUM1, Blimp1, LMP2A, pSTAT6, and EBV EBER expression.
Results
Patient characteristics and HGAL staining
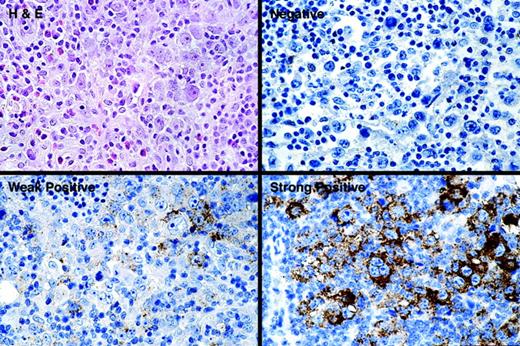
Overall, 145 informative cHL patients whose age ranged from 19 to 84 years (median, 32 years) were studied. All patients were treated with curative intent: 45% received combination chemotherapy and radiation therapy, 38% received only chemotherapy, while 17% received only radiation therapy. The follow-up period for the 145 patients ranged from 1 month to 17.6 years (median, 74.6 months). Forty patients (28%) failed initial treatment, and 31 (21%) died during the follow-up. HGAL protein was expressed in 109 (75%) cHL specimens, whereas 36 (25%) tumors lacked staining for HGAL. Of those with staining for HGAL, 71 (65%) showed strong staining, while 38 patients (35%) showed weak staining. HGAL staining was localized to the cytoplasm of large atypical cells corresponding to Hodgkin cells in all cases. Representative examples of strong, weak, and lack of HGAL staining are shown in Figure 1.
HGAL protein expression in classic Hodgkin lymphoma. The spectrum of HGAL staining in cHL cases is shown. Staining is localized to the cytoplasm of large atypical cells. Staining in more than 5% but less than 30% of Hodgkin cells was defined as weak, whereas staining in 30% or more Hodgkin cells was defined as strong.
HGAL protein expression in classic Hodgkin lymphoma. The spectrum of HGAL staining in cHL cases is shown. Staining is localized to the cytoplasm of large atypical cells. Staining in more than 5% but less than 30% of Hodgkin cells was defined as weak, whereas staining in 30% or more Hodgkin cells was defined as strong.
Comparative immunohistologic analysis and EBV RNA expression
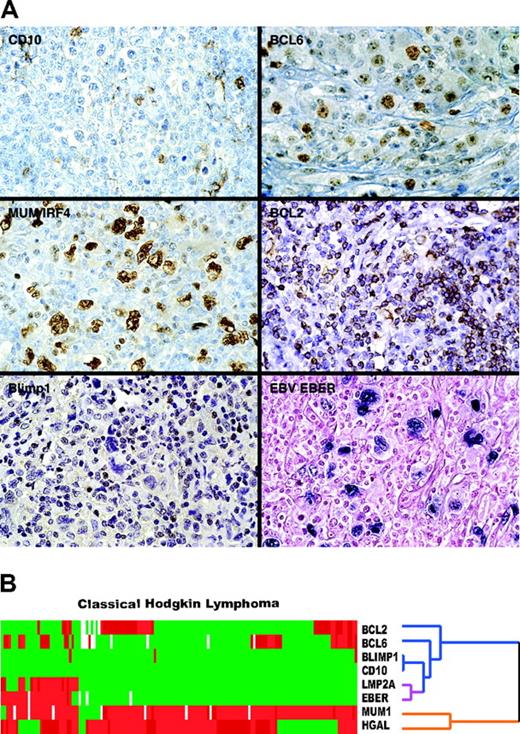
CD10 was not expressed in Hodgkin cells. Occasional cases with only cytoplasmic or Golgi staining were present; however, none of the tumors exhibited membrane reactivity for CD10. BCL6 was positive in 36 of 136 (26%) cases. MUM1/IRF4 was highly expressed in most cases (127 of 138; 92%). BCL2 was positive in 44 of 140 (31%) cases. Significant staining for Blimp1 was found in 0.02% of cases (3 of 139). In several additional cases only occasional Hodgkin cells showed staining for Blimp1, but the numbers of positive cells did not warrant a positive score as defined in Table 1 In situ hybridization for EBV using the EBER probe detected EBV-specific RNA in 28 of 140 (20%) cases. Because LMP2A has been suggested to play a role in the survival of EBV-associated lymphoma cells, we analyzed LMP2A staining and found it present in 22 of 28 (79%) EBER-positive cases but in only 2 of the remaining 140 (0.02%) EBER-negative Hodgkin lymphomas. Representative images of cHL cases stained for CD10, BCL6, BCL2, MUM1, Blimp1, and EBV EBER RNA are shown (Figure 2A)
Reagents and conditions for immunohistochemistry and in situ hybridization
| Antibody/probe . | Source* . | Dilution . | Pretreatment† . | Predominant localization . | Scoring scale, % . | ||
|---|---|---|---|---|---|---|---|
| Negative . | Weak . | Strong . | |||||
| HGAL | 115 | 1:20 | Citrate | Cytoplasmic | < 5 | > 5-30 | ≥ 30 |
| BCL6 | 2 | 1:40 | TRIS | Nuclear | < 5 | > 5-30 | ≥ 30 |
| CD10 | 3 | 1:40 | EDTA | Membrane | < 5 | > 5-30 | ≥ 30 |
| MUM1/IRF4 | 2 | 1:50 | EDTA | Nuclear | < 5 | > 5-30 | ≥ 30 |
| BCL2 | 2 | 1:1000 | TRIS | Cytoplasmic | < 20 | > 20-50 | ≥ 50 |
| Blimp1 | 418 | 1:10 | EDTA | Nuclear | < 5 | > 5-< 30 | ≥ 30 |
| pSTAT6 | 5 | 1:50 | Citrate | Nuclear | < 20 | ≥ 20 low‡ | ≥ 20 high‡ |
| EBV EBER | 6 | NA | NA | Nuclear | < 5 | > 5-30 | ≥ 30 |
| LMP2A | 719 | 1:100 | Citrate | Membrane | 0 | 1-10 | ≥ 10 |
| Antibody/probe . | Source* . | Dilution . | Pretreatment† . | Predominant localization . | Scoring scale, % . | ||
|---|---|---|---|---|---|---|---|
| Negative . | Weak . | Strong . | |||||
| HGAL | 115 | 1:20 | Citrate | Cytoplasmic | < 5 | > 5-30 | ≥ 30 |
| BCL6 | 2 | 1:40 | TRIS | Nuclear | < 5 | > 5-30 | ≥ 30 |
| CD10 | 3 | 1:40 | EDTA | Membrane | < 5 | > 5-30 | ≥ 30 |
| MUM1/IRF4 | 2 | 1:50 | EDTA | Nuclear | < 5 | > 5-30 | ≥ 30 |
| BCL2 | 2 | 1:1000 | TRIS | Cytoplasmic | < 20 | > 20-50 | ≥ 50 |
| Blimp1 | 418 | 1:10 | EDTA | Nuclear | < 5 | > 5-< 30 | ≥ 30 |
| pSTAT6 | 5 | 1:50 | Citrate | Nuclear | < 20 | ≥ 20 low‡ | ≥ 20 high‡ |
| EBV EBER | 6 | NA | NA | Nuclear | < 5 | > 5-30 | ≥ 30 |
| LMP2A | 719 | 1:100 | Citrate | Membrane | 0 | 1-10 | ≥ 10 |
For scoring scale, negative, weak, and strong staining is indicated numerically as 0, 2, and 3, respectively.
NA indicates not applicable.
Sources were as follows: 1, generated in our laboratory15; 2, DAKO; 3, Novocastra, New Castle-Upon-Tyne, United Kingdom; 4, kindly provided by Prof Katheryn Calame, Columbia University, New York, NY18; 5, Cell Signaling Technology, Danvers, MA; 6, INFORM EBER probe, Ventana; 7, kindly provided by Prof Gerald Niedobitek, Friedrich Alexander University, Erlangen, Germany.19
Pretreatment for heat-induced antigen retrieval consisted of microwaving with 1 of the following buffers: citrate (10 mM, pH 6.0, for 10 minutes), Tris (5 mM, pH 10.0, for 20 minutes), or EDTA (1 mM, pH 8.0, for 15 minutes).
pSTAT6 staining was scored based on low- and high-intensity nuclear labeling in at least 20% of Hodgkin/Reed-Sternberg cells.
The comparative immunohistologic and in situ hybridization data are summarized in Table 2 We performed hierarchic cluster analysis to address whether there is correlation among the expression patterns of these proteins and with EBV RNA (Figure 2B). In contrast with our previous report of coclustering of HGAL with other GC markers in GC lymphocytes and DLBCL,15 HGAL clustered here on a separate branch of the dendrogram from CD10 and BCL6 proteins, whose expression is lost in many cases of cHL. Consequently, HGAL clustered on the same branch of the dendrogram as MUM1/IRF4, which is expressed in most cHLs. Both markers used to detect EBV, EBER and LMP2A, coclustered on the dendrogram shown in Figure 2B; however, there was no relationship between the expression of HGAL and EBV in these cases.
Summary of immunohistologic results
| Antibody . | Total no. of cases . | Score . | Total positive . | % positive . | ||
|---|---|---|---|---|---|---|
| 0 . | 2 . | 3 . | ||||
| HGAL | 145 | 36 | 38 | 71 | 109 | 75 |
| BCL6 | 136 | 100 | 23 | 13 | 36 | 26 |
| CD10 | 145 | 145 | 0 | 0 | 0 | 0 |
| MUM1/IRF4 | 138 | 11 | 8 | 119 | 127 | 92 |
| BCL2 | 140 | 96 | 30 | 14 | 44 | 31 |
| Blimp1 | 139 | 136 | 3 | 0 | 3 | 0.02 |
| pSTAT6 | 142 | 23 | 25 | 94 | 119 | 83 |
| EBV EBER | 140 | 112 | 1 | 27 | 28 | 20 |
| LMP2A | 140 | 115 | 6 | 19 | 25 | 18 |
| Antibody . | Total no. of cases . | Score . | Total positive . | % positive . | ||
|---|---|---|---|---|---|---|
| 0 . | 2 . | 3 . | ||||
| HGAL | 145 | 36 | 38 | 71 | 109 | 75 |
| BCL6 | 136 | 100 | 23 | 13 | 36 | 26 |
| CD10 | 145 | 145 | 0 | 0 | 0 | 0 |
| MUM1/IRF4 | 138 | 11 | 8 | 119 | 127 | 92 |
| BCL2 | 140 | 96 | 30 | 14 | 44 | 31 |
| Blimp1 | 139 | 136 | 3 | 0 | 3 | 0.02 |
| pSTAT6 | 142 | 23 | 25 | 94 | 119 | 83 |
| EBV EBER | 140 | 112 | 1 | 27 | 28 | 20 |
| LMP2A | 140 | 115 | 6 | 19 | 25 | 18 |
Scores 0, 2, and 3 are defined in the footnotes to Table 1.
Representative immunostains and EBV expression in classic Hodgkin lymphoma. (A) Typical examples of TMA cores of cHL stained for BCL6, CD10, BCL2, MUM1/IRF4, and Blimp1 and in situ hybridization for EBV EBER RNA are shown. (B) Hierarchic cluster analysis of immunohistologic and in situ hybridization data shows that HGAL clusters with MUM1/IRF4 on 1 branch of the dendrogram (orange) but not with BCL2, CD10, and BCL6 (blue) or with the 2 EBV-specific markers, LMP2A and EBER (purple), which cocluster with each other.
Representative immunostains and EBV expression in classic Hodgkin lymphoma. (A) Typical examples of TMA cores of cHL stained for BCL6, CD10, BCL2, MUM1/IRF4, and Blimp1 and in situ hybridization for EBV EBER RNA are shown. (B) Hierarchic cluster analysis of immunohistologic and in situ hybridization data shows that HGAL clusters with MUM1/IRF4 on 1 branch of the dendrogram (orange) but not with BCL2, CD10, and BCL6 (blue) or with the 2 EBV-specific markers, LMP2A and EBER (purple), which cocluster with each other.
Cytokine signaling and pSTAT6
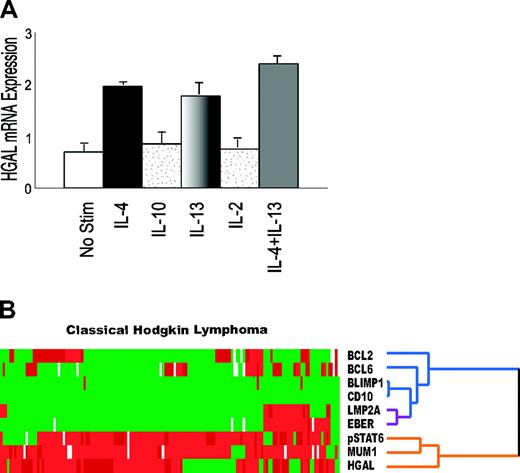
The cytokine milieu in cHL tissue is important for both the proliferation of Hodgkin cells and maintenance of the characteristic inflammatory environment. Because we previously demonstrated that HGAL expression is induced by IL-4,13 we next evaluated whether cytokines implicated in cHL pathogenesis may induce the expression of the HGAL gene. Enriched peripheral-blood lymphocytes were stimulated with IL-4, IL-10, IL-2, IL-13, or a combination of IL-4 and IL-13 for 6 hours. Only IL-4 and IL-13 were found to induce HGAL mRNA expression (Figure 3A) A combination of IL-4 and IL-13 resulted in a slightly additive induction of HGAL mRNA expression. However, incubation with higher concentrations of IL-2 and IL-10 did not lead to induction of HGAL mRNA expression (data not shown). Because pSTAT6 is a mediator of IL-4 and IL-13 cytokine signaling and is commonly implicated in induction of their target genes, we further investigated whether pSTAT6 expression correlated with cHL cases with HGAL protein expression by immunohistologic staining. Staining for pSTAT6 was present in 119 of 142 (83%) cases and was predominantly localized to the nucleus. Reclustering of pSTAT6 staining results with the previously analyzed markers revealed that pSTAT6 clustered on the same branch of the dendrogram as HGAL and MUM1/IRF4 (Figure 3B).
Cytokine signaling and pSTAT6 in classic Hodgkin lymphoma. (A) Enriched peripheral-blood lymphocytes stimulated with 100 U/mL IL-4, 50 ng/mL IL-10, 100 U/mL IL-2, 10 ng/mL IL-13, or a combination of IL-4 and IL-13 for 6 hours are shown. Only IL-4 and IL-13 induced HGAL mRNA expression, and the combination of IL-4 and IL-13 resulted in a slightly additive induction of HGAL mRNA expression. Error bars indicate SD among 3 experiments. (B) Hierarchic cluster analysis shows that pSTAT6 expression coclusters with HGAL and MUM1/IRF4 on the same branch of the dendrogram (orange), indicating a similar pattern of staining for these 3 proteins across the 145 cHL cases.
Cytokine signaling and pSTAT6 in classic Hodgkin lymphoma. (A) Enriched peripheral-blood lymphocytes stimulated with 100 U/mL IL-4, 50 ng/mL IL-10, 100 U/mL IL-2, 10 ng/mL IL-13, or a combination of IL-4 and IL-13 for 6 hours are shown. Only IL-4 and IL-13 induced HGAL mRNA expression, and the combination of IL-4 and IL-13 resulted in a slightly additive induction of HGAL mRNA expression. Error bars indicate SD among 3 experiments. (B) Hierarchic cluster analysis shows that pSTAT6 expression coclusters with HGAL and MUM1/IRF4 on the same branch of the dendrogram (orange), indicating a similar pattern of staining for these 3 proteins across the 145 cHL cases.
HGAL identifies a subset of cHL with improved survival
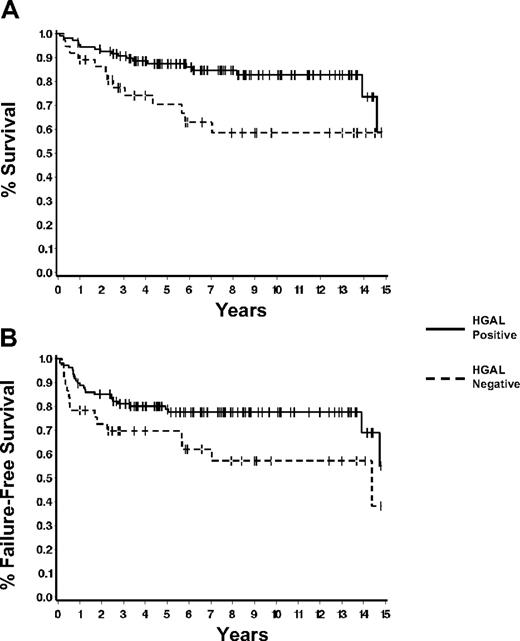
We examined the relationships between HGAL staining and patient and disease characteristics (Table 3) HGAL staining tended to correlate with younger patient age (less than 45 years, P = .05) but not with sex, stage, IPS, presence or absence of bulky disease, or histologic subtype. Among patients with early-stage disease (I or II), there was also no association with the presence or absence of B symptoms or the number of involved sites. Correlation between clinical prognostic factors, HGAL staining, and patients' survival are summarized in Table 4 The 5-year overall survival and failure-free survival rates of our cohort of 145 patients were 83% ± 3% and 76% ± 4%, respectively. The overall survival correlated with young age (less than 45 years, P < .001), low stage (stage I and II, P = .04), and low IPS (presence of 2 or fewer adverse prognostic factors, P = .002). Failure-free survival correlated with younger age (P < .001) and low IPS (P = .003). HGAL expression by the tumor cells correlated with both overall survival (P = .01) and failure-free survival (P = .05) (Figure 4A) . The estimated 5-year survival for HGAL-positive patients was 87% (95% confidence interval, 81% to 93%) compared with 70% for HGAL-negative patients (95% confidence interval, 54% to 86%).
Correlation of HGAL expression with clinical variables
| Variable . | HGAL expression† . | ||
|---|---|---|---|
| Negative, % . | Positive, % . | Fisher exact P . | |
| Sex | .85 | ||
| Male | 24 | 76 | |
| Female | 27 | 73 | |
| Age, y | .05 | ||
| Less than 45 | 21 | 79 | |
| 45 or more | 39 | 61 | |
| Stage | .70 | ||
| I, II | 24 | 76 | |
| III, IV | 28 | 72 | |
| IPS | .83 | ||
| 2 or below | 26 | 74 | |
| More than 2 | 29 | 71 | |
| Bulky disease | .83 | ||
| No | 27 | 73 | |
| Yes | 24 | 76 | |
| Histology | > .99 | ||
| Nodular sclerosis | 26 | 74 | |
| Other | 25 | 75 | |
| B symptoms* | > .99 | ||
| No | 24 | 76 | |
| Yes | 25 | 75 | |
| No. of involved sites* | .75 | ||
| 1 to 3 | 25 | 75 | |
| More than 3 | 18 | 82 | |
| Variable . | HGAL expression† . | ||
|---|---|---|---|
| Negative, % . | Positive, % . | Fisher exact P . | |
| Sex | .85 | ||
| Male | 24 | 76 | |
| Female | 27 | 73 | |
| Age, y | .05 | ||
| Less than 45 | 21 | 79 | |
| 45 or more | 39 | 61 | |
| Stage | .70 | ||
| I, II | 24 | 76 | |
| III, IV | 28 | 72 | |
| IPS | .83 | ||
| 2 or below | 26 | 74 | |
| More than 2 | 29 | 71 | |
| Bulky disease | .83 | ||
| No | 27 | 73 | |
| Yes | 24 | 76 | |
| Histology | > .99 | ||
| Nodular sclerosis | 26 | 74 | |
| Other | 25 | 75 | |
| B symptoms* | > .99 | ||
| No | 24 | 76 | |
| Yes | 25 | 75 | |
| No. of involved sites* | .75 | ||
| 1 to 3 | 25 | 75 | |
| More than 3 | 18 | 82 | |
Restricted to patients with stage I or II disease.
Negative, n = 37 (N = 37); positive, n = 108 (N = 108).
Patient and disease characteristics and univariate analysis
| Variable . | Overall survival . | Failure-free survival . | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of patients . | No. of deaths . | 5-y overall survival, % . | Wilcoxon P . | No. of patients . | No. of deaths . | 5-y failure-free survival, % . | Wilcoxon P . | |
| Age, y | < .001 | < .001 | ||||||
| Less than 45 | 109 | 13 | 90 ± 3 | 108 | 19 | 84 ± 4 | ||
| 45 or more | 36 | 18 | 62 ± 8 | 36 | 21 | 55 ± 8 | ||
| Sex | .24 | .40 | ||||||
| Male | 74 | 20 | 82 ± 5 | 73 | 23 | 73 ± 5 | ||
| Female | 71 | 11 | 85 ± 4 | 71 | 17 | 80 ± 5 | ||
| Stage | .04 | .11 | ||||||
| I, II | 83 | 13 | 88 ± 4 | 83 | 20 | 81 ± 4 | ||
| III, IV | 61 | 18 | 76 ± 6 | 60 | 20 | 69 ± 6 | ||
| IPS | .002 | .003 | ||||||
| 2 or below | 104 | 17 | 83 ± 3 | 103 | 24 | 82 ± 4 | ||
| More than 2 | 35 | 13 | 67 ± 8 | 35 | 15 | 60 ± 8 | ||
| Bulky disease | .65 | .79 | ||||||
| No | 101 | 24 | 81 ± 4 | 100 | 30 | 76 ± 4 | ||
| Yes | 37 | 7 | 86 ± 6 | 37 | 10 | 74 ± 8 | ||
| HGAL | .01 | .05 | ||||||
| Negative | 37 | 13 | 70 ± 8 | 37 | 15 | 70 ± 8 | ||
| Positive | 108 | 18 | 87 ± 3 | 107 | 25 | 79 ± 4 | ||
| B symptoms* | .48 | .14 | ||||||
| No | 73 | 12 | 87 ± 4 | 72 | 16 | 85 ± 4 | ||
| Yes | 46 | 10 | 84 ± 6 | 46 | 15 | 69 ± 7 | ||
| No. of sites* | .79 | .25 | ||||||
| 1-3 | 72 | 13 | 87 ± 4 | 72 | 17 | 83 ± 4 | ||
| More than 3 | 44 | 9 | 82 ± 6 | 43 | 13 | 72 ± 7 | ||
| Variable . | Overall survival . | Failure-free survival . | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of patients . | No. of deaths . | 5-y overall survival, % . | Wilcoxon P . | No. of patients . | No. of deaths . | 5-y failure-free survival, % . | Wilcoxon P . | |
| Age, y | < .001 | < .001 | ||||||
| Less than 45 | 109 | 13 | 90 ± 3 | 108 | 19 | 84 ± 4 | ||
| 45 or more | 36 | 18 | 62 ± 8 | 36 | 21 | 55 ± 8 | ||
| Sex | .24 | .40 | ||||||
| Male | 74 | 20 | 82 ± 5 | 73 | 23 | 73 ± 5 | ||
| Female | 71 | 11 | 85 ± 4 | 71 | 17 | 80 ± 5 | ||
| Stage | .04 | .11 | ||||||
| I, II | 83 | 13 | 88 ± 4 | 83 | 20 | 81 ± 4 | ||
| III, IV | 61 | 18 | 76 ± 6 | 60 | 20 | 69 ± 6 | ||
| IPS | .002 | .003 | ||||||
| 2 or below | 104 | 17 | 83 ± 3 | 103 | 24 | 82 ± 4 | ||
| More than 2 | 35 | 13 | 67 ± 8 | 35 | 15 | 60 ± 8 | ||
| Bulky disease | .65 | .79 | ||||||
| No | 101 | 24 | 81 ± 4 | 100 | 30 | 76 ± 4 | ||
| Yes | 37 | 7 | 86 ± 6 | 37 | 10 | 74 ± 8 | ||
| HGAL | .01 | .05 | ||||||
| Negative | 37 | 13 | 70 ± 8 | 37 | 15 | 70 ± 8 | ||
| Positive | 108 | 18 | 87 ± 3 | 107 | 25 | 79 ± 4 | ||
| B symptoms* | .48 | .14 | ||||||
| No | 73 | 12 | 87 ± 4 | 72 | 16 | 85 ± 4 | ||
| Yes | 46 | 10 | 84 ± 6 | 46 | 15 | 69 ± 7 | ||
| No. of sites* | .79 | .25 | ||||||
| 1-3 | 72 | 13 | 87 ± 4 | 72 | 17 | 83 ± 4 | ||
| More than 3 | 44 | 9 | 82 ± 6 | 43 | 13 | 72 ± 7 | ||
Restricted to patients with stage I or II disease.
HGAL protein expression correlates with overall survival. (A) HGAL protein expression correlates with overall survival (P = .01). (B) HGAL protein expression correlates with improved failure-free survival (P = .05).
HGAL protein expression correlates with overall survival. (A) HGAL protein expression correlates with overall survival (P = .01). (B) HGAL protein expression correlates with improved failure-free survival (P = .05).
Multivariate analyses were performed including HGAL and the clinical parameters listed in Table 4. The primary analysis included all 145 patients as a single cohort and assessed the relative prognostic value of HGAL staining, IPS, and its components in the prediction of cHL outcome. However, because non-IPS parameters are commonly used for subclassification into favorable- and unfavorable-risk groups in patients with early-stage disease (I or II), we performed separate multivariate analyses in early- and advanced-stage (III or IV) HL patients with the National Comprehensive Cancer Network guidelines and the IPS prognostic factors. All analyses were stratified by medical center to adjust for any possible inherent differences between the Cleveland Clinic Foundation and the Nebraska Lymphoma Study Group. Because age and stage are integral parts of the IPS, separate multivariate analyses based on HGAL staining, age, and stage or HGAL staining and IPS risk groups were performed. Based on the analysis of all 145 patients, age or IPS were independent predictors of both overall and failure-free survival (P < .01 in all cases). There was some indication that stage may also be an independent predictor of overall survival (P = .07). HGAL expression did not have an independent impact on overall or failure-free survival in this analysis (P > .05 in all cases). It also was not an independent predictor of either outcome when early-stage disease and advanced-stage disease patients were assessed separately, even after adjusting for factors such as the presence or absence of bulky disease, B symptoms, and the number of involved sites (early-stage disease) or age or IPS (advanced-stage disease) (P ≥ .08 in all cases). Although all patients in our study were treated with curative intent, 25 patients (17%) received radiation treatment only. We therefore repeated the primary statistical analysis excluding these 25 patients and found that HGAL expression showed a similar trend in correlation with overall survival in univariate analysis (P = .05) but was not associated with failure-free survival (P = .19); it again did not have an impact on overall or failure-free survival in multivariate analysis.
Discussion
The derivation and pathogenesis of classic Hodgkin lymphoma remains controversial. Unlike other lymphomas derived from GC B cells, Hodgkin cells typically lack expression of the leukocyte common antigen, CD45RB, and B-lineage markers CD20, CD19, and CD79 are infrequently expressed.20,21,22 That cHL cells are unlike any normal hematopoietic cells in their immunophenotype is underscored by the frequent expression of markers unrelated or not specific to the B lineage. Two such examples are CD30 and CD15, both of which are not B-lineage specific but whose expression is most often used in confirming a histopathologic diagnosis of cHL.23 In addition, the expression of MUMI/IRF4 and CD138 have led to speculations by some that cHL may be derived from post–germinal center B cells.24,25,26 Deleterious mutations of Ig genes are found in only 25% of cHLs, and most cHL cases lack Ig transcripts due to a combination of defects in Ig-specific transcriptional machinery and epigenetic phenomena including promoter methylation of B-lineage–specific genes.6,27 Data from oligonucleotide microarrays demonstrate loss of B-lineage–specific gene expression as well as abrogation of major signaling pathways and transcription factors in cHL cell lines.7
We previously cloned and characterized the HGAL gene and showed that the HGAL mRNA and its cognate protein are specifically expressed in normal GC and in GC-derived B-cell lymphoma.13,15 Lymphomas of GC derivation include follicular, Burkitt, and lymphocyte-predominant Hodgkin lymphoma among others, which are disparate in their clinicopathologic manifestations. Despite this heterogeneity, B-cell non-Hodgkin lymphomas typically retain B-lineage markers and features of differentiation that are related to normal B-cell ontogeny. Although the World Health Organization classification of Hodgkin lymphoma includes lymphocyte-predominant Hodgkin lymphoma and classic Hodgkin lymphoma,23 it is recognized that these 2 entities are significantly different from one another in their gene expression, transcriptional regulation, intracellular signaling, and immunophenotype.7,28,29,30,31,32 Although important transcription factors in normal B-cell development such as PU.1, Oct2, and BOB.1 are down-regulated in cHL,32 PAX5, a transcription factor imperative for B-lineage commitment and maintenance, is not.33 Similarly, the transcription factor ATF3 is constitutively overexpressed in cHL and is considered a key factor for proliferation and survival of Hodgkin cells.34 Our results show that HGAL protein is expressed in a significant number of cHLs (75%) in a cohort of cases separate from our original report.15 This finding, similar to the expression of PAX5, suggests that there are exceptions to the general down-regulation of B-lineage–specific genes and supports the hypothesis that at least some cHLs may be derived from GC B cells.
In our prior characterization of HGAL in DLBCL we found that HGAL protein expression correlated closely with other GC-specific markers, BCL6 and CD10, but not with BCL2 or MUM1/IRF4.15 These antibodies were selected because their corresponding genes had previously been found to contribute to the distinction of prognostic subclasses of DLBCL and represented the GC subtype (BCL6, CD10, and HGAL) and the non-GC subtype (MUM1/IRF4 and BCL2).14,35 In addition, the expression of Blimp1, a transcriptional repressor required for terminal B-cell differentiation, was studied because the expression of Blimp1 is repressed by BCL6, a factor required for GC formation.18,24,36 Our results show that GC markers are not uniformly expressed in cHL, and the expression pattern in our cHL cases is similar to those previously reported in the literature.18,24,37,38,39,40 Although BCL6 was expressed in 26% of cases, CD10 staining was completely lacking. In addition, as described previously, MUM1/IRF4 is highly expressed in most cHL cases24,37,39 while BCL2 reactivity was seen in a small subset (31%) of cases.12 The close correlation of MUM1/IRF4, a marker expressed in late GC B cells and in post-GC plasma cells, with HGAL expression suggests that there is a significant overlap in the expression of these 2 markers in the continuum of GC B-cell differentiation. In contrast, Blimp1, another marker of late GC B cells and post-GC plasma cells, was found only in rare cHL cases. Very few of our cHL cases were scored positive for Blimp1, although rare Hodgkin cells stained positive in additional cases that did not meet our scoring criteria for positivity. These findings confirm the low levels of expression of Blimp1 in cHL and, as recently reported, may represent abortive initiation of terminal B-cell differentiation in cHL.24
The enigma of Hodgkin cells that escape from apoptosis despite lacking the B-cell receptor has generated much interest. Recently, 3 groups of investigators reported that EBV plays an important role in rescuing B-cell receptor–negative GC B cells from apoptosis and that these B cells are capable of entering the cell cycle upon infection by EBV to yield clonal lymphoblastoid cell lines that remain B-cell receptor deficient.3,4,5 The rescue of preapoptotic GC B cells by the EBV-encoded LMP2A protein implies that it may have a role in a transforming event that overrides negative selection in the GC and offers an intriguing model of lymphomagenesis applicable to a subset of cHL.3,4,5 We tested whether the cHL cases expressing HGAL harbored EBV. Although 20% of our cases showed EBV-specific EBER RNA in neoplastic cells, and most EBER-positive cases showed LMP2A expression by immunohistologic analysis, there was no correlation with expression of HGAL protein, suggesting that these cases most likely do not overlap with those containing deleterious mutations of Ig genes.
HGAL protein expression is highly specific to GC-derived lymphomas, and our results indicate that it could serve as a marker of GC derivation in cHL. However, in the cytokine-rich environment of cHL, HGAL expression may also be triggered by cytokine signaling, which prompted us to investigate this possibility. Our results showed that HGAL expression is triggered by IL-4 and IL-13 signaling, 2 pathways known to be active in cHL.41,42 Although the intracellular signaling mechanism that induces HGAL expression by IL-4 and IL-13 is unknown, the expression of most IL-4– and IL-13–inducible genes is mediated by STAT6. This led us to examine the status of phosphorylated STAT6 expression in our cHL cases. We found that pSTAT6 expression, as assessed by immunohistochemistry, correlated with HGAL and MUM1/IRF4 expression in hierarchic cluster analysis. However, this coclustering is most likely due to expression of HGAL, MUM1/IRF4, and pSTAT6 proteins in most cHL cases rather than to a specific correlation of pSTAT6 with HGAL expression. Indeed, a subset of HGAL-positive cases did not demonstrate expression of pSTAT6, and no correlation between pSTAT6 and HGAL expression in cHL samples was observed by statistical analysis (data not shown). Thus, although cytokine-induced signaling may contribute to HGAL expression, it is more likely that it represents retention of expression of a GC-specific marker in classic Hodgkin lymphoma.
The expression of HGAL protein in cHL led us to further investigate whether its expression could identify distinct subgroups of cHL that may be associated with different prognoses. In univariate analysis our results show that HGAL expression correlates with improved outcome and is more common in younger patients (less than 45 years). The lack of impact of HGAL expression in multivariate analysis in our study most likely represents the fact that HGAL expression may not have additional prognostic value over already known clinical variables of superior outcome such as young age, low stage, and low IPS. It also suggests that there may be underlying differences in the biology of cHL tumor patients with favorable clinical characteristics.
In conclusion, our results confirm that HGAL protein is expressed in most cHLs. Comparative immunohistologic analyses show that HGAL protein expression is not correlated with the expression of CD10 and BCL6. Thus, it appears that HGAL expression is retained in the absence of other GC-specific proteins and may function as a marker of GC derivation in classic Hodgkin lymphoma. Our results further show that in addition to IL-4 cytokine, HGAL expression is also induced by IL-13, both of which are implicated in cHL pathogenesis. The lack of correlation between HGAL and pSTAT6 staining in our cohort of patients suggests that although deregulated cytokine signaling may contribute to HGAL expression, it may also function as a marker of GC derivation in a subset of cHL tumors. Lastly, we show that HGAL protein expression distinguishes a biologically distinct subgroup of cHL associated with young age and improved overall survival.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: B.P. and M.B. compiled the clinical data; Y.N., E.D.H., P.A., H.N., R.L., and I.S.L. contributed to study design, data analysis, and writing the manuscript; S.Z. performed immunohistochemistry and TMA construction; and P.E. performed the statistical analyses.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants CA109335 and CA34233 and The Dwoskin Family Foundation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal