Abstract
Gfi1 is a transcriptional repressor essential during myeloid differentiation. Gfi1−/− mice exhibit a block in myeloid differentiation resulting in the accumulation of an immature myelo-monocytic cell population and the complete absence of mature neutrophils. Even though mRNA levels of Gfi1 appear to be very low in monocytes, Gfi1 might play a role in the monocytic lineage as Gfi1−/− mice exhibit diminished monocyte-derived dendritic cells and disturbed cytokine production by macrophages in response to LPS. We show here that Gfi1 protein levels are mainly regulated by the ubiquitin-proteasome system. Upon forced monocytic differentiation of U937 cells, Gfi1 mRNA levels dropped but protein levels increased due to diminished proteasomal turnover. Similarly, Gfi1 mRNA levels are low in primary monocytes whereas the protein is clearly detectable. Conversely, Gfi1 mRNA levels are high in granulocytes but the protein is swiftly degraded by the proteasome in these cells. Chromatin immunoprecipitation experiments showed that Gfi1 binds to the promoter of several granulocyte-specific genes in primary monocytes, including C/EBPα, neutrophil elastase, and Gfi1 itself. The binding of the repressor Gfi1 to these promoters correlated with low expression of these genes in monocytes compared with granulocytes. Our data fit a model in which Gfi1 protein levels are induced in primary monocytes, due to diminished proteasomal degradation, to repress genes that play a role in granulocytic differentiation.
Introduction
The generation of mature myeloid cells from hematopoietic stem cells (HSCs) is a tightly regulated process. HSCs may differentiate in common myeloid progenitors (CMPs) that in turn differentiate toward common granulocyte/monocyte progenitor cells (GMPs). These latter cells may differentiate into mature, functional myeloid blood cells. Different regulatory mechanisms are responsible for maintaining the correct levels of mature blood cells. Lineage-determining factors stimulate specific transcription factors that direct progenitor cells toward a specific cell type, while simultaneously they may actively suppress alternative lineage programs. In myelopoiesis, several transcriptional regulators have been found to play an essential role including PU.1,1,2 C/EBPα,3 and C/EBPϵ.4
Recently, the transcription factor growth factor independence 1 (Gfi1) has also been shown to play an essential role during myelopoiesis. Gfi1−/− knockout mice are severely neutropenic and exhibit a block in myeloid differentiation resulting in the accumulation of an atypical immature myeloid cell population.5,6 This is in line with reported heterozygous GFI1 missense mutations in patients suffering from hereditary neutropenia.7 Gfi1 may function as a transcriptional repressor and regulate genes that play a role in cell-cycle regulation (eg, E2F5 and cMyc) and granulopoiesis (eg, C/EBPα, C/EBPϵ, and neutrophil elastase).8 Furthermore, Gfi1 can repress its own promoter and the promoter of its paralogue Gfi1B.9,10
Gfi1 was originally identified as a proviral insertion site resulting in interleukin 2 (IL2)–independent growth of T-cells.11 Overexpressed Gfi1 acts as a dominant oncogene and cooperates with known oncoproteins such as Myc and Pim-1 in lymphoma development.12,13 In normal hematopoiesis, Gfi1 is essential for T-cell development and for self-renewal and long-term reconstituting potential of HSCs.5,14,15 Recently, Gfi1 was also shown to be important for lymphocyte-derived dendritic cell (DC) development.16
Whether Gfi1 plays a role in the monocytic lineage is under debate. Although the immature atypical myeloid population in Gfi1−/− mice exhibits both granulocytic and monocytic characteristics, it has been speculated that Gfi1 is not relevant in monocytic differentiation. This is mainly based on low Gfi1 mRNA expression patterns in the monocytic lineage as measured by reverse transcriptase–polymerase chain reaction (RT-PCR) or indirectly by measuring Gfi1 promoter activity using GFP:Gfi1 knock-in mice. During granulocytic and monocytic differentiation, Gfi1 mRNA expression is strongly induced at the GMP stage and expression is detected in mature granulocytes, whereas low expression is found in monocytic/macrophage cells.6,15,17 However, recent studies suggest a role for Gfi1 in monocyte-derived DC differentiation. Gfi1−/− progenitor cells fail to differentiate into DCs in in vitro assays.16 In addition, Gfi1−/− macrophages exhibit aberrant cytokine expression profiles and are hyperresponsive to LPS.6,18 These data suggest that Gfi1 may be important in the monocytic lineage. To clarify this issue, we analyzed Gfi1 protein expression and found that Gfi1 is very efficiently targeted for ubiquitin-proteasomal degradation in immature myeloid cells and that diminished proteasomal degradation in primary monocytes results in significant Gfi1 protein levels, despite low RNA levels.
Materials and methods
Cell culture and selection
U937, NB4, and HL60 cells were grown in RPMI 1640 medium (Life Technologies, Bethesda, MD) supplemented with 10% fetal calf serum (Invitrogen, Carlsbad, CA), 50 IU/mL penicillin, and 50 μg/mL streptomycin (ICN). Cells were differentiated for indicated time points with 10−6 M ATRA (all-trans retinoic acid; Sigma, St Louis, MO) or 10 ng/mL PMA (phorbol 12-myristate 13-acetate; Sigma). The 26S proteasome activity was inhibited overnight with 5 μM MG132. For Gfi1 half-life determination, cycloheximide was used at a concentration of 25 μg/mL. COS-1 and HEK293 cells were grown in Iscove modified Dulbecco medium (IMDM; Life Technologies) supplemented as described for U937 cells. All cells were grown at 37°C at 5% CO2. Differentiation of U937 cells was assessed by the expression of CD11c (Immunotech, Marseille, France). Apoptosis was determined by flow cytometer using Annexin V (Molecular Probes, Eugene, OR) and propidium iodide staining, respectively. Primary cells were obtained from healthy volunteers. Human monocytes, used for RNA isolation, were stained with CD14 antibodies and isolated by fluorescence activated cell sorting (FACS) and granulocytes were FACS sorted based on CD45 positivity and scatter characteristics. For chromatin immunoprecipitation (ChIP), degradation assays and Western blot analysis monocytes were sorted by magnetic activated cell sorting (MACS) using CD14 magnetic beads and granulocytes were isolated using Ficoll 1077 density gradient centrifugation followed by erythrocyte lysis using NH4Cl. Cell purities were confirmed to be more than 95% by flow cytometric analysis.
Plasmids
Gfi1 expression plasmids containing FLAG-Gfi1, FLAG-Gfi1ΔZn, FLAG-Gfi1-Zn (provided by Dr M. Osawa19 ), Gfi-GFP and Gfi1-FLAG (provided by Dr T. Möröy), and Gfi1 wild type and Gfi1 A382S and K403R point mutants (provided by Dr M. Horwitz7 ) were used as indicated. Expression plasmids for His-tagged ubiquitin (His-Ub) and FLAG-tagged ubiquitin (FLAG-Ub) were obtained from Dr M. Scheffner.
Quantitative PCR
RNA from cell lines was isolated at different time points during differentiation using RNA-bee (ISO-TEX Diagnostics, Friendswood, TX). At all time points cell death was less than 10% as assessed by Annexin V and propidium positivity measured by flow cytometer. RNA from primary cells was isolated using the mini RNA isolation II kit (Zymo Research, Orange, CA). cDNA was generated in a reverse transcription reaction as previously described.20 Gfi1 mRNA levels were measured with real-time quantitative PCR using the following primers: Gfi1-F, 5′-GAGCCTGGAGCAGCACAAAG-3′; Gfi1-R, 5′-GTGGATGACCTCTTGAAGCTCTTC-3′. Ela2 mRNA levels were measured with real-time quantitative PCR using the following primers: Ela2-F, 5′-CACTGCGTGGCGAATGTAAA-3′; Ela2-R, 5′-CGACGTGCCGCTTGTGG-3′. C/EBPα mRNA expression was measured using predeveloped TaqMan Gene Expression Assay no. Hs00269 972_s1 (Applied Biosystems, Foster City, CA). PCRs were performed in universal master mix using SYBR-green (Roche, Milan, Italy). Conditions were as follows: 10 minutes at 95°C followed by 45 cycles of 15 seconds at 95°C and 1 minute at 62°C. For normalization, β-actin or 18S rRNA was used as a reference gene (Applied Biosystems).21
Coimmunoprecipitation and immunoblotting assays
For coimmunoprecipitations, COS-1 AND HEK293 cells were grown in 10-cm culture dishes and transiently transfected with FLAG-Ub and GFP-Gfi1 using the calcium phosphate precipitation method. Cells were lysed 36 hours after transfection in RIPA buffer with Complete protease inhibitors (Roche) and lysates were incubated with green fluorescence protein (GFP)–antibody and Prot-A beads for 4 hours at 4°C with gentle rotation. Beads were washed 5 times with RIPA. Bound proteins were eluted in loading buffer by heating for 5 minutes at 95°C. The immunoprecipitates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to PVDF membranes, blocked with 2% ELK (Campina, Eindhoven, The Netherlands) and 0.5% bovine serum albumin (BSA; Sigma) and immunoblotted with an α-FLAG antibody (M2; Sigma). Proteins were visualized using enhanced chemiluminescence (ECL; Bio-Rad, Hercules, CA). Protein levels on Western blot were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). For detection of endogenous Gfi1, C/EBPα, C/EBPϵ, actin, lamin, and ubiquitin Western blots were stained with α-Gfi1 (N-20; Santa Cruz Biotechnology, Santa Cruz, CA), C/EBPα (Santa Cruz Biotechnology), C/EBPϵ (Santa Cruz Biotechnology), α-actin (Sigma), α-lamin (Becton Dickinson, San Jose, CA), and α-ubiquitin (6C1; Sigma) antibodies, respectively. For preparation of lysates, granulocytes were treated with 10% trichloroacetic acid (Sigma) for 30 minutes at 4°C, followed by lysis in 50 μL 2× loading buffer (including 5 μL 1 M Tris) and heated at 95°C for 5 minutes.22
In vitro degradation assay
Hematopoietic cell lines, granulocytes, and MACS-sorted CD14+ cells were lysed in RIPA buffer supplemented with Complete protease inhibitor mix (Roche). Cell lysates taken at different time points during differentiation were normalized for equal protein content using the Bradford protein quantification assay. 35 S methionine–labeled, in vitro–translated Gfi1 (2 μL), made using the rabbit reticulocyte lysate TnT Quick Coupled Transcription/Translation System (Promega, Madison, WI), was added to cell lysate containing 100 μg protein (Bio-Rad protein quantification assay) in a total volume of 35 μL. MG132 (100 μM), Velcade (100 μM; PS341/bortezomib), or DMSO (1 μL/mL vehicle) was added prior to incubation at 37°C. Reactions were stopped at the indicated time points by the addition of loading buffer and heating the samples for 5 minutes at 95°C. Samples were resolved by SDS-PAGE, gels were fixed, treated with NAMP-100 (Amersham, Arlington Heights, IL), dried, and exposed.
Proteasome activity assay
Proteasome activity was determined in U937 cell lysates after PMA-induced differentiation. Protein (67.5 μg; concentration determined using Bio-Rad protein quantification assay) was incubated with the proteasome substrate Suc-Leu-Leu-Val-Tyr-AMC (Boston Biochem, Cambridge, MA) for 2 hours at 37°C as described.21,23 AMC liberated from the substrate by proteasome activity was determined by measuring fluorescence (Fluorstar, BMG; Isogen Life Sciences, ÿsselstein, The Netherlands).
In vivo ubiquitination assay.
COS-1 cells grown in 10-cm dishes were transfected with 10 μg His-Ub and 10 μg Gfi1-FLAG expression constructs, using calcium phosphate precipitation. At 36 hours after transfection, cells from each dish were collected in 2 aliquots. One aliquot (10%) was used in Western blotting to assess the expression of transfected proteins, while the remaining cells (90%) were used for purification of His6-tagged proteins by His-select (NTA) beads (Sigma). The cell pellet was lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4, 10 mM imidazole, 10 mM β-mercaptoethanol, Complete protease inhibitor [Roche]) and incubated with His-Select beads (Sigma) for 4 hours at 4°C. The beads were washed 2 times with buffer A, 2 times with a mixture of 4 parts buffer A and 1 part buffer B (50 mM Tris pH 6.8, 20 mM imidazole), and 2 times with buffer B. Bound proteins were eluted with 200 mM imidazole in loading buffer. Ub-conjugated Gfi1 was visualized on Western blot using an α-FLAG antibody.
Electrophoretic mobility-shift assay (EMSA).
FLAG-Gfi1 was synthesized in vitro by using the TnT SP6 transcription/translation system (Promega). Double-stranded oligonucleotides (5′-ACCATCACCACATAAATCACTGCCTATCCTGTG-3′) were 32P end-labeled using T4 polynucleotide kinase. Binding reactions (10 μL vol) contained labeled DNA probes in 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM MgCl2, 1 mM ZnSO4, 0.5 mM DTT, 0.5 mM EDTA, 10% glycerol, 0.05% Nonidet P-40, and 1 μg poly(dI-dC), with 1 μL of the synthesized protein and were performed for 20 minutes at room temperature.
Chromatin immunoprecipitation.
Chromatin immunoprecipitations (ChIPs) were executed as described previously.24 Cells were cross-linked for 30 minutes at 37°C by adding formaldehyde (1%) to the culture medium. Cross-linking was stopped by the addition of glycine to a final concentration of 125 mM. Cells were washed with cold phosphate-buffered saline, buffer B (10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100, 20 mM HEPES [pH 7.6]), buffer C (1 mM EDTA, 0.5 mM EGTA, 0.15 M NaCl, 50 mM HEPES [pH 7.6]) and incubated in buffer D (0.15% SDS, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6] and Complete protease inhibitor [Roche]) at 33 × 106 cells/mL. Chromatin was sonicated using the Bioruptor (Cosmo Bio, Tokyo, Japan), high setting, for 15 minutes with a 0.5-minute interval. Insoluble material was removed by centrifugation at 11 000 g at 4°C for 15 minutes. Chromatin (100 μL) was incubated with 15 μL protein G sepharose beads (Amersham), 0.1% BSA, 36 μL 5× buffer D, complete protease inhibitors (Roche), and 10 μL antibody and rocked at 4°C for 16 hours. The beads were harvested by centrifugation and washed twice with wash-buffer 1 (0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, and 20 mM HEPES [pH 7.6]), once with wash-buffer 2 (0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.5 M NaCl, 1 mM EDTA, 0.5 mM EGTA, and 20 mM HEPES [pH 7.6]), once with wash-buffer 3 (0.25 M LiCl, 0.5% NaDOC, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, and 20 mM HEPES [pH 7.6]), and twice with wash-buffer 4 (1 mM EDTA, 0.5 mM EGTA, and 20 mM HEPES [pH 7.6]). Chromatin antibody complexes were eluted from the protein G sepharose beads by addition of 1% SDS and 0.1 M NaHCO3 to the pellet and incubated for 20 minutes at room temperature. Cross-linking was reversed by addition of NaCl (0.44 M final concentration) and incubation of the eluted samples for 4 hours at 65°C. DNA was recovered by a phenol-chloroform-isoamylalcohol extraction followed by a chloroform-isoamylalcohol extraction and precipitation with sodium acetate (pH 5.2) and ethanol. Precipitated DNA was subject to quantitative PCR. Total DNA from the IP input was included in the PCR and all PCR signals were corrected for this. Quantitative PCR following ChIP was done using SYBR green PCR (Applied Biosystems). Genomic primer sequences for the Gfi1 target genes were as follows: Gfi1-F2, 5′-TTCTCTCGCTGCGGAGTCT-3′; Gfi1-R2, 5′-AGGCACTAGAAATGACTTGAAAGAAAA-3′; C/EBPα-F, 5′-AGATCAGAGCTAGGAGACGCAGA-3′; C/EBPα-R, 5′-ATTCTCTTTCAAAGCCAGAACCA-3′; ELA2-F2, 5′-CAAGTCCCTCAGGTCTAGGTTTG-3′; ELA2-R2, 5′-GGGCTGGTCTCGACGTTTT-3′. Quantitave PCR for the albumin promoter was performed using probe ALB-VIC 5′-TGCTGAAACATTCACCTTCCATGCAGA-3′ with the primers ALB-F 5′-TGAAACATACGTTCCCAAAGAGTTT-3′ and ALB-R 5′-CTCTCCTTCTCAGAAAGTGTGCATAT-3′.
Results
Gfi1 protein levels increase during monocytic differentiation by posttranscriptional mechanisms
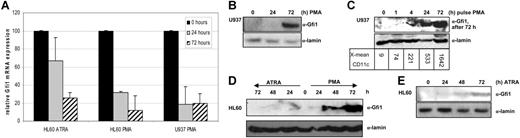
Although Gfi1 may play a role in the monocytic lineage, the mRNA expression is very low in macrophages. To study this issue in more detail, we analyzed Gfi1 mRNA and protein expression during forced differentiation of U937 and HL60 cells. During PMA-induced monocytic differentiation of these cells, Gfi1 mRNA levels dropped as measured with quantitative RT-PCR (Figure 1A) Also, during ATRA-induced granulocytic differentiation of HL60 cells, Gfi1 mRNA levels decreased, though to a lesser extent, as was observed during PMA-induced differentiation. Remarkably, upon terminal monocytic differentiation of PMA-treated U937 cells, Gfi1 protein levels increased (Figure 1B). To determine the length of time required for PMA to induce Gfi1 and monocytic differentiation, we exposed U937 cells for various time intervals to PMA. This showed that a pulse with PMA for only 4 hours already resulted in Gfi1 protein induction after 72 hours of culturing. The Gfi1 protein induction correlated with the extent of differentiation as determined by CD11c staining (Figure 1C).
Gfi1 mRNA levels decrease, Gfi1 protein levels increase during monocytic differentiation. (A) Gfi1 quantitative RT-PCR was performed on RNA samples of U937 and HL60 cells during PMA- or ATRA-induced differentiation (n = 3). Gfi1 expression levels in untreated HL60 and U937 cells were set at 100% and values were normalized for β-actin expression. Error bars indicate standard deviation. (B) Cell lysates of PMA-differentiated U937 cells taken at indicated time points were immunoblotted and stained with an α-Gfi1 antibody (N20; Santa Cruz Biotechnology). Lamin staining shows equal loading. (C) U937 cells were incubated with PMA. After the indicated pulse times cells were washed and harvested 72 hours after the beginning of the experiment. Numbers below the blot indicate the x-mean of CD11c expression. (D) Gfi1 levels in HL60 cell lysates taken after 24-, 48-, and 72-hour treatments with ATRA or PMA were compared with untreated cells showing that Gfi1 is strongly induced upon monocytic differentiation. Lamin staining shows equal loading. (E) A 5-times-more protein input of lysates from ATRA-treated HL60 cells resulted in the detection of a modest increase in Gfi1 protein levels, although the levels are significantly lower compared with monocytic differentiation (see panel D).
Gfi1 mRNA levels decrease, Gfi1 protein levels increase during monocytic differentiation. (A) Gfi1 quantitative RT-PCR was performed on RNA samples of U937 and HL60 cells during PMA- or ATRA-induced differentiation (n = 3). Gfi1 expression levels in untreated HL60 and U937 cells were set at 100% and values were normalized for β-actin expression. Error bars indicate standard deviation. (B) Cell lysates of PMA-differentiated U937 cells taken at indicated time points were immunoblotted and stained with an α-Gfi1 antibody (N20; Santa Cruz Biotechnology). Lamin staining shows equal loading. (C) U937 cells were incubated with PMA. After the indicated pulse times cells were washed and harvested 72 hours after the beginning of the experiment. Numbers below the blot indicate the x-mean of CD11c expression. (D) Gfi1 levels in HL60 cell lysates taken after 24-, 48-, and 72-hour treatments with ATRA or PMA were compared with untreated cells showing that Gfi1 is strongly induced upon monocytic differentiation. Lamin staining shows equal loading. (E) A 5-times-more protein input of lysates from ATRA-treated HL60 cells resulted in the detection of a modest increase in Gfi1 protein levels, although the levels are significantly lower compared with monocytic differentiation (see panel D).
To compare Gfi1 proteins levels during monocytic and granulocytic differentiation we analyzed Gfi1 expression in HL60 cells that can be forced along the monocytic and granulocytic lineages with, respectively, PMA and ATRA. As in U937 cells, upon monocytic differentiation of HL60 cells a clear Gfi1 protein induction was observed (Figure 1D). During ATRA-forced differentiation, Gfi1 was not detectable. However, when 5 times more protein was used in Western blotting a modest increase in Gfi1 protein levels became detectable upon granulocytic differentiation of HL60 cells (Figure 1E). This indicates that the Gfi1 protein induction is most prominent during monocytic differentiation. The observed differences between the expression of Gfi1 mRNA and protein levels suggests that Gfi1 protein expression is regulated at the posttranscriptional level during myeloid cell differentiation.
Gfi1 is targeted for degradation by the ubiquitin-proteasome pathway
The ubiquitin-proteasome system is an important system through which the half-life of various proteins is regulated, including transcription factors.25 To study whether Gfi1 is targeted for degradation by the ubiquitin-proteasome system we transfected HEK293 cells with a Gfi1-GFP expression construct. The mean fluorescence of these cells is a measure for the amount of GFP-tagged Gfi1. FACS analysis of cells treated with the proteasome inhibitor MG132 resulted in a 4-fold increase in the mean fluorescence, showing an accumulation of Gfi1-GFP protein levels compared with untreated cells (Figure 2A). As proteasome inhibition of cells transfected with GFP alone did not result in an increase in fluorescence (data not shown), these data suggest that Gfi1 protein levels are regulated by 26S proteasome activity.
Gfi1 is degraded by the ubiquitin-proteasome system. (A) HEK293 cells were split after transfection with Gfi1-GFP and were grown overnight with or without MG132. Cells were measured by flow cytometer and the mean fluorescence index (MFI) of untreated cells was set at 100%. After proteasome inhibition (5 μM MG132), a clear increase of Gfi1-GFP levels per cell was observed, indicated as MFI. (B) COS-1 cells were transfected with FLAG-Ub and Gfi1-GFP or with empty vector, followed by immunoprecipitation with an α-GFP antibody. Subsequent immunoblotting with an α-FLAG antibody resulted in the detection of a high molecular smear of proteins [(Ub)n], indicating that Gfi1 is present in ubiquitinated complexes, or is ubiquitinated itself. → Indicates the height of unmodified Gfi1-GFP. Bottom panel with α-GFP staining of whole-cell extract (WCE) shows a clear distinct Gfi1-GFP band. (C) COS-1 cells were transfected with His-Ub and FLAG-Gfi1, and MG123 was added as indicated. Ubiquitinated proteins were selected with His-select beads under denaturing conditions, and immunoblotted for α-FLAG. Clearly visible are the ubiquitinated forms of Gfi1 [(Gfi1-Ub)n], especially after proteasome inhibition. Whole-cell lysate was stained for FLAG, confirming equal loading. * Indicates α-specific binding of unmodified Gfi1 to His-select beads.
Gfi1 is degraded by the ubiquitin-proteasome system. (A) HEK293 cells were split after transfection with Gfi1-GFP and were grown overnight with or without MG132. Cells were measured by flow cytometer and the mean fluorescence index (MFI) of untreated cells was set at 100%. After proteasome inhibition (5 μM MG132), a clear increase of Gfi1-GFP levels per cell was observed, indicated as MFI. (B) COS-1 cells were transfected with FLAG-Ub and Gfi1-GFP or with empty vector, followed by immunoprecipitation with an α-GFP antibody. Subsequent immunoblotting with an α-FLAG antibody resulted in the detection of a high molecular smear of proteins [(Ub)n], indicating that Gfi1 is present in ubiquitinated complexes, or is ubiquitinated itself. → Indicates the height of unmodified Gfi1-GFP. Bottom panel with α-GFP staining of whole-cell extract (WCE) shows a clear distinct Gfi1-GFP band. (C) COS-1 cells were transfected with His-Ub and FLAG-Gfi1, and MG123 was added as indicated. Ubiquitinated proteins were selected with His-select beads under denaturing conditions, and immunoblotted for α-FLAG. Clearly visible are the ubiquitinated forms of Gfi1 [(Gfi1-Ub)n], especially after proteasome inhibition. Whole-cell lysate was stained for FLAG, confirming equal loading. * Indicates α-specific binding of unmodified Gfi1 to His-select beads.
Proteins destined for proteasomal degradation are usually tagged by covalently linked poly-ubiquitin chains. To test whether Gfi1 can be ubiquitinated, FLAG-Ub was cotransfected with Gfi1-GFP in COS-1 cells, followed by GFP immunoprecipitation and FLAG staining. This revealed a smear of ubiquitinated proteins representing ubiquitinated Gfi1 species or Gfi1 in a complex with other ubiquitinated proteins (Figure 2B). In addition, we studied the ubiquitination of Gfi1 in an in vivo ubiquitination experiment. COS-1 cells were transfected with Gfi1-FLAG and His-tagged ubiquitin (Ub-His). Under denaturing conditions, proteins with covalently linked ubiquitin were isolated using His-select beads. Subsequent staining for Gfi1 in Western blot analysis detected a protein smear consisting of (poly-) ubiquitinated forms of Gfi1. An increase of ubiquitinated Gfi1 forms was observed after proteasome inhibition with MG132, indicating that ubiquitinated Gfi1 is targeted for degradation by the 26S proteasome. The latter finding was confirmed by a higher total amount of Gfi1 in the whole-cell lysate after proteasome inhibition (Figure 2C). Similar results were obtained with Gfi1-FLAG in HEK293 cells (data not shown).
Gfi1 is degraded in U937 cells in a proteasome-dependent manner
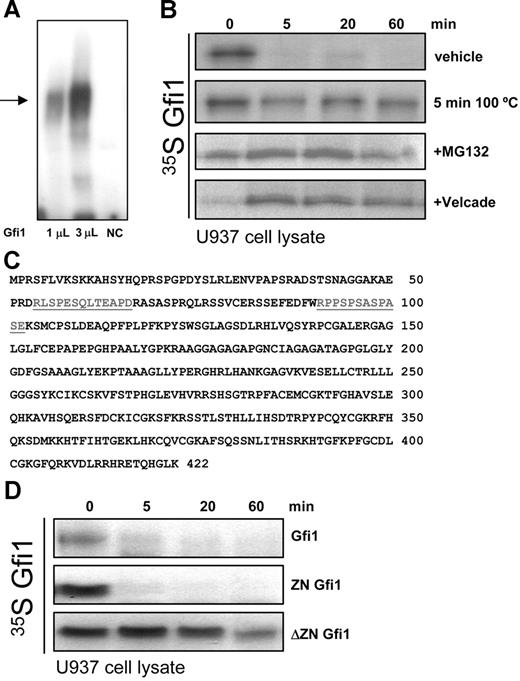
To investigate whether proteasomal degradation of Gfi1 could explain the lower Gfi1 protein levels in immature hematopoietic cells despite high mRNA levels, we developed an in vitro degradation assay. In this assay the stability of a fixed amount of 35 S-labeled, in vitro–translated Gfi1 was studied by incubation for different time points with cell lysates derived from hematopoietic cell lines.26,27 First, the correct folding of Gfi1 was verified by testing the DNA-binding capacity of in vitro–translated Gfi1 in an EMSA using a Gfi1 binding consensus sequence8 as probe (Figure 3A) Gfi1 stability was next assayed using lysates of undifferentiated U937 cells. After 5 minutes most of the Gfi1 was degraded, suggesting that Gfi1 has a short half-life in immature blood cells. To show that the observed degradation was caused by enzymatic factors present in U937 lysate and not by factors from the rabbit reticulocyte lysate, the same assay was performed with U937 extracts heated at 100°C for 5 minutes. In these lysates Gfi1 was not degraded, indicating that the degradation of Gfi1 is dependent on protein activity present in U937 cells. When degradation experiments were performed in the presence of the proteasome inhibitors MG132 or Velcade, the degradation of Gfi1 was almost completely blocked. This showed that the transcriptional repressor Gfi1 is rapidly degraded by the 26S proteasome in lysates from hematopoietic cell lines (Figure 3B).
Gfi1 is degraded by the 26S proteasome in hematopoietic cells. (A) The DNA binding activity of in vitro–translated Gfi1 was checked using EMSA after incubation with 32 P-labeled oligos containing a consensus Gfi1 binding site. A clear band shift (→) was found only in the lanes containing 1 μL or 3 μL Gfi1-programmed reticulocyte lysate. Unprogrammed reticulocyte lysate was used as negative control (NC). (B) An in vitro degradation assay using 35S-labeled in vitro–translated Gfi1 incubated with U937 lysates at 37°C in the presence of vehicle, or the proteasome inhibitors MG132 or Velcade or denatured cell lysate (5 minutes at 100°C). At the indicated time points, samples were inactivated in loading buffer and resolved by SDS-PAGE. (C) The localization of 2 putative PEST domains in Homo sapiens Gfi1 is underlined. (D) 35 S-labeled in vitro–translated full-length Gfi1, the zinc finger domain of Gfi1 (ZN Gfi1), and the non–zinc finger domain of Gfi1 (ΔZN Gfi1) were used in an in vitro degradation assay as described in panel B.
Gfi1 is degraded by the 26S proteasome in hematopoietic cells. (A) The DNA binding activity of in vitro–translated Gfi1 was checked using EMSA after incubation with 32 P-labeled oligos containing a consensus Gfi1 binding site. A clear band shift (→) was found only in the lanes containing 1 μL or 3 μL Gfi1-programmed reticulocyte lysate. Unprogrammed reticulocyte lysate was used as negative control (NC). (B) An in vitro degradation assay using 35S-labeled in vitro–translated Gfi1 incubated with U937 lysates at 37°C in the presence of vehicle, or the proteasome inhibitors MG132 or Velcade or denatured cell lysate (5 minutes at 100°C). At the indicated time points, samples were inactivated in loading buffer and resolved by SDS-PAGE. (C) The localization of 2 putative PEST domains in Homo sapiens Gfi1 is underlined. (D) 35 S-labeled in vitro–translated full-length Gfi1, the zinc finger domain of Gfi1 (ZN Gfi1), and the non–zinc finger domain of Gfi1 (ΔZN Gfi1) were used in an in vitro degradation assay as described in panel B.
The Gfi1 protein consists of an N-terminal SNAG repressor domain followed by a central domain with unknown function and a C-terminal DNA binding domain containing 6 zinc fingers. Computer-based prediction (https://emb1.bcc.univie.ac.at/toolbox/pestfind/pestfind-analysis-webtool.htm) identified 2 putative polypeptide enriched in proline (P), glutamic acid (E), serine (S), and threonine (T) (PEST) regions in Gfi1, located in the N-terminal non–zinc finger part at amino acid positions 54-66 and 90-102 (Figure 3C). PEST domains are associated with ubiquitin-proteasome–mediated turnover and are found in several short-lived transcription factors.28 To determine the involvement of the putative PEST domains in Gfi1 degradation, 2 independent Gfi1 constructs containing either the zinc fingers (ZN Gfi1) or the SNAG and non–zinc finger domain (ΔZN Gfi1) were used in the degradation assay. This showed that the zinc finger part of Gfi1 is degraded at a similar rate as the full-length protein, whereas the non–zinc finger part of Gfi1 was not affected by the ubiquitin-proteasome system (Figure 3D). This suggests that the proteasomal degradation of Gfi1 takes place at the zinc finger domain of Gfi1 and is independent of the putative PEST domains.
Gfi1 degradation is diminished upon monocytic differentiation of U937 cells
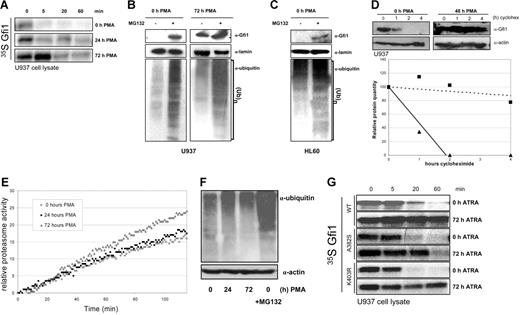
To examine whether the induction of Gfi1 protein levels during myeloid differentiation could be explained by a difference in Gfi1 turnover, the degradation of Gfi1 was examined with the in vitro degradation assay using lysates from 0-, 24-, and 72-hour PMA-stimulated U937 cells. As is shown in Figure 4A, the turnover rate of Gfi1 was clearly diminished after induction of monocytic differentiation of U937 cells with PMA. This indicates that the degradation of Gfi1 depends on the differentiation status of the hematopoietic cell. To investigate whether the observed increase in half-life of Gfi1 was generally seen during differentiation and not a differentiation-independent effect of PMA, U937 cells were also differentiated toward the monocytic lineage with ATRA. Lysates from U937 cells induced with ATRA showed a similar prolonged half-life of Gfi1, comparable with PMA-stimulated cells (data not shown).
Proteasomal Gfi1 degradation is diminished during differentiation. (A) U937 cells were differentiated with PMA and collected at the indicated time points. Cell lysates were normalized using Bradford protein quantification assay and used in an in vitro degradation assay with 35 S-labeled in vitro–translated full-length Gfi1. (B) Differentiated and undifferentiated U937 cells were treated overnight with MG132. Endogenous Gfi1, ubiquitin, and lamin levels were quantified by Western blot. Note the clear increase in Gfi1 levels in undifferentiated U937 cells after proteasome inhibition. Lamin staining was used as a control for equal loading. Ubiquitin staining was performed to confirm the global effectiveness of MG132. (C) In undifferentiated HL60 cells proteasome inhibition results in an accumulation of Gfi1 protein levels. The same controls were used as in panel B. (D) The stability of endogenous Gfi1 was studied in undifferentiated and 48-hour PMA-treated U937 cells. Cells were treated with 25 μg/mL cycloheximide for the indicated times. Lamin staining was used to check for equal loading and the density of Gfi1 bands was quantified using ImageJ software. The relative protein amount was plotted against the time of cycloheximide treatment. ▴ indicate the relative amount of Gfi1 in untreated cells and ▪ indicate the relative amount of Gfi1 in 48-hour PMA-differentiated cells. (E) 26S proteasome function in lysates from differentiated U937 cells was analyzed by measuring the cleavage of Suc-Leu-Leu-Val-Tyr-AMC in the lysate for 2 hours at 37°C. (F) U937 cell lysates were immunoblotted and stained for ubiquitin. Cell lysates treated overnight with MG132 were used as a positive control for decreased proteasome activity. Equal loading was confirmed with actin staining. (G) In vitro–translated wild-type Gfi1 (WT), and the Gfi1 point mutants A382S and K403R were used in an in vitro degradation assay with 72-hour ATRA-treated and untreated U937 cell lysates for the indicated time points. Cell lysates were normalized using the Bio-Rad protein quantification assay.
Proteasomal Gfi1 degradation is diminished during differentiation. (A) U937 cells were differentiated with PMA and collected at the indicated time points. Cell lysates were normalized using Bradford protein quantification assay and used in an in vitro degradation assay with 35 S-labeled in vitro–translated full-length Gfi1. (B) Differentiated and undifferentiated U937 cells were treated overnight with MG132. Endogenous Gfi1, ubiquitin, and lamin levels were quantified by Western blot. Note the clear increase in Gfi1 levels in undifferentiated U937 cells after proteasome inhibition. Lamin staining was used as a control for equal loading. Ubiquitin staining was performed to confirm the global effectiveness of MG132. (C) In undifferentiated HL60 cells proteasome inhibition results in an accumulation of Gfi1 protein levels. The same controls were used as in panel B. (D) The stability of endogenous Gfi1 was studied in undifferentiated and 48-hour PMA-treated U937 cells. Cells were treated with 25 μg/mL cycloheximide for the indicated times. Lamin staining was used to check for equal loading and the density of Gfi1 bands was quantified using ImageJ software. The relative protein amount was plotted against the time of cycloheximide treatment. ▴ indicate the relative amount of Gfi1 in untreated cells and ▪ indicate the relative amount of Gfi1 in 48-hour PMA-differentiated cells. (E) 26S proteasome function in lysates from differentiated U937 cells was analyzed by measuring the cleavage of Suc-Leu-Leu-Val-Tyr-AMC in the lysate for 2 hours at 37°C. (F) U937 cell lysates were immunoblotted and stained for ubiquitin. Cell lysates treated overnight with MG132 were used as a positive control for decreased proteasome activity. Equal loading was confirmed with actin staining. (G) In vitro–translated wild-type Gfi1 (WT), and the Gfi1 point mutants A382S and K403R were used in an in vitro degradation assay with 72-hour ATRA-treated and untreated U937 cell lysates for the indicated time points. Cell lysates were normalized using the Bio-Rad protein quantification assay.
The observed stabilization upon differentiation was further studied by analyzing the proteasomal turnover of endogenous Gfi1 protein levels in U937 cells. Gfi1 protein levels could only be detected upon proteasome inhibition of immature U937 cells. This indicates that Gfi1 is degraded in a proteasome-dependent manner (Figure 4B). The same result was observed in immature HL60 cells (Figure 4C). However, in U937 cells treated for 72 hours with PMA, Gfi1 levels were not significantly induced after MG132 treatment, whereas an α-ubiquitin staining showed that the 26S proteasome was successfully inhibited. This indicates that in differentiated U937 cells endogenous Gfi1 is no longer subject to efficient proteasomal degradation. Subsequently, the half-life of endogenous Gfi1 protein was determined in undifferentiated and 48-hour PMA-treated U937 cells using the protein translation inhibitor cycloheximide. In immature U937 cells Gfi1 has a half-life shorter than 1 hour, whereas in differentiated cells the half-life is more than 4 hours (after 4 hours 80% of the original amount of Gfi1 was still present, Figure 4D). Together these data confirm the result obtained with the in vitro degradation assays and also explain the observed up-regulation of Gfi1 protein levels during differentiation, in spite of decreasing mRNA levels. We conclude that Gfi1 is rapidly degraded by the proteasome in immature myeloid cells, whereas in mature monocytic cells Gfi1 degradation is diminished.
The decreased turnover of Gfi1 might be due to decreased overall proteasomal activity during myeloid differentiation. To investigate this, we measured the chymotrypsin activity of the 26S proteasome in U937 cell lysates by measuring the cleavage of the 26S proteasome substrate Suc-Leu-Leu-Val-Tyr-AMC. The proteasomal activity did not decrease but rather slightly increased during differentiation (Figure 4E). These results were confirmed by ubiquitin staining of whole-cell lysates of PMA-differentiated U937 cells. No significant differences in the total amount of high-molecular-mass ubiquitinated proteins could be detected between differentiated and undifferentiated cells, whereas proteasome inhibition of these cells resulted in a clear accumulation of ubiquitinated proteins (Figure 4F). Together, these observations show that the difference in Gfi1 turnover during differentiation is not due to overall differences in proteasomal activity, indicating that Gfi1 is specifically targeted by the ubiquitin-proteasome system.
Two Gfi1 missense mutations have recently been identified in patients suffering from neutropenia.7 The 2 mutations, N382S and K403R, occur in the zinc fingers of Gfi1, the region that is targeted for degradation. The K403R mutation results in the loss of a lysine in the zinc finger region. Since lysine residues are the primary targets for ubiquitin modification, the lysine mutation might especially affect the protein half-life. Therefore, the turnover of the 2 mutated Gfi1 proteins were compared with the wild-type protein in an in vitro degradation assay. No clear differences in the degradation of normal and mutated Gfi1 in both undifferentiated and ATRA-differentiated U937 cells were observed. This suggests that these mutations have no effect on the proteasomal degradation of Gfi1 (Figure 4G).
Gfi1 is stable in mature primary monocytes and rapidly degraded in granulocytes
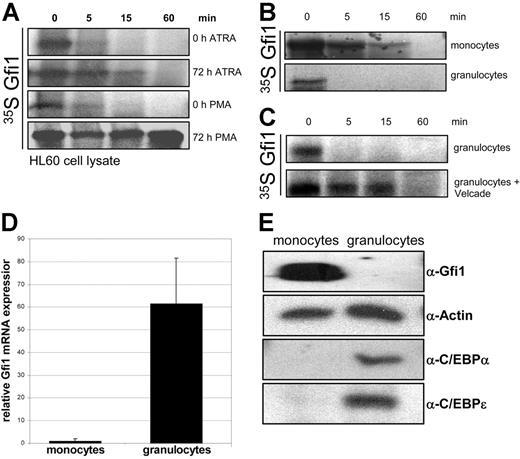
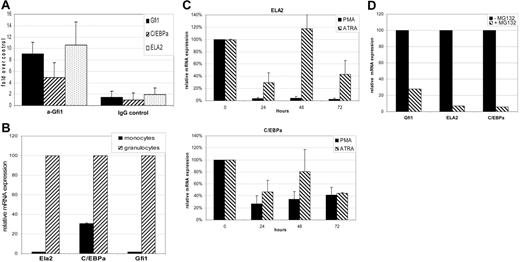
To study the differences in Gfi1 stability during monocytic and granulocytic differentiation, lysates from bipotential HL60 cells differentiated toward monocytes with PMA and toward granulocytes with ATRA were compared. Gfi1 was stabilized during both monocytic and granulocytic differentiation. However, the increase in stability of Gfi1 during granulocytic differentiation is significantly less compared with monocytic differentiation (Figure 5A). This explains the higher amount of Gfi1 protein levels after monocytic differentiation compared with granulocytic differentiation in these cells (Figure 1C). These data also suggest that the observed diminished Gfi1 degradation is playing a more important role during monocytic differentiation than during granulocytic differentiation. We tested whether this difference in Gfi1 turnover was also present in primary monocytes compared with granulocytes. To this end, cell lysates of monocytes and granulocytes were used in an in vitro Gfi1 degradation assay. Gfi1 was more stable in lysates from CD14+-sorted monocytes compared with granulocytes (Figure 5B). Gfi1 degradation in lysates from primary granulocytes could be inhibited by the addition of Velcade, indicating that the observed degradation of Gfi1 was due to 26S proteasomal activity (Figure 5C). In accordance with earlier studies,6,10,15,17 we found a 60-fold-higher Gfi1 mRNA expression in granulocytes compared with monocytes (Figure 5D). Strikingly, at the protein level we could detect Gfi1 only in lysates from monocytes and not from granulocytes (Figure 5E). As control, the same lysates were stained for C/EBPα and C/EBPϵ, indicating that other nuclear transcription factors could be correctly detected in granulocytes. We conclude that due to diminished proteasomal degradation, Gfi1 protein levels are high in primary monocytes despite low mRNA levels.
Gfi1 protein levels are high in monocytes compared with granulocytes due to decreased proteasomal degradation. (A) HL60 cells were differentiated with ATRA toward granulocytes for 72 hours. PMA was used to stimulate HL60 cells for monocytic differentiation. Cell lysates were used in an in vitro degradation assay. (B) Lysates of primary monocytes and granulocytes were used in an in vitro degradation assay with 35 S-labeled Gfi1. (C) The Gfi1 turnover in granulocyte lysate in an in vitro degradation assay depended on 26S proteasome activity. (D) Quantitative Gfi1 RT-PCR was performed on RNA from primary monocytes and granulocytes (n = 3). Error bars indicate standard deviation. (E) Cell lysates of isolated monocytes and granulocytes were immunoblotted and stained with α-Gfi1 antibody, and α-actin staining was used to check for equal loading. α-C/EBPα and α-C/EBPϵ protein levels were stained as positive control for the used protein lysates. Cell lysates were normalized using the Bio-Rad protein quantification assay.
Gfi1 protein levels are high in monocytes compared with granulocytes due to decreased proteasomal degradation. (A) HL60 cells were differentiated with ATRA toward granulocytes for 72 hours. PMA was used to stimulate HL60 cells for monocytic differentiation. Cell lysates were used in an in vitro degradation assay. (B) Lysates of primary monocytes and granulocytes were used in an in vitro degradation assay with 35 S-labeled Gfi1. (C) The Gfi1 turnover in granulocyte lysate in an in vitro degradation assay depended on 26S proteasome activity. (D) Quantitative Gfi1 RT-PCR was performed on RNA from primary monocytes and granulocytes (n = 3). Error bars indicate standard deviation. (E) Cell lysates of isolated monocytes and granulocytes were immunoblotted and stained with α-Gfi1 antibody, and α-actin staining was used to check for equal loading. α-C/EBPα and α-C/EBPϵ protein levels were stained as positive control for the used protein lysates. Cell lysates were normalized using the Bio-Rad protein quantification assay.
Gfi1 may repress specific target genes in mature monocytes
Gfi1 is known to repress its own promoter,9,10,16 and is present on the promoter of several granulocyte-specific genes.8 To study whether Gfi1 binds to the promoter of known Gfi1 target genes we performed ChIP experiments using primary monocytes. Of the analyzed genes GFI1, ELA2, and CEBPA, all promoters could be immunoprecipitated with the α-Gfi1 antibody (Figure 6A). In line with this, the mRNA expression of these genes was lower in monocytes compared with granulocytes (Figures 5D and 6B). Together these data indicate that Gfi1 is present on the promoters of granulocyte-specific genes in monocytes, suggesting that Gfi1 might function to repress the granulocytic phenotype in these cells. This hypothesis is in line with the observation that during monocytic (PMA) differentiation of HL60 cells the expression of the Gfi1 target genes ELA2 and C/EBPα are lower when compared with granulocytic (ATRA) differentiation (Figure 6C). This is also the case for Gfi1 itself (Figure 1A). Also, when Gfi1 protein levels increase after proteasome inhibition of HL60 cells, the mRNA expression of ELA2, C/EBPα, and Gfi1 are decreased (Figure 6D). The inverse correlation between Gfi1 protein levels and the expression of its target genes might imply an important role for the here-observed proteasomal regulation mechanism of Gfi1 during monocytic differentiation.
Gfi1 is present on Gfi1 target genes in monocytes. (A) ChIP analysis on sonicated chromatin of primary monocytes was performed using an α-Gfi1 antibody or a goat–IgG antibody as control (n = 2). Quantitative PCR was used to measure the amount of immunoprecipitated promoter, which was compared with the control genomic sequence of albumin. The percentage of recovery of the ChIP promoters was plotted as the fold increase over the percentage of recovery of albumin. Error bars indicate standard deviation. (B) The mRNA levels of Gfi1-regulated genes were measured using quantitative RT-PCR on mRNA from monocytes (n = 5) and granulocytes (n = 6). Expression in granulocytes was set at 100%. (C) ELA2 (top panel) and C/EBPα (bottom panel) quantitative RT-PCR was performed on RNA samples of HL60 cells during PMA- or ATRA-induced differentiation (n = 2). ELA2 and C/EBPα expression levels in untreated HL60 and U937 cells were set at 100% and values were normalized for β-actin expression. (D) ELA2, C/EBPα, and Gfi1 mRNA expression was measured using quantitative RT-PCR and were normalized for 18S rRNA expression. The levels in untreated cells (−MG132) were set at 100% and compared with MG132-treated HL60 cells.
Gfi1 is present on Gfi1 target genes in monocytes. (A) ChIP analysis on sonicated chromatin of primary monocytes was performed using an α-Gfi1 antibody or a goat–IgG antibody as control (n = 2). Quantitative PCR was used to measure the amount of immunoprecipitated promoter, which was compared with the control genomic sequence of albumin. The percentage of recovery of the ChIP promoters was plotted as the fold increase over the percentage of recovery of albumin. Error bars indicate standard deviation. (B) The mRNA levels of Gfi1-regulated genes were measured using quantitative RT-PCR on mRNA from monocytes (n = 5) and granulocytes (n = 6). Expression in granulocytes was set at 100%. (C) ELA2 (top panel) and C/EBPα (bottom panel) quantitative RT-PCR was performed on RNA samples of HL60 cells during PMA- or ATRA-induced differentiation (n = 2). ELA2 and C/EBPα expression levels in untreated HL60 and U937 cells were set at 100% and values were normalized for β-actin expression. (D) ELA2, C/EBPα, and Gfi1 mRNA expression was measured using quantitative RT-PCR and were normalized for 18S rRNA expression. The levels in untreated cells (−MG132) were set at 100% and compared with MG132-treated HL60 cells.
Discussion
Transcription factors play a crucial role during hematopoiesis by regulating specific differentiation programs. Gfi1 knock-out studies have shown that Gfi1 plays important roles during different stages of hematopoiesis. Gfi1−/− mice show a block in myeloid cell differentiation and lack neutrophils, indicating that Gfi1 is essential for neutrophilic differentiation.5,6 Recent studies also showed abnormal monocyte-derived DCs and macrophages in Gfi1−/− mice, implying a role for Gfi1 in the monocytic lineage.6,16,18
We show here that Gfi1 protein levels are mainly regulated by the ubiquitin-proteasome pathway during monocytic differentiation. The proteasomal degradation of Gfi1 is diminished upon monocytic differentiation of U937 and HL60 cells, resulting in an accumulation of protein levels (Figures 1 and 4). The dimished proteasomal degradation during monocytic differentiation is not caused by a decrease in general proteasomal activity (Figure 4C-D). This strongly suggests that a specific mechanism exists that regulates Gfi1 protein turnover. The stability of proteins targeted for 26S proteasomal degradation can be regulated at different levels, for example by de-ubiquitination enzymes or by other posttranslational modifications with, for example, small ubiquitin-related modifier protein (SUMO).29 Alternatively, alterations in E3 ubiquitin ligase activity may cause Gfi1 stabilization during monocytic differentiation. Which E3 ubiquitin ligase(s) specifically mark Gfi1 for proteasomal degradation is unknown. It was recently shown that Ataxin-1 expression correlates with an increase in proteasomal degradation of Gfi1.30 While Ataxin-1 has no known E3 ligase activity by itself, it may be part of a multiprotein complex containing ubiquitin ligase activity.
We show that the zinc finger domain of Gfi1 is specifically targeted for proteasomal degradation (Figure 3D). The Gfi1 paralog Gfi1B is an essential transcription factor for erythroid and megakarocyte differentiation.31,32 Especially, the zinc finger–containing DNA-binding region of Gfi1B is very homologous to Gfi1 (97%). Therefore, it may be speculated that Gfi1B is also subject to proteasomal degradation and it would be interesting to study whether Gfi1B protein levels are regulated in a similar way during erythroid/megakaryocytic differentiation.
Gfi1−/− mice exhibit a block in neutrophilic differentiation, resulting in the accumulation of an immature myelo-monocytic population featuring both monocytic as well as granulocytic characteristics. The observed differentiation block results in the absence of mature neutrophils and takes place just after the promyelocyte stage, demonstrating that Gfi1 is essential during the first steps of GMP differentiation toward the promyelocyte stage. Whether Gfi1 is required after this stage of differentiation is currently not known. In primary monocytes Gfi1 is more stable compared with primary granulocytes, in which Gfi1 is swiftly degraded by the 26S proteasome. This results in higher Gfi1 protein levels in monocytes compared with granulocytes, despite 60-fold-higher mRNA levels in granulocytes (Figure 5). Whether the observed rapid degradation of Gfi1 in mature granulocytes is essential for terminal granulocytic function remains to be elucidated. A possible role for the proteasomal down-regulation of Gfi1 in mature granulocytes may be the release of Gfi1-dependent repression of granulocyte-specific genes, such as C/EBPA, C/EBPE, and ELA2.
The presence of Gfi1 in primary monocytes suggests a role for Gfi1 in these cells. Gfi1 might serve to repress granulocyte-specific genes, thereby inhibiting granulocytic traits in monocytes. This is in line with the observation that Gfi1 target genes (C/EBPα and ELA-2) were expressed at higher levels in granulocytes compared with monocytes (Figure 6B). Moreover, ChIP experiments showed that Gfi1 is present on the promoter of these genes in primary monocytes (Figure 6A). The ChIP experiments also showed that Gfi1 is present on its own promoter, suggesting that Gfi1 is capable of regulating its own expression in myeloid cells. If so, this could explain the high Gfi1 mRNA levels in granulocytes, due to the lack of repression by Gfi1, whereas in mature monocytes Gfi1 represses its own promoter, resulting in low mRNA expression.
The accumulated myelomonocytic population in Gfi1−/− mice featuring both monocytic and granulocytic characteristics was originally assumed to consist of arrested differentiating neutrophils. However, a partial block during monocytic differentiation could not be dismissed.5,6 This latter explanation was suggested to be unlikely since Gfi1 mRNA was not detected in macrophages and Gfi1 mRNA levels are down-regulated during monocytic differentiation of HL60 cells.33 As our results show that the Gfi1 protein is present in monocytes, it will be interesting to readdress this question and study whether the mixed myeloid population in Gfi1−/− mice also consists of arrested monocytes. As Gfi1 functions in many steps during hematopoiesis but also in the development of the intestine34 and inner ear,35,36 it will be interesting to test whether the here-described degradation of Gfi1 also plays an important regulatory role in these processes.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: the authors declare no competing financial interests.
Acknowledgments
We would like to thank Dr T. Möröy for valuable discussions and for providing us with Gfi1-FLAG and Gfi1-GFP constructs, Dr M. Osawa for the FLAG-Gfi1 plasmids, Dr M. Horwitz for the Gfi1 point mutants, and Dr M. Scheffner for His-Ub and FLAG-Ub plasmids. We thank W. Wissink and P. Jansen for technical assistance, E. de Grouw for cDNA samples of monocytes and granulocytes, and L. van der Locht for primer design. We appreciate the assistance of S. van Wageningen with the ChIP experiments.
This work was supported by grants from the Dutch Cancer Society project code KUN2001-2395 (J.A.F.M., L.V.E.), the Royal Netherlands Academy of Sciences KNAW (J.H.J.), and the Vanderes Foundation (B.A.v.d.R.).

![Figure 2. Gfi1 is degraded by the ubiquitin-proteasome system. (A) HEK293 cells were split after transfection with Gfi1-GFP and were grown overnight with or without MG132. Cells were measured by flow cytometer and the mean fluorescence index (MFI) of untreated cells was set at 100%. After proteasome inhibition (5 μM MG132), a clear increase of Gfi1-GFP levels per cell was observed, indicated as MFI. (B) COS-1 cells were transfected with FLAG-Ub and Gfi1-GFP or with empty vector, followed by immunoprecipitation with an α-GFP antibody. Subsequent immunoblotting with an α-FLAG antibody resulted in the detection of a high molecular smear of proteins [(Ub)n], indicating that Gfi1 is present in ubiquitinated complexes, or is ubiquitinated itself. → Indicates the height of unmodified Gfi1-GFP. Bottom panel with α-GFP staining of whole-cell extract (WCE) shows a clear distinct Gfi1-GFP band. (C) COS-1 cells were transfected with His-Ub and FLAG-Gfi1, and MG123 was added as indicated. Ubiquitinated proteins were selected with His-select beads under denaturing conditions, and immunoblotted for α-FLAG. Clearly visible are the ubiquitinated forms of Gfi1 [(Gfi1-Ub)n], especially after proteasome inhibition. Whole-cell lysate was stained for FLAG, confirming equal loading. * Indicates α-specific binding of unmodified Gfi1 to His-select beads.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/1/10.1182_blood-2006-02-003590/4/m_zh80010705870002.jpeg?Expires=1767845181&Signature=p~FJWAn8Hz-6rvzullk6HR80vRLlcLiKtpiHG3eyPxcc6kcCSBAuYUfWuk-kL2cT3stAFb98tRByPyQgBx2BfN6cKBNUgr5NrKD-AS2EHDJ5V4JeaevjxzrAeqal11nWTW6Q73Rrve~-sRPCTpbVz4WpAP91uygwZ8x352-W1zdNMEg8XOEDhH0ZTN2COaTcxDwc~TWjiH4g6GmVEYcQaNhWZLq3YHIEOrp~IND7pDd0wH4KpA9Xj~Vj2WxMsEankvyNvx3n6FJDZBwGFUskAujlper8ufBp6lxkDbca3vX~pAOl50N3ZAkh3teLBTRtuU2WD43EFcdiG7a97bMG7Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal