Abstract
To obtain insight into human CD4+ T cell differentiation and selection in vivo, we longitudinally studied cytomegalovirus (CMV)–specific CD4+ T cells after primary infection. Early in infection, CMV-specific CD4+ T cells have the appearance of interferon γ (IFNγ)–producing T-helper 1 (TH1) type cells, whereas during latency a large population of CMV-specific CD4+CD28– T cells emerges with immediate cytotoxic capacity. We demonstrate that CD4+CD28– T cells could lyse CMV antigen–expressing target cells in a class II–dependent manner. To clarify the clonal relationship between early and late CMV-specific CD4+ T cells, we determined their Vβ usage and CDR3 sequences. The T-cell receptor β (TCRβ) diversity in the early CMV-specific CD4+ T-cell population was high in contrast to the use of a very restricted set of TCRβ sequences in latent infection. T-cell clones found in the late CMV-specific CD4+ T-cell population could not be retrieved from the early CD4+ T-cell population, or were present only at a low frequency. The observation that dominant CMV-specific CD4+ clones during latency were only poorly represented in the acute phase suggests that after the initial control of the virus strong selection and/or priming of novel clones takes place in persistent infections in humans.
Introduction
CD4+ T cells play a central role in the defense against pathogens such as viruses, mostly by helping other cells to acquire and execute their effector functions.1-5 Moreover, they may impede the spread of viruses through the secretion of cytokines such as interferon γ (IFNγ) that block viral replication.6 Last, cytotoxic functions have also been attributed to CD4+ T cells,7 but as yet it is unclear whether recognition of viral antigens can activate the cytolytic machinery.
Human cytomegalovirus (CMV)–specific CD4+ T cells have been studied both during their development in primary CMV infection and during the latency phase. In the acute response, CMV-specific CD4+ T cells are highly activated and proliferating, have a CD45RA+CD45R0+CD28+CD27+ phenotype, and produce IFNγ after stimulation with CMV antigen in vitro.8 Late after primary infection, CMV-specific CD4+ T cells have returned to a resting stage, and the percentage of IFNγ-producing CMV-specific CD4+ T cells is considerably lower than during the acute infection. Moreover, a considerable number of CMV-specific CD4+ T cells have progressed toward a further differentiated phenotype, characterized by the loss of both CD27 and CD28 and the acquisition of cytolytic molecules such as perforin and granzyme B (grB).8,9 CD4+CD28– grB+ T cells appear to be characteristic for CMV infection because they emerge after primary CMV infection and can be found only in CMV-seropositive individuals.9 Consequently, proliferation and cytokine production by CD4+CD28– T cells can be induced by CMV but not by other viral antigens.
How virus-specific memory CD4+ T cells differentiate in humans is largely unknown. This can be attributed to the difficulty of tracking primary infections and the limitations in measuring antigen-specific CD4+ T cells. From murine studies, it can be deduced that after acute infection, numbers of CD4+ memory T cells, in contrast to CD8+ T cells, decline over time.10 For CD8+ T cells in persistent viral infection, the hierarchy of immunodominant epitopes during the late persistence phase is completely altered, compared with the acute phase of the infection.11-14 Furthermore, it has been shown that during the latency phase, virus-specific cells can still be primed and develop into memory cells.15 These last 2 findings indicate that during acute infection, different types of T cells may be needed compared with the phase of viral persistence. Moreover, different viral antigens may be presented to the T cells during the persistence phase, which can contribute to shaping the clonality of the memory T-cell population.
We studied CMV-specific CD4+ T cells during primary CMV infection to examine whether CMV-specific CD4+ T cells found early in infection differentiated and acquired the features of the late CMV-specific cells, or, alternatively, whether new CMV-specific CD4+ T cells emerged after acute infection and (partially) replaced the early cells. Analysis and comparison of the T-cell receptor (TCR) Vβ CDR3 regions of the different early and late CMV-specific CD4+ T-cell populations allowed us to unravel the interrelationship between the virus-specific cells.
Materials and methods
Subjects
Peripheral blood mononuclear cells (PBMCs) were isolated from renal transplant recipients treated with basic immunosuppressive therapy (cyclosporine A, prednisolone, and mycophenolate mofetil). All subjects gave written informed consent, and the medical ethics committee of the Academic Medical Center, Amsterdam, approved the study.
Transduction of LCLs
CMV Ag and peptides
CMV antigen (Ag) (Microbix Biosystems, Toronto, ON, Canada) and human leukocyte antigen (HLA)–DR4–binding CMV pp65-derived peptide IIKPGKISHIMLDVA18 and HLA-A201–binding CMV pp65-derived peptide NLVPMVATV were used to stimulate PBMCs directly or loaded on LCL.
Intracellular cytokine staining
PBMCs were incubated for 6 hours with control LCL, LCL pp65, LCL pulsed with CMV Ag or CMV DR4 peptide, or just CMV Ag, CMV A2 peptide or as a positive control Staphylococcus aureus enterotoxin B (SEB; ICN/Fluka, Buchs SG, Switzerland). For the final 5 hours of culture, brefeldin A (Sigma Chemical, St Louis, MO) was added. Cells were fixed and permeabilized, followed by (intracellular) staining with different combinations of IFNγ-FITC, CD3–peridinin chlorophyll protein (PerCP) Cy5.5, CD4-PerCP Cy5.5, CD4-allophycocyanin, CD8-PerCP Cy5.5, CD8-allophycocyanin, CD28-PE, CD69-PE (all from BD Biosciences, San Jose, CA) and CD69-allophycocyanin (Invitrogen, Carlsbad, CA), and analysis by fluorescence-activated cell sorter (FACS).
Cytotoxicity assay
Ex vivo cytotoxicity was assessed by incubating 51Cr-labeled autologous LCLs empty or pulsed with CMV Ag or CMV DR4 peptide with PBMCs at effector-target (E/T) ratios of 1:1, calculated based on absolute numbers IFNγ-producing CD4+ T cells. HLA-DR–blocking antibody R3E2 was used (ascitis, 25 μL/mL). Percentage of specific lysis was calculated from the following formula: percentage of specific release = [(experimental release – spontaneous release)/(maximum release – spontaneous release)] × 100%.
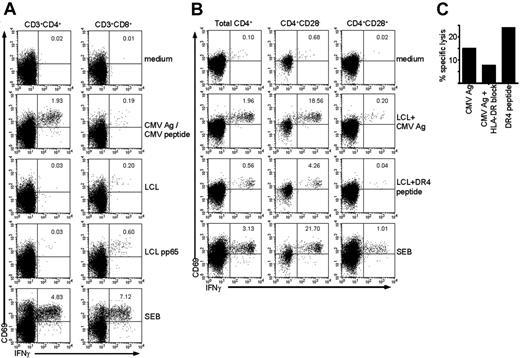
CD4+CD28– T cells are class II–restricted cytotoxic CMV-specific T cells. (A) Dot plots gated on either CD4+ T cells (left column) or CD8+ T cells (right column) show intracellular cytokine staining for IFNγ in unstimulated cells (medium), cells stimulated with CMV Ag (for CD4+ T cells), or CMV peptide (for CD8+ T cells), with a control autologous LCL, or with an autologous LCL transduced with a pp65 construct. SEB is the positive control. Numbers indicate the percentage of CD69+ IFNγ+ cells within total CD4+ or CD8+ T cells, respectively. (B) Dot plots gated on either total CD4+ lymphocytes, CD4+CD28– cells, or CD4+CD28+ cells (from left to right) show intracellular cytokine staining for IFNγ in unstimulated cells (medium), cells stimulated with autologous LCLs loaded with CMV Ag, LCLs loaded with a CMV HLA-DR4–restricted peptide, or with SEB as a positive control. Numbers indicate the percentage of CD69+ IFNγ+ cells within the population shown. (C) Graph shows the percentage of specific lysis measured in a 51Cr-release assay of total PBMCs using either CMV Ag or CMV HLA-DR4–restricted peptide–loaded autologous LCLs as target cells. The second bar shows the percentage of lysis of CMV Ag–loaded autologous LCLs in the presence of a class II–blocking antibody.
CD4+CD28– T cells are class II–restricted cytotoxic CMV-specific T cells. (A) Dot plots gated on either CD4+ T cells (left column) or CD8+ T cells (right column) show intracellular cytokine staining for IFNγ in unstimulated cells (medium), cells stimulated with CMV Ag (for CD4+ T cells), or CMV peptide (for CD8+ T cells), with a control autologous LCL, or with an autologous LCL transduced with a pp65 construct. SEB is the positive control. Numbers indicate the percentage of CD69+ IFNγ+ cells within total CD4+ or CD8+ T cells, respectively. (B) Dot plots gated on either total CD4+ lymphocytes, CD4+CD28– cells, or CD4+CD28+ cells (from left to right) show intracellular cytokine staining for IFNγ in unstimulated cells (medium), cells stimulated with autologous LCLs loaded with CMV Ag, LCLs loaded with a CMV HLA-DR4–restricted peptide, or with SEB as a positive control. Numbers indicate the percentage of CD69+ IFNγ+ cells within the population shown. (C) Graph shows the percentage of specific lysis measured in a 51Cr-release assay of total PBMCs using either CMV Ag or CMV HLA-DR4–restricted peptide–loaded autologous LCLs as target cells. The second bar shows the percentage of lysis of CMV Ag–loaded autologous LCLs in the presence of a class II–blocking antibody.
Isolation of CMV-specific cells
IFNγ-producing CD4+ cells were isolated using IFN-γ Secretion Assay Detection Kit (PE) (Miltenyi Biotec, Amsterdam, the Netherlands) and subsequently sorted using FACsARIA (BD). For isolation of CD4+CD28– T cells, CD4-PerCP Cy5.5 (BD Biosciences, San Jose, CA), CD28-FITC (Sanquin, Amsterdam, the Netherlands) were used to stain the cells.
Cloning and sequencing
RNA isolated from sorted cells was subjected to template switch-anchored reverse transcriptase–polymerase chain reaction (RT-PCR) by using Super Smart PCR cDNA Synthesis Kit (BD Biosciences Clontech, Palo Alto, CA). Vβ PCR was performed on amplicons as previously described.19 For the spectratyping, samples were mixed with Genescan-500 ROX size standards and run on an ABI 3100 capillary sequencer (Applied Biosystems, Warrington, United Kingdom) in Genescan mode. Vβ PCR products were purified, ligated into pGEM-T Easy vector (Promega, Madison, WI), cloned by transformation of competent DH5α Escherichia coli, and sequenced with an M13 forward primer.
Junctional region–specific PCR
Sequence-specific primers (5′-TTCTGTGCCAGCAGAAAACA and 5′-AGAAGGCTCTAGGCTGACC) were designed based on Jβ and CDR3 sequence found in the Vβ8 of CD4+CD28– T cells (week 36) from patient 1 (5′-TTCTGTGCCAGCAGAAAACAGGAGACCGGGGAGCTGTTTTTTGGAGAAGGCTCTAGGCTGACC) to determine whether this could be traced back in early IFNγ-producing CD4+ T cells (week 9). PCR products were confirmed by cloning and sequencing.
Results
CD4+CD28– T cells recognize CMV-derived peptides in a class II–restricted fashion
We have previously shown that CD4+CD28– T cells appear after primary CMV infection and produce cytokines upon stimulation with CMV but not other antigens.9 Interestingly, these CMV-specific CD4+CD28– T cells have characteristics normally attributed to CD8+ T cells, such as the expression of the class I–binding natural killer (NK) receptors CD158b, NKB1, CD94 (dim), and NKG2D (data not shown and Groh et al20 ). This corresponds to the finding that circulating KIR+ CD4+ T cells are differentiated antigen-primed cells that can produce IFNγ.21 Since the CD4+CD28– T cells have properties associated with CD8+ T cells, and CD4+ T cells recognizing HLA class I have been described, we assessed whether the CD4+CD28– T-cell population contains cells that can recognize CMV-derived peptides presented in HLA class I. To this end, we used autologous EBV-LCLs that were transduced with a retroviral vector inducing expression of either GFP alone (LCL) or GFP in combination with pp65 (LCL pp65), an immunodominant protein of human CMV. As can be seen in Figure 1A, in contrast to CD8+ T cells from the same donor, CD4+ T cells did not produce IFNγ upon stimulation with LCL pp65. Still, CMV-specific cells were present in both CD4+ and CD8+ T cells, as can be seen from the response upon stimulation with CMV Ag or CMV peptide, respectively (Figure 1A). This showed that the transduction of the LCLs induced presentation of pp65 peptides in HLA class I, but that these endogenously processed peptides were not recognized by CD4+ T cells. We therefore did not find indications that CD4+CD28– T cells can recognize CMV peptides in a class I–presented manner.
CD4+CD28– T cells contain grB and perforin and have cytotoxic capacity in redirected killing assays.7 In order to examine whether these cells are able to lyse targets expressing CMV epitopes, we aimed to load CMV peptides on EBV-LCL in class II. Therefore we tested whether CD4+CD28– T cells recognized peptides from autologous LCLs loaded with CMV Ag overnight. As shown in Figure 1B, from the total CD4+ T-cell population, especially the CD4+CD28– T cells produced high amounts of IFNγ when stimulated with LCLs pulsed with CMV Ag. Recently, a CMV pp65–derived peptide (IIKPGKISHIMLDVA) has been described18 that was recognized by CD4+ T cells from HLA-DR4–typed individuals. In our assay, CD4+ T cells from an HLA-DR4–typed donor also produced very high quantities of IFNγ upon stimulation with LCLs loaded with this peptide. Notably, CD4+CD28+ T cells did not respond to this peptide, although the total percentage of CD4+CD28+ T cells responding to CMV Ag is already low so a response to one peptide might be difficult to observe (Figure 1B). Concerning CD8+ T cells, no IFNγ was produced upon stimulation with either CMV Ag or DR4 peptide–loaded LCL (data not shown).
Now that it was determined that CD4+ T cells and specifically the CD4+CD28– T cells recognized LCLs presenting CMV epitopes, a chromium release cytotoxicity assay was performed to examine direct antigen-specific cytolysis. Efficient cytotoxicity toward target cells loaded with CMV Ag was observed (Figure 1C). Cytolysis could be blocked by an HLA-DR antibody confirming the HLA class II restriction of the response (Figure 1C). Since recognition of the CMV DR4 peptide also resulted in lysis of the target cells, these findings demonstrate that CD4+CD28– T cells can lyse in a class II–restricted, CMV-specific manner.
That CD4+ T cells have cytotoxic potential has been described before, but these conclusions were either based on CD4+ T-cell lines or clones, which run the risk of being functionally modulated by in vitro culture,22-24 or the lysis was non–antigen specific.7 Here, for the first time we demonstrate direct ex vivo antigen-specific killing by human CD4+ T cells. The specific contribution of these cytotoxic CD4+ T cells to the maintenance of CMV latency is unknown and cannot be easily assessed in humans.25 Still, a major mechanism of CMV to avoid recognition by the T-cell immune system is down-regulation of class I expression, which will impede elimination of infected cells by class I restricted CD8+ cytotoxic T lymphocytes (CTLs).26 Because class II–expressing monocytes are the major reservoir of CMV during latency, CD4+CD28– cytotoxic T cells may remove productively infected cells that escape elimination through the CD8+ effector T-cell compartment in a class II–dependent fashion.
Early virus-specific CD4+ T cells have a much broader TCR Vβ repertoire than late virus-specific cells
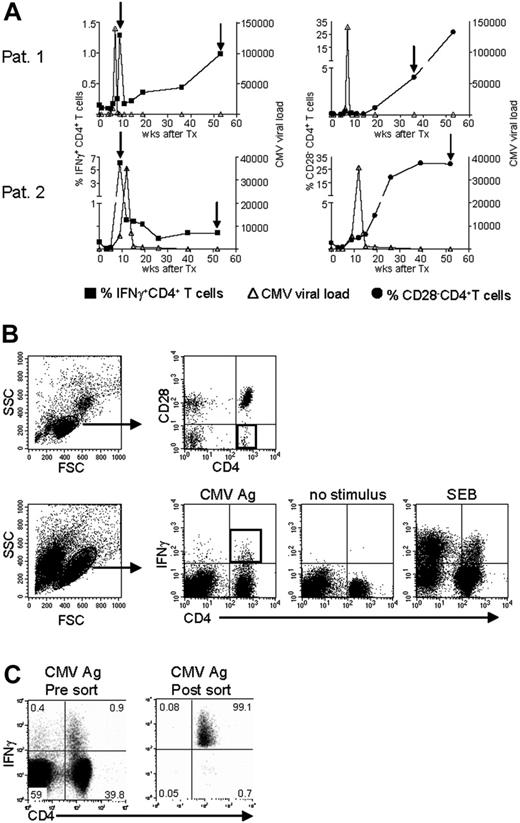
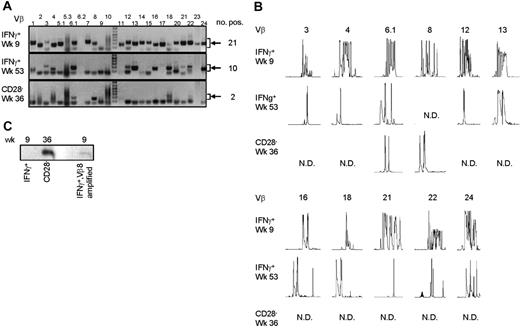
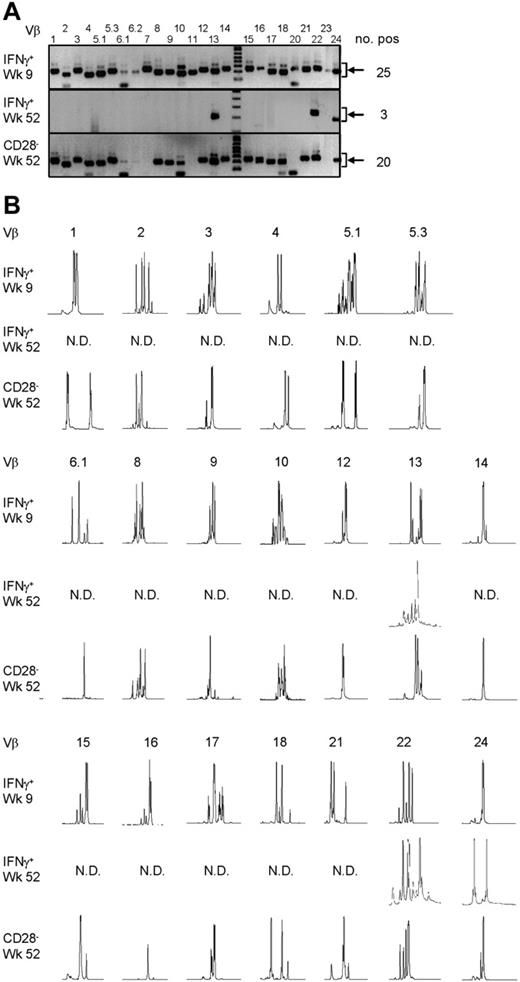
To answer the question of whether the CMV-specific CD4+ T cells found late after CMV infection originated from the early CMV-specific CD4+ T cells or developed independently, we longitudinally studied 2 patients experiencing a primary CMV infection. The kinetics of the viral load and the CMV-specific CD4+ T cells from both patients are shown in Figure 2A. We isolated CMV-specific IFNγ-producing CD4+ T cells at the peak of the response (week 9) and around 1 year later. Moreover, the cytotoxic CD4+CD28– T cells were isolated from both patients at late time points (Figure 2A). As can be seen from the sorting gates shown in Figure 2B, we were able to accurately isolate the populations of interest. Figure 2C shows an example of the purity of sorted IFNγ-producing CD4+ T cells at a late time point. The percentage of IFNγ-producing CD4+ T cells was 0.9% before sort and this increased to over 99% after sorting. From all these different cell populations, RNA was isolated and amplified, and cDNA was made, after which the TCR Vβ repertoire usage was determined. The first analysis for the presence of Vβ families already clarified that CMV-specific cells found early in primary CMV infection in both patients had a much broader repertoire than the late cells (Figure 3A). In patient 1, this restriction was the most prominent in the CD4+CD28– T cells where only Vβ6.1 and Vβ8 could be retrieved. Vβ6.1 was the only Vβ family found in all 3 CMV-specific CD4+ cell populations studied (Figure 3A). In the second patient, especially the late IFNγ-producing CD4+ T cells were extremely restricted, as only Vβ families 13, 22, and 24 could be detected (Figure 4A). The restriction in the CD4+CD28– CMV-specific cells from the second patient was less pronounced but the number of Vβs used was less than in the early cells (20 vs 25 respectively; Figure 4A). The broader repertoire of CMV-specific memory CD4+ T cells in patient 2 compared with patient 1 could be attributed to the difference in height of the viral load. A higher viral load might induce more clonal expansion of the virus-specific cells resulting in clonal exhaustion, as has been described for EBV-specific memory CD8+ T cells.27 Along the same line of reasoning, it might be expected that in healthy individuals the narrowing of the CMV-specific T-cell repertoire will be slower than in renal transplant recipients, like we studied here, who usually have higher viral loads because of the immunosuppressive therapy. This would also explain why high percentages of oligoclonal CD28– T cells are, in nonimmunocompromised individuals, mostly found in the elderly.9,28
Isolation of CMV-specific CD4+ T cells found early and late during primary CMV infection. (A) Top panels show patient 1; bottom panels, patient 2. In the left-hand panels, kinetics of CMV viral load as measured by quantitative PCR (right y-axis) are plotted together with the percentage of CMV-specific IFNγ-producing CD4+ T cells (left y-axis). In the right-hand panels, the CMV viral load is plotted together with the percentage of CD4+CD28– T cells (left y-axis). On the x-axes, the time is indicated as weeks after transplantation, which coincides with the moment of infection. The arrows indicate the time points at which CMV-specific IFNγ-producing or CD28–CD4+ T cells were isolated. (B) Sorting gates of CMV-specific CD4+ T cells in samples at a late time point after infection. Top panels show gating on lymphocytes and subsequently on CD4+CD28– T cells. Bottom panels show gating on (activated) lymphocytes followed by gating IFNγ-producing CD4+ T cells upon stimulation with CMV Ag. Unstimulated cells and SEB-stimulated cells were used to set the gates. Black squares indicate the populations of CMV-specific cells that were isolated. (C) Example of purity of sorted IFNγ+ CD4+ T cells. Left panel shows sample before sort; right panel shows the result after sorting. Numbers indicate the percentages within the corresponding quadrants.
Isolation of CMV-specific CD4+ T cells found early and late during primary CMV infection. (A) Top panels show patient 1; bottom panels, patient 2. In the left-hand panels, kinetics of CMV viral load as measured by quantitative PCR (right y-axis) are plotted together with the percentage of CMV-specific IFNγ-producing CD4+ T cells (left y-axis). In the right-hand panels, the CMV viral load is plotted together with the percentage of CD4+CD28– T cells (left y-axis). On the x-axes, the time is indicated as weeks after transplantation, which coincides with the moment of infection. The arrows indicate the time points at which CMV-specific IFNγ-producing or CD28–CD4+ T cells were isolated. (B) Sorting gates of CMV-specific CD4+ T cells in samples at a late time point after infection. Top panels show gating on lymphocytes and subsequently on CD4+CD28– T cells. Bottom panels show gating on (activated) lymphocytes followed by gating IFNγ-producing CD4+ T cells upon stimulation with CMV Ag. Unstimulated cells and SEB-stimulated cells were used to set the gates. Black squares indicate the populations of CMV-specific cells that were isolated. (C) Example of purity of sorted IFNγ+ CD4+ T cells. Left panel shows sample before sort; right panel shows the result after sorting. Numbers indicate the percentages within the corresponding quadrants.
Late CMV-specific CD4+ T cells have a very restricted TCR Vβ usage. (A) Pictures show the bands on gel retrieved after PCRs for the different Vβ families of patient 1. Analysis was performed on cDNA obtained after amplification of total RNA isolated from the different sorted CMV-specific CD4+ T-cell populations. Arrows on the right side indicate the height at which the specific bands for the Vβ families can be found. Smaller bands at the bottom of the picture are primer dimers and should therefore not be considered specific. Numbers on the right indicate the number of Vβs present in that sample. (B) Results of the spectratype of Vβ families which were found in both early and late CMV-specific CD4+ T-cell populations of patient 1. Each peak represents a CDR3 region with a certain length spaced by 3 nucleotides. Numbers indicate the corresponding Vβ; on the left side, the different cell populations are mentioned. N.D. indicates not detectable in that cell population. (C) Figure shows bands obtained after specific PCR based on sequence found in Vβ8 of CD4+CD28– T cells of patient 1.
Late CMV-specific CD4+ T cells have a very restricted TCR Vβ usage. (A) Pictures show the bands on gel retrieved after PCRs for the different Vβ families of patient 1. Analysis was performed on cDNA obtained after amplification of total RNA isolated from the different sorted CMV-specific CD4+ T-cell populations. Arrows on the right side indicate the height at which the specific bands for the Vβ families can be found. Smaller bands at the bottom of the picture are primer dimers and should therefore not be considered specific. Numbers on the right indicate the number of Vβs present in that sample. (B) Results of the spectratype of Vβ families which were found in both early and late CMV-specific CD4+ T-cell populations of patient 1. Each peak represents a CDR3 region with a certain length spaced by 3 nucleotides. Numbers indicate the corresponding Vβ; on the left side, the different cell populations are mentioned. N.D. indicates not detectable in that cell population. (C) Figure shows bands obtained after specific PCR based on sequence found in Vβ8 of CD4+CD28– T cells of patient 1.
From the Vβs that were found in both patients, spectratyping was performed, in which each peak represents a different CDR3 length. In contrast to the early cells that showed a skewed but still quite broad distribution, very few peaks were present in the samples from the late CMV-specific cells, corroborating the profound restriction in these cell populations (Figure 3B for patient 1 and Figure 4B for patient 2). On average in patient 1, the late IFNγ-producing cells had only 40% of the number of peaks present in the early IFNγ-producing cells; the number of different CDR3 lengths in the CD28–CD4+ T cells was even reduced to 27% compared with the early CMV-specific cells. For patient 2 these numbers were 90% and 64%, respectively. Markedly, in some cases peak lengths were found in the late CMV-specific cells that were not apparent in the early CMV-specific cells. An example can be seen in Figure 3B for Vβ16 in case of the late IFNγ-producing CMV-specific CD4+ T cells and for Vβ8 in case of the CMV-specific CD4+CD28– T cells. From this we could conclude that certain clones may be abundantly present at later time points but not early in primary infection. This already indicated that there is a very strong selection within the CMV-specific CD4+ T cells and that new clones may arise after the acute response to the viral infection. From patient 2, we also isolated CD4+CD28– T cells 2 years after infection, but these hardly differed from the cells retrieved after 1 year, implying that during the latency stage the CD4+ T-cell population remained relatively stable in this patient (data not shown).
Late CMV-specific CD4+ T cells have a very restricted TCR Vβ usage. (A) Pictures show the bands on gel retrieved after PCRs for the different Vβ families of patient 2. Analysis was performed on cDNA obtained after amplification of total RNA isolated from the different sorted CMV-specific CD4+ T-cell populations. Arrows on the right side indicate the height at which the specific bands for the Vβ families can be found. Smaller bands at the bottom of the picture are primer dimers and should therefore not be considered specific. Numbers on the right indicate the number of Vβs present in that sample. (B) Results of the spectratype of Vβ families which were found in both early and late CMV-specific CD4+ T-cell populations of patient 2. Each peak represents a CDR3 region with a certain length spaced by 3 nucleotides. Numbers indicate the corresponding Vβ; on the left side, the different cell populations are mentioned. N.D. indicates not detectable in that cell population.
Late CMV-specific CD4+ T cells have a very restricted TCR Vβ usage. (A) Pictures show the bands on gel retrieved after PCRs for the different Vβ families of patient 2. Analysis was performed on cDNA obtained after amplification of total RNA isolated from the different sorted CMV-specific CD4+ T-cell populations. Arrows on the right side indicate the height at which the specific bands for the Vβ families can be found. Smaller bands at the bottom of the picture are primer dimers and should therefore not be considered specific. Numbers on the right indicate the number of Vβs present in that sample. (B) Results of the spectratype of Vβ families which were found in both early and late CMV-specific CD4+ T-cell populations of patient 2. Each peak represents a CDR3 region with a certain length spaced by 3 nucleotides. Numbers indicate the corresponding Vβ; on the left side, the different cell populations are mentioned. N.D. indicates not detectable in that cell population.
Selection of CMV-specific CD4+ T-cell clones after the acute response
For more extensive comparison between the early and late CMV-specific CD4+ T cells, we sequenced the CDR3 regions. This was done from Vβs in which identical clones could be expected based on the presence of peaks with a similar length in both early and late cells. The sequences that were found are depicted in Tables 1 and 2 for both patients. Analysis of the different sequences again substantiated the wide variability in clones found in the CMV-specific CD4+ T cells present early in primary infection, whereas particularly in patient 1 the number of clones found in the late CMV-specific CD4+ T-cell population was very limited. Multiple identical clones were found when late CMV-specific IFNγ-producing and CD28–CD4+ T-cell populations were compared. This was to be expected because most IFNγ-producing CD4+ T cells have lost expression of CD28, and both populations thus overlap. However, most of the clones retrieved from either one of the late CMV-specific CD4+ T-cell populations could not be detected in the early virus-specific CD4+ T cells. Despite the selection for Vβ families with corresponding CDR3 lengths, only a few identical sequences were found in early and late CMV-specific CD4+ T cells in patient 2 (Table 2). In patient 1, not a single sequence from the late IFNγ-producing or CD28–CD4+ T cells matched the early CMV-specific CD4+ T cells (Table 1). A specific PCR was developed based on the sequence found in all Vβ8 clones of the CD4+CD28– T cells in this patient. No specific product could be detected in the total cDNA of the early IFNγ-producing CD4+ T cells, only in material that was amplified with Vβ8-specific primers (Figure 3C), indicating that the particular clone is present in the early population but only at an extreme low frequency. Since we did not perform such a PCR for all clones found in the late CMV-specific CD4+ T-cell populations, we cannot exclude that others are present in the early populations. However, it is clear that the frequency of these clones within early CMV-specific CD4+ T cells can only be very low. In conjunction with the observed strong selection of CMV-specific CD4+ T cells present early in infection, new naive T cells also might be primed during the latency phase, as has been reported in mice.15 In any case, it is apparent that the virus-specific cells late in infection are different from the population of CD4+ T cells responding early.
Overview of CDR3 sequences of CMV-specific CD4+ T cells from patient 1
Specific Vβ family and CDR3* . | Jβ† . | IFNγ+, wk 9‡ . | IFNγ+, wk 53‡ . | CD28-, wk 36‡ . |
|---|---|---|---|---|
| 3 | ||||
| CASSPTSNWGGWTEAFF | 1.101 | 1/4 | — | — |
| CASSLFGLASTDTQYF | 2.301 | — | 7/7 | — |
| CASSLGPAGLAKNIQYF | 2.401 | 1/4 | — | — |
| CASRRGKETQYF | 2.501 | 1/4 | — | — |
| CASKAPLGEQYF | 2.701 | 1/4 | — | — |
| 6.1 | ||||
| CASSKVDTEAFF | 1.101 | — | — | 2/23 |
| CASSTRQGGTEAFF | 1.101 | — | — | 9/23 |
| CASSFGGGGTEAFF | 1.101 | 6/60 | — | — |
| CASSIRGGTLNTEAFF | 1.101 | 2/60 | — | — |
| CASSVRTLDGNTEAFF | 1.101 | 11/60 | — | — |
| CASSPRGASIYF§ | 1.301 | — | 3/10 | 6/23 |
| CASSLARSISGNTIYF | 1.301 | 1/60 | — | — |
| CASSLVGWGQPQHF | 1.501 | 13/60 | — | — |
| CASSLGWGDPNQPQHF | 1.501 | — | 7/10 | — |
| CASSPRTGGSPLHF | 1.602 | — | — | 1/23 |
| CASSMTGELRSPLHF | 1.602 | 2/60 | — | — |
| CASSPGLPGANEQFF | 2.101 | 9/60 | — | — |
| CASSVETGELFF | 2.201 | — | — | 5/23 |
| CASSLPIGTSGGQNIQYF | 2.401 | 6/60 | — | — |
| CASSSQSYEQYF | 2.701 | 5/60 | — | — |
| CASSREVAGGTTSEQYF | 2.701 | 1/60 | — | — |
| CASSLEGGVGRNYEQYF | 2.701 | 3/60 | — | — |
| CASSSQSYEQYF | 2.701 | 1/60 | — | — |
| 8 | ||||
| CASCWRDQYF | 1.101 | 1/15 | — | — |
| CASFGREHYF | 1.101 | 1/15 | — | — |
| CASSAQILDTEAFF | 1.101 | 1/15 | — | — |
| CASGSGPQGYYGYTF | 1.201 | 1/15 | — | — |
| CASSPGTSGSRGEQFF | 2.101 | 4/15 | — | — |
| CASRKQETGELFF | 2.201 | —∥ | — | 10/10 |
| CATGGRASQNTGELFF | 2.201 | 1/15 | — | — |
| CASITVPNTGELFF | 2.201 | 1/15 | — | — |
| CASSLEETQYF | 2.701 | 2/15 | — | — |
| CASSEDTNEQYF | 2.701 | 3/15 | — | — |
| 22 | ||||
| CASSPDRGLNYGYTF | 1.201 | — | 4/9 | — |
| CASSERQGLGYGYTF | 1.201 | — | 1/9 | — |
| CASSIREKSSQPQHF | 1.501 | 1/8 | — | — |
| CASRRLEPDQPQHF | 1.501 | — | 2/9 | — |
| CASREGLAGGNEQFF | 2.101 | 3/8 | — | — |
| CASSGLAGGEDEQFF | 2.101 | 1/8 | — | — |
| CASRVGLVVNTDTQYF | 2.301 | 1/8 | — | — |
| CASSEGTGGSGANVLTF | 2.601 | 1/8 | — | — |
| CASGQGAIFGSYEQYF | 2.701 | — | 2/9 | — |
| CASRMDNYEQYF | 2.701 | 1/8 | — | — |
Specific Vβ family and CDR3* . | Jβ† . | IFNγ+, wk 9‡ . | IFNγ+, wk 53‡ . | CD28-, wk 36‡ . |
|---|---|---|---|---|
| 3 | ||||
| CASSPTSNWGGWTEAFF | 1.101 | 1/4 | — | — |
| CASSLFGLASTDTQYF | 2.301 | — | 7/7 | — |
| CASSLGPAGLAKNIQYF | 2.401 | 1/4 | — | — |
| CASRRGKETQYF | 2.501 | 1/4 | — | — |
| CASKAPLGEQYF | 2.701 | 1/4 | — | — |
| 6.1 | ||||
| CASSKVDTEAFF | 1.101 | — | — | 2/23 |
| CASSTRQGGTEAFF | 1.101 | — | — | 9/23 |
| CASSFGGGGTEAFF | 1.101 | 6/60 | — | — |
| CASSIRGGTLNTEAFF | 1.101 | 2/60 | — | — |
| CASSVRTLDGNTEAFF | 1.101 | 11/60 | — | — |
| CASSPRGASIYF§ | 1.301 | — | 3/10 | 6/23 |
| CASSLARSISGNTIYF | 1.301 | 1/60 | — | — |
| CASSLVGWGQPQHF | 1.501 | 13/60 | — | — |
| CASSLGWGDPNQPQHF | 1.501 | — | 7/10 | — |
| CASSPRTGGSPLHF | 1.602 | — | — | 1/23 |
| CASSMTGELRSPLHF | 1.602 | 2/60 | — | — |
| CASSPGLPGANEQFF | 2.101 | 9/60 | — | — |
| CASSVETGELFF | 2.201 | — | — | 5/23 |
| CASSLPIGTSGGQNIQYF | 2.401 | 6/60 | — | — |
| CASSSQSYEQYF | 2.701 | 5/60 | — | — |
| CASSREVAGGTTSEQYF | 2.701 | 1/60 | — | — |
| CASSLEGGVGRNYEQYF | 2.701 | 3/60 | — | — |
| CASSSQSYEQYF | 2.701 | 1/60 | — | — |
| 8 | ||||
| CASCWRDQYF | 1.101 | 1/15 | — | — |
| CASFGREHYF | 1.101 | 1/15 | — | — |
| CASSAQILDTEAFF | 1.101 | 1/15 | — | — |
| CASGSGPQGYYGYTF | 1.201 | 1/15 | — | — |
| CASSPGTSGSRGEQFF | 2.101 | 4/15 | — | — |
| CASRKQETGELFF | 2.201 | —∥ | — | 10/10 |
| CATGGRASQNTGELFF | 2.201 | 1/15 | — | — |
| CASITVPNTGELFF | 2.201 | 1/15 | — | — |
| CASSLEETQYF | 2.701 | 2/15 | — | — |
| CASSEDTNEQYF | 2.701 | 3/15 | — | — |
| 22 | ||||
| CASSPDRGLNYGYTF | 1.201 | — | 4/9 | — |
| CASSERQGLGYGYTF | 1.201 | — | 1/9 | — |
| CASSIREKSSQPQHF | 1.501 | 1/8 | — | — |
| CASRRLEPDQPQHF | 1.501 | — | 2/9 | — |
| CASREGLAGGNEQFF | 2.101 | 3/8 | — | — |
| CASSGLAGGEDEQFF | 2.101 | 1/8 | — | — |
| CASRVGLVVNTDTQYF | 2.301 | 1/8 | — | — |
| CASSEGTGGSGANVLTF | 2.601 | 1/8 | — | — |
| CASGQGAIFGSYEQYF | 2.701 | — | 2/9 | — |
| CASRMDNYEQYF | 2.701 | 1/8 | — | — |
— indicates not detected.
Amino acid sequence of the retrieved CDR3 sequence.
Corresponding Jβ.
Frequency in which the specific sequence was found in proportion to the total number of clones analyzed for that specific cell population.
Sequences similar in different cell populations.
Similar sequence found in early CMV-specific cells after specific PCR based on sequence found in late CD4+ CD28- population of patient 1.
Overview of CDR3 sequences of CMV-specific CD4+ T cells from patient 2
Specific Vβ family and CDR3* . | Jβ† . | IFNγ+, wk 9‡ . | IFNγ+, wk 52‡ . | CD28-, wk 52‡ . |
|---|---|---|---|---|
| 8 | ||||
| CASSLRGGPAEAFF | 1.101 | 3/32 | — | — |
| CASSRGRIGTEASF | 1.101 | — | — | 11/26 |
| CASSRGRINTEAFF | 1.101 | — | — | 1/26 |
| CASKRDAEAFF | 1.101 | — | — | 2/26 |
| CASSRGPFYGYTF | 1.201 | — | — | 2/26 |
| CASSRSVGTGIYGYTF | 1.201 | 1/32 | — | — |
| CASSSEPLREKLFF | 1.401 | 1/32 | — | — |
| CASGLVTGFSSPLHF | 1.601 | 12/32 | — | — |
| CASSLARQEEFF | 2.101 | 3/32 | — | — |
| CASSFFLNTGELFF | 2.201 | 4/32 | — | — |
| CASSLRTGTTDTQYF | 2.301 | — | — | 1/26 |
| CASSPRGRTFQYF | 2.401 | 8/32 | — | — |
| CASTRIDTSGANVLTF | 2.601 | — | — | 9/26 |
| 9 | ||||
| CASSQVGGQPQHF | 1.501 | 3/23 | — | — |
| CASSPGGPVGNEQFF | 2.101 | 5/23 | — | — |
| CASSKLAGGRTGELFF | 2.201 | — | — | 1/8 |
| CASSLLDANTQYF | 2.301 | — | — | 7/8 |
| CASSQEAGQYEQYF | 2.701 | 15/23 | — | — |
| 12 | ||||
| CAITPGQGGGDTQYF | 2.301 | 14/16 | — | — |
| CAISPGGTGAEQYF § | 2.701 | 2/16 | — | 5/8 |
| CAIMAGGSCYEQYF | 2.701 | — | — | 1/8 |
| CAIMGGGSSYEQYF | 2.701 | — | — | 1/8 |
| CAISPGGTSAEQYF | 2.701 | — | — | 1/8 |
| 14 | ||||
| CASSLGDTNQPQHF | 1.501 | — | — | 1/9 |
| CASSLGDSNQPQHF | 1.501 | — | — | 6/9 |
| CASSAGQERKPQHF § | 1.501 | 15/20 | — | 2/9 |
| CASSSAGAQGQPQHF | 1.501 | 2/20 | — | — |
| CASSPQGADSPLHF | 1.601 | 3/20 | — | — |
| 22 | ||||
| CASSEDISGNTIYF | 1.301 | — | — | 6/27 |
| CASNTYQGINEKLFF § | 1.401 | 4/26 | 8/17 | 8/27 |
| CASNTVGGDQPQHF | 1.501 | 1/26 | — | |
| CASRKGQRSNSPLHF | 1.601 | — | — | 2/27 |
| CASSTLAGGGNEQFF | 2.101 | 4/26 | — | — |
| CASSEEGSGYQGEQFF | 2.101 | 6/26 | — | — |
| CASSKDYYNEQFF | 2.101 | 4/26 | — | 3/27 |
| CASSVRPGPGAEQFF | 2.101 | — | — | 1/27 |
| CASREDLSGTEQFF | 2.101 | — | — | 1/27 |
| CASNRDHTDTQYF | 2.301 | 4/26 | — | — |
| CAGILDYATDTQYF | 2.301 | — | 1/17 | — |
| CASGREPGIQYF § | 2.401 | 2/26 | 5/17 | — |
| CASGFNQETQYF § | 2.501 | — | 3/17 | 6/27 |
| CASSRDYHNEQYF | 2.701 | 1/26 | — | — |
Specific Vβ family and CDR3* . | Jβ† . | IFNγ+, wk 9‡ . | IFNγ+, wk 52‡ . | CD28-, wk 52‡ . |
|---|---|---|---|---|
| 8 | ||||
| CASSLRGGPAEAFF | 1.101 | 3/32 | — | — |
| CASSRGRIGTEASF | 1.101 | — | — | 11/26 |
| CASSRGRINTEAFF | 1.101 | — | — | 1/26 |
| CASKRDAEAFF | 1.101 | — | — | 2/26 |
| CASSRGPFYGYTF | 1.201 | — | — | 2/26 |
| CASSRSVGTGIYGYTF | 1.201 | 1/32 | — | — |
| CASSSEPLREKLFF | 1.401 | 1/32 | — | — |
| CASGLVTGFSSPLHF | 1.601 | 12/32 | — | — |
| CASSLARQEEFF | 2.101 | 3/32 | — | — |
| CASSFFLNTGELFF | 2.201 | 4/32 | — | — |
| CASSLRTGTTDTQYF | 2.301 | — | — | 1/26 |
| CASSPRGRTFQYF | 2.401 | 8/32 | — | — |
| CASTRIDTSGANVLTF | 2.601 | — | — | 9/26 |
| 9 | ||||
| CASSQVGGQPQHF | 1.501 | 3/23 | — | — |
| CASSPGGPVGNEQFF | 2.101 | 5/23 | — | — |
| CASSKLAGGRTGELFF | 2.201 | — | — | 1/8 |
| CASSLLDANTQYF | 2.301 | — | — | 7/8 |
| CASSQEAGQYEQYF | 2.701 | 15/23 | — | — |
| 12 | ||||
| CAITPGQGGGDTQYF | 2.301 | 14/16 | — | — |
| CAISPGGTGAEQYF § | 2.701 | 2/16 | — | 5/8 |
| CAIMAGGSCYEQYF | 2.701 | — | — | 1/8 |
| CAIMGGGSSYEQYF | 2.701 | — | — | 1/8 |
| CAISPGGTSAEQYF | 2.701 | — | — | 1/8 |
| 14 | ||||
| CASSLGDTNQPQHF | 1.501 | — | — | 1/9 |
| CASSLGDSNQPQHF | 1.501 | — | — | 6/9 |
| CASSAGQERKPQHF § | 1.501 | 15/20 | — | 2/9 |
| CASSSAGAQGQPQHF | 1.501 | 2/20 | — | — |
| CASSPQGADSPLHF | 1.601 | 3/20 | — | — |
| 22 | ||||
| CASSEDISGNTIYF | 1.301 | — | — | 6/27 |
| CASNTYQGINEKLFF § | 1.401 | 4/26 | 8/17 | 8/27 |
| CASNTVGGDQPQHF | 1.501 | 1/26 | — | |
| CASRKGQRSNSPLHF | 1.601 | — | — | 2/27 |
| CASSTLAGGGNEQFF | 2.101 | 4/26 | — | — |
| CASSEEGSGYQGEQFF | 2.101 | 6/26 | — | — |
| CASSKDYYNEQFF | 2.101 | 4/26 | — | 3/27 |
| CASSVRPGPGAEQFF | 2.101 | — | — | 1/27 |
| CASREDLSGTEQFF | 2.101 | — | — | 1/27 |
| CASNRDHTDTQYF | 2.301 | 4/26 | — | — |
| CAGILDYATDTQYF | 2.301 | — | 1/17 | — |
| CASGREPGIQYF § | 2.401 | 2/26 | 5/17 | — |
| CASGFNQETQYF § | 2.501 | — | 3/17 | 6/27 |
| CASSRDYHNEQYF | 2.701 | 1/26 | — | — |
— indicates not detected.
Amino acid sequence of the retrieved CDR3 sequence.
Corresponding Jβ.
Frequency in which the specific sequence was found in proportion to the total number of clones analyzed for that specific cell population.
Sequences similar in different cell populations.
An interesting question is what factors determine which clones remain present, which clones disappear over time, and even which clones arise later during infection. For CMV-specific CD8+ T cells it has been shown elegantly that the avidity for antigen plays a major role in shaping the clonal dominance.29 It is therefore possible that the same will hold true for CD4+ T cells, but since we do not know the specific peptides these cells recognize it is difficult to clarify this now. Other factors that might be involved in determining the dominance of CMV-specific clones are the efficiency of presentation of different peptides by antigen-presenting cells (APCs) and the abundance of the epitopes. In line with the last, it is not unlikely that the epitopes presented to T cells will change from the acute to the persistent phase of infection, which will probably be reflected in the composition of the T-cell population.
Discussion
In this study, we wanted to clarify whether the late emerging cytotoxic CD4+CD28– T cells originated and differentiated from the IFNγ-producing CMV-specific CD4+ T cells found early in primary CMV infection. Altogether our data show that the CMV-specific cytotoxic CD4+ T cells present during the persistence stage of CMV infection have developed through very strong selection from the virus-specific cells early in primary viral infection. After the acute response to CMV infection, the clonal diversity of virus-specific CD4+ T cells contracts and dominant clones can be found that were hardly, if at all, present in the early phase. Also in the late CMV-specific CD4+ T cells, CDR3 lengths can be found that differ from those found in early responding cells. This means that some of the late virus-specific cells did not originate from virus-specific cells present early in infection but developed independently in a later phase. To our knowledge, this is the first study describing the development of virus-specific CD4+ T cells from the initial response to primary infection until the persistence phase of the virus. We conclude that the selection of virus-specific CD4+ T cells in persistent infections does not occur only during the acute phase of the response but continues when the virus has become latent. This leads to a CD4+ memory T-cell population in the persistence phase that is not merely a reflection of the virus-specific cells found early in primary infection.
Prepublished online as Blood First Edition Paper, July 13, 2006; DOI 10.1182/blood-2006-03-006809.
Supported by a Vici grant from the Netherlands Organization for Scientific Research (NWO) to R.A.W.v.L.
E.M.M.v.L. designed research, performed research, analyzed data, and wrote the paper; E.B.M.R. performed research and analyzed data; M.H.M.H. contributed vital new reagents and analytical tools; I.J.M.t.B. designed research and wrote the paper; and R.A.W.v.L. designed research and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Berend Hooibrink (Department of Cell Biology and Histology, Academic Medical Center, Amsterdam, the Netherlands) for sorting the different cell populations, technicians from the Department of Clinical Virology for performing CMV PCRs and CMV serology, and Dr P. Baars for critical reading of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal