Abstract
Acute myeloid leukemia (AML) is a clonal disease characterized by heterogeneous involvement of hematopoietic stem cell/progenitor cell populations. Using FLT3 internal tandem duplication (FLT3/ITD) as a molecular marker, we tested the hypothesis that clinical outcome in AML correlates with disease involvement of CD34+/CD33- precursors. Diagnostic specimens from 24 children with FLT3/ITD-positive AML were sorted by fluorescence-activated cell sorting (FACS), and resultant CD34+/CD33- and CD34+/CD33+ progenitors were analyzed directly and after colony-forming cell (CFC) assay for the presence of FLT3/ITD. FLT3/ITD was present in all CD34+/CD33+ patient samples. In contrast, FLT3/ITD was detected in CD34+/CD33- progenitors in only 19 of 24 samples. A bipotent progenitor was affected in a subset of patients, as evidenced by the presence of FLT3/ITD in both granulocyte-macrophage colony-forming unit (CFU-GM) and erythroid burst-forming unit (BFU-E) colonies. Those patients in whom CD34+/CD33- precursors harbored the FLT3/ITD had worse clinical outcome; actuarial event-free survival (EFS) at 4 years from study entry for those patients with and without FLT3/ITD detection in CD34+/CD33- progenitors was 11% ± 14% versus 100% ± 0%, respectively (P = .002). This study suggests that FLT3/ITD involvement in CD34+/CD33- precursors is heterogeneous and that detection of the mutation in the less-mature progenitor population may be associated with disease resistance.
Introduction
Current evidence indicates that involvement of hematopoietic stem cells/progenitor cells in acute myeloid leukemia (AML) is heterogeneous. Classic studies by Fialkow et al1 using the X chromosome-linked enzyme glucose-6-phosphate dehydrogenase as a marker of clonality demonstrated that some cases of AML originate in a highly immature multipotent precursor cell with differentiative potential for the myeloid, erythroid, and megakaryocytic lineages, whereas in other cases leukemic involvement is limited to the granulocytic pathway. Furthermore, analysis of fluorescence-activated cell sorted diagnostic AML samples demonstrated that colony-forming cells (CFCs) derived from CD34+/CD33- progenitors are largely nonclonal in origin, whereas colonies derived from more mature CD34+/CD33+ precursors are predominantly clonally derived.2 Evidence that disease involvement of CD34+/CD33- progenitors might correlate with response to chemotherapy came from the observation that patients with monosomy 7 AML carrying the cytogenetic abnormality in immature CD34+/CD33- progenitor cells had higher rates of induction failure than patients lacking the mutation in this early progenitor population.3
In the present study we tested whether the clinical heterogeneity seen in AML might reflect differences in disease involvement of an immature CD34+/CD33- progenitor population and whether such heterogeneity has therapeutic implications. Specifically, are patients with high-risk disease more likely to have requisite mutations detected in less-mature CD34+/CD33- hematopoietic progenitors than patients with favorable outcome? We used FLT3 internal tandem duplication (FLT3/ITD) as a disease marker and evaluated for the presence of FLT3/ITD in CD34+/CD33- and CD34+/CD33+ hematopoietic progenitors isolated from patients with FLT3/ITD-positive AML. FLT3/ITD is present in approximately 15% of pediatric and 30% of adult patients with AML, and its presence is associated with poor clinical response.4-11 However, nearly 25% of patients with FLT3/ITD have favorable outcome,6 suggesting that the clinical variation seen may reflect differences in the underlying biology of the mutation. We show here that FLT3/ITD involvement appears to be heterogeneous in a CD34+/CD33- progenitor population and that detection of the mutation in this early precursor population may correlate with disease resistance.
Patients, materials, and methods
Patients and treatment
Pediatric patients with previously identified FLT3/ITD-positive AML and enrollment in Children's Cancer Group (CCG) AML clinical protocols CCG-2891, -2941, and -2961 were candidates for this study. Details of the aforementioned protocols are described in detail elsewhere.12-14 CCG-2961 and its preceding pilot CCG-2941 treated 988 patients with de novoAML. Of this group, 630 patients had diagnostic specimens available for analysis, 77 of which (12%) were found to be FLT3/ITD positive.29 CCG-2891 treated 888 patients, of which 91 patient samples were tested for FLT3/ITD. Prevalence of the mutation in this population was 16.5%.6
Available diagnostic bone marrow (n = 26) or peripheral blood (n = 1) specimens from 27 pediatric patients identified as having de novo FLT3/ITD-positive AML were obtained from the Children's Oncology Group (COG) AML reference laboratory for our study. Three of the 27 specimens either lacked viable cells (n = 1) or the requisite CD34+/CD33- progenitor population (n = 2) necessary for analysis. The remaining 24 specimens were included in our study. This study was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board and the COG Myeloid Disease Biology Committee.
FACS purification of CD34+/CD33- and CD34+/CD33+ progenitors
Cells (1 × 107-5 × 107) were suspended in 10 mL RPMI medium (Invitrogen, Carlsbad, CA), 20% fetal bovine serum (FBS; Gemini Bio-Products, Woodland, CA), and 100 U/mL DNAse (Sigma, St Louis, MO) prior to staining for flow sorting. Diagnostic samples were stained with immunofluorescent antibody and separated using fluorescence-activated cell sorting (FACS) as described.15 All staining was done at concentrations of 107 cells/mL with cells suspended in sterile phosphate-buffered saline supplemented with 2% human AB serum (PBS/HABS; Gemini Bio-Products) to minimize nonspecific binding. For 2-color staining, untreated cells were incubated with anti-CD34-fluorescein isothiocyanate (FITC) and anti-CD33-phycoerythrin (PE; Becton Dickinson, San Jose, CA). Control cells were incubated with (1) mouse control IgG1-FITC and mouse IgG1-PE (Becton Dickinson); (2) mouse control IgG1-FITC and anti-CD33-PE (Becton Dickinson); or (3) anti-CD34-FITC and mouse control IgG1-PE (Becton Dickinson). Control and experimental cells were incubated with the primary antibodies for 20 minutes at 4°C. After addition of propidium iodide (PI; Sigma) to a concentration of 12.5 μg/mL for use as a marker for nonviable cells, cells were further incubated for 5 minutes at 4°C then washed twice with PBS/HABS, suspended in PBS/HABS, and sorted with a Vantage flow cytometer (Becton Dickinson). Cells that stained with PI were not sorted. Cells were considered CD34+ if their FITC fluorescence intensity was greater than 99% of cells stained with FITC control antibody. Expression of CD33 was evaluated on the CD34+ cells. Cells were considered CD33+ if their PE fluorescence intensity was greater than that of 99% of cells stained with PE control antibody. Cells were considered CD33- if their fluorescence intensity was less than that of the top 15% of PE control antibody-stained cells. Sorted CD34+/CD33- and CD34+/CD33+ cell populations were collected in Iscoves modified Dulbecco medium (IMDM; Invitrogen).
Culture of CD34+/CD33- and CD34+/CD33+ progenitors
After FACS isolation of CD34+/CD33- and CD34+/CD33+ progenitors, an aliquot of approximately 2000 freshly sorted cells from each population was set aside, in most instances, for direct FLT3/ITD analysis. The remaining cells were placed in methylcellulose culture containing 40% methylcellulose (Stem Cell Technologies, Vancouver, BC, Canada), 20% FBS, 20% bovine serum albumin (Intergen, Purchase, NY) in 10% IMDM, 10% IMDM, 0.1 M 2-mercaptoethanol (Sigma), 100 ng/mL each of granulocyte colony-stimulating factor, granulocyte-monocyte colony-stimulating factor, stem cell factor, interleukin-3, interleukin-6 (PeproTech, Rocky Hill, NJ), and 2 U/mL erythropoietin (Amgen, Thousand Oaks, CA). Culture plates were incubated for 14 days at 37°C in a 5% CO2 humidified atmosphere. Granulocyte-macrophage colony-forming unit (CFU-GM) colonies of at least 50 cells were plucked with a sterile pipette tip and placed in 20 μL diethylpyrocarbonate-treated water (Ambion, Austin, TX).16 When total colony growth per patient cell population (eg, colonies derived from CD34+/CD33- cells or CD34+/CD33+ cells) exceeded 75 colonies, each culture plate was divided into quartiles, and one quartile was randomly selected for colony harvest. Similar techniques were used to harvest erythroid burst-forming unit (BFU-E) colonies.
FLT3/ITD analysis of freshly sorted CD34+/CD33- and CD34+/CD33+ cells and resultant colonies
Pediatric patient specimens were screened for FLT3/ITD mutations, as described previously, prior to inclusion in our study.4,6,10,29 Following FACS, freshly sorted CD34+/CD33- and CD34+/CD33+ cells as well as resultant CFU-GM and BFU-E colonies were analyzed for the presence of the patient-specific FLT3/ITD using similar techniques.4,6,10,29 Whole-genome amplification was conducted, based on described techniques,17 if polymerase chain reaction (PCR) amplification of the FLT3 gene was insufficient. PCR products were resolved by 2% agarose gel electrophoresis and Genescan analysis. Colonies that failed to yield wild-type (WT) FLT3 or FLT3/ITD signal despite multiple attempts at whole-genome amplification or genomic PCR amplification were excluded from analysis.
FLT3/ITD analysis of freshly sorted lymphocytes
Lymphocytes were isolated from a subset of patients (n = 12) by flow cytometry after incubation of diagnostic samples with murine anti-CD3 and anti-CD20 antibodies, followed by FITC-conjugated goat antimurine antibody (Kirkegaard and Perry Laboratories, Gaithersburg, MD) as described.6,18 DNA was extracted from the sorted cells using the Puregene protocol (Gentra Systems, Minneapolis, MN) and subjected to FLT3/ITD analysis as referenced in “FLT3/ITD analysis of freshly sorted CD34+/CD33- and CD34+/CD33+ cells and resultant colonies.”
Statistical methods
Data from CCG-2891, -2941 and -2961 were analyzed through February 2004, April 2005, and June 2005, respectively. Pearson chi-square test was used to test for differences in the distribution of categoric variables. Fisher exact test was used when data were sparse. The Mann-Whitney test was used to analyze differences of medians. The Kaplan-Meier method was used for nonparametric survival curve analyses.19 A patient was defined as being in complete remission (CR) if morphologic evaluation of the bone marrow at the end of induction demonstrated no more than 5% blasts, absolute neutrophil count (ANC) of 1 × 109/L (1000/μL) or greater, and platelet count of 50 × 109/L (50 000/μL) or greater. Partial remission (PR) was defined as no more than 5% blasts and ANC less than 1 × 109/L (1000/μL) or platelet count less than 50 × 109/L (50 000/μL). Overall survival (OS) was defined as time from study entry until death. Event-free survival (EFS) was defined as time from study entry until remission failure, relapse, or death. Patients lost to follow-up were censored at their date of last known contact or at a cutoff of 6 months prior to the study data creation date to compensate for the tendency of deaths and relapses to be reported sooner than ongoing follow-up. Differences between Kaplan-Meier survival curves were tested using the log-rank statistic.20 Confidence intervals for survival estimates were calculated using Greenwood estimate of the standard error.
Results
Patients and treatment
Twenty-four patients with FLT3/ITD-positive AML were selected as described under “Patients and treatment.” Twenty-two of the total 24 patients were registered in CCG protocol 2961 (n = 21) or its pilot CCG-2941 (n = 1) and the remaining 2 patients were enrolled in CCG-2891. To determine whether our study population was representative of the overall FLT3/ITD population, we compared laboratory and clinical characteristics for the 22 study patients enrolled on CCG-2941 and -2961 to those FLT3/ITD-positive patients enrolled on CCG-2941 and -2961 but not included in our study (n = 55). There was no significant difference in white blood cells (WBCs) at presentation (P = .207), sex (P > .99), OS (P = .832), or relapse-free survival (P = .767) between the 2 groups. Patients in our study were found to be significantly older (median age, 14.3 years versus 11.3 years; P = .011) and to have lower percentage of bone marrow blasts (median, 74.5% versus 90%, P = .005) than the FLT3/ITD-positive population at large.
Selection of CD34+/CD33- and CD34+/CD33+ progenitors and resultant CFU-GM growth
Nucleated cells (1 × 107-5 × 107) from the 24 diagnostic patient samples included in our study were initially stained and sorted by FACS for CD34+/CD33- and CD34+/CD33+ progenitors. Median percentage of viability was 96% with a range of 79% to 99%. Median CD34+/CD33- progenitor cell count was 3.14 × 104 cells per patient sample with a range of 0.05 × 104 to 150 × 104 cells per sample. CD34+/CD33+ progenitor cell counts were significantly higher (P < .001), with a median of 34.5 × 104 cells per patient sample (range, 2.4 × 104-580 × 104 cells per sample). CD34+/CD33- and CD34+/CD33+ progenitors were plated on semisolid medium at 1 × 103 or 1 × 104 cells/plate. Resultant CFU-GM colonies were counted and harvested after 2 weeks of growth. Samples from 23 of 24 patients yielded CFU-GM colonies derived from CD34+/CD33- cells; growth varied between 3 and 273 colonies (median, 21 colonies per patient sample), with a corresponding colony growth of 0.001% to 2.35% (median, 0.1%) of total CD34+/CD33- cells plated. CFU-GM colonies derived from CD34+/CD33+ cells were seen in 20 of 24 patient samples, with a range of 1 to 238 colonies per patient sample (median, 9.5 colonies per patient sample) and a corresponding colony growth of 0.0001% to 0.1% (median, 0.002%) of total CD34+/CD33+ cells plated. This growth was significantly less than that seen for the CD34+/CD33- population (P < .001).
FLT3/ITD analysis of CD34+/CD33- and CD34+/CD33+ progenitors and resultant CFU-GM
FACS-purified CD34+/CD33- progenitors and their resultant colonies were tested for the presence of FLT3/ITD. A particular population was defined as being FLT3/ITD-positive if the patient-specific ITD was detected in freshly sorted CD34+/CD33- cells or in CFU-GM colonies derived from that particular population if FACS-purified cells were not available. For a population to be considered FLT3/ITD negative, freshly sorted cells were to be negative or, if FACS-purified cells were unavailable, all CFU-GM colonies tested negative (minimum of 10 colonies required). Of the 24 patient specimens tested, 19 (79%) of 24 were found to be FLT3/ITD positive in CD34+/CD33- precursors; all 19 demonstrated the mutation in freshly sorted CD34+/CD33- cells. The remaining 5 (21%) of 24 patients lacked FLT3/ITD detection in CD34+/CD33- precursors. This was demonstrated in 4 of 5 cases by direct analysis of CD34+/CD33- progenitors. A fifth patient lacked FACS-purified cells for analysis but demonstrated WT FLT3 in 11 CFU-GM colonies analyzed (Table 1).
Disease characteristics and FLT3/ITD detection in CD34+/CD33- and CD34+/CD33+ progenitors and their resultant CFU-GM and BFU-E colonies
. | Disease characteristics . | . | . | FLT3 status in CD34+/CD33- cells/colonies . | . | . | FLT3 status in CD34+/CD33+ cells/colonies . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | BM blasts, % . | CD34+/CD33- cell count, × 104 (% total) . | Allelic ratio . | Freshly sorted cells . | CFU-GM, n (%) . | BFU-E, n (%) . | Freshly sorted cells . | CFU-GM, n (%) . | BFU-E, n (%) . | ||||||
| 1* | 96 | 0.05 (0.005) | 0.2 | NA | 0/11 (0) | NG | ITD | NG | NG | ||||||
| 2* | 91 | 0.2 (0.01) | 0.19 | WT | 0/8 (0) | 0/2 (0) | ITD | 0/8 (0) | 0/4 (0) | ||||||
| 3* | 36 | 32 (4) | 0.12 | WT | 0/51 (0) | 0/16 (0) | ITD | 0/1 (0) | 0/1 (0) | ||||||
| 4* | 60 | 0.1 (0.01) | 0.29 | WT | 0/17 (0) | 0/2 (0) | ITD | NG | NG | ||||||
| 5* | 81 | 0.3 (0.006) | 0.52 | WT | 0/30 (0) | 0/12 (0) | ITD | 3/10 (30) | NG | ||||||
| 6 | 44 | 28 (1) | 0.56 | ITD | NG | NG | ITD | NG | NG | ||||||
| 7 | 68 | 0.08 (0.003) | 1.6 | ITD | 0/6 (0) | 0/5 (0) | ITD | 0/1 (0) | NG | ||||||
| 8 | 92 | 34 (2) | 0.8 | ITD | 0/5 (0) | NG | ITD | 0/1 (0) | 0/4 (0) | ||||||
| 9 | 41 | 15 (1) | 0.73 | ITD | 0/3 (0) | NG | ITD | NG | NG | ||||||
| 10 | 85 | 0.3 (0.03) | 0.76 | ITD | 1/47 (2) | 1/13 (8) | ITD | 0/3 (0) | 1/1 (100) | ||||||
| 11 | 59 | 0.08 (0.01) | 0.7 | ITD | 1/16 (6) | 0/4 (0) | ITD | 0/3 (0) | 0/1 (0) | ||||||
| 12 | 90 | 150 (16) | 0.48 | ITD | 1/17 (6) | 0/20 (0) | ITD | 0/1 (0) | 0/2 (0) | ||||||
| 13 | 85 | 1 (0.02) | 0.58 | ITD | 4/33 (12) | 0/21 (0) | NA | 9/10 (90) | 0/2 (0) | ||||||
| 14 | 66 | 7 (0.4) | 0.62 | ITD | 5/35 (14) | 0/12 (0) | ITD | 7/8 (88) | NG | ||||||
| 15 | 71 | 82 (8) | 0.18 | ITD | 3/19 (16) | 0/15 (0) | ITD | 3/3 (100) | 0/7 (0) | ||||||
| 16 | 48 | 10 (1) | 0.13 | ITD | 2/9 (22) | 0/5 (0) | ITD | 0/2 (0) | NG | ||||||
| 17 | 78 | 0.8 (0.08) | 2 | ITD | 15/56 (27) | 1/21 (5) | NA | 2/11 (18) | 1/1 (100) | ||||||
| 18 | 74 | 2 (0.2) | 0.53 | ITD | 11/19 (58) | 0/5 (0) | ITD | 2/6 (33) | NG | ||||||
| 19 | 70 | 8 (0.3) | 0.75 | ITD | 28/42 (67) | 0/10 (0) | NA | 12/15 (80) | NG | ||||||
| 20 | 88 | 3 (0.1) | 0.4 | ITD | 28/37 (76) | 0/9 (0) | ITD | 39/46 (85) | NG | ||||||
| 21 | 61 | 3 (0.4) | 0.6 | ITD | 7/9 (78) | NG | ITD | 12/12 (100) | NG | ||||||
| 22 | 89 | 17 (0.4) | 2.8 | ITD | 18/22 (82) | 9/9 (100) | ITD | 7/7 (100) | 7/7 (100) | ||||||
| 23 | 75 | 3 (0.3) | 0.62 | ITD | 19/22 (86) | 3/3 (100) | ITD | 4/4 (100) | NG | ||||||
| 24 | 93 | 2 (0.1) | 0.82 | ITD | 3/3 (100) | 1/1 (100) | ITD | 3/3 (100) | 4/4 (100) | ||||||
. | Disease characteristics . | . | . | FLT3 status in CD34+/CD33- cells/colonies . | . | . | FLT3 status in CD34+/CD33+ cells/colonies . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | BM blasts, % . | CD34+/CD33- cell count, × 104 (% total) . | Allelic ratio . | Freshly sorted cells . | CFU-GM, n (%) . | BFU-E, n (%) . | Freshly sorted cells . | CFU-GM, n (%) . | BFU-E, n (%) . | ||||||
| 1* | 96 | 0.05 (0.005) | 0.2 | NA | 0/11 (0) | NG | ITD | NG | NG | ||||||
| 2* | 91 | 0.2 (0.01) | 0.19 | WT | 0/8 (0) | 0/2 (0) | ITD | 0/8 (0) | 0/4 (0) | ||||||
| 3* | 36 | 32 (4) | 0.12 | WT | 0/51 (0) | 0/16 (0) | ITD | 0/1 (0) | 0/1 (0) | ||||||
| 4* | 60 | 0.1 (0.01) | 0.29 | WT | 0/17 (0) | 0/2 (0) | ITD | NG | NG | ||||||
| 5* | 81 | 0.3 (0.006) | 0.52 | WT | 0/30 (0) | 0/12 (0) | ITD | 3/10 (30) | NG | ||||||
| 6 | 44 | 28 (1) | 0.56 | ITD | NG | NG | ITD | NG | NG | ||||||
| 7 | 68 | 0.08 (0.003) | 1.6 | ITD | 0/6 (0) | 0/5 (0) | ITD | 0/1 (0) | NG | ||||||
| 8 | 92 | 34 (2) | 0.8 | ITD | 0/5 (0) | NG | ITD | 0/1 (0) | 0/4 (0) | ||||||
| 9 | 41 | 15 (1) | 0.73 | ITD | 0/3 (0) | NG | ITD | NG | NG | ||||||
| 10 | 85 | 0.3 (0.03) | 0.76 | ITD | 1/47 (2) | 1/13 (8) | ITD | 0/3 (0) | 1/1 (100) | ||||||
| 11 | 59 | 0.08 (0.01) | 0.7 | ITD | 1/16 (6) | 0/4 (0) | ITD | 0/3 (0) | 0/1 (0) | ||||||
| 12 | 90 | 150 (16) | 0.48 | ITD | 1/17 (6) | 0/20 (0) | ITD | 0/1 (0) | 0/2 (0) | ||||||
| 13 | 85 | 1 (0.02) | 0.58 | ITD | 4/33 (12) | 0/21 (0) | NA | 9/10 (90) | 0/2 (0) | ||||||
| 14 | 66 | 7 (0.4) | 0.62 | ITD | 5/35 (14) | 0/12 (0) | ITD | 7/8 (88) | NG | ||||||
| 15 | 71 | 82 (8) | 0.18 | ITD | 3/19 (16) | 0/15 (0) | ITD | 3/3 (100) | 0/7 (0) | ||||||
| 16 | 48 | 10 (1) | 0.13 | ITD | 2/9 (22) | 0/5 (0) | ITD | 0/2 (0) | NG | ||||||
| 17 | 78 | 0.8 (0.08) | 2 | ITD | 15/56 (27) | 1/21 (5) | NA | 2/11 (18) | 1/1 (100) | ||||||
| 18 | 74 | 2 (0.2) | 0.53 | ITD | 11/19 (58) | 0/5 (0) | ITD | 2/6 (33) | NG | ||||||
| 19 | 70 | 8 (0.3) | 0.75 | ITD | 28/42 (67) | 0/10 (0) | NA | 12/15 (80) | NG | ||||||
| 20 | 88 | 3 (0.1) | 0.4 | ITD | 28/37 (76) | 0/9 (0) | ITD | 39/46 (85) | NG | ||||||
| 21 | 61 | 3 (0.4) | 0.6 | ITD | 7/9 (78) | NG | ITD | 12/12 (100) | NG | ||||||
| 22 | 89 | 17 (0.4) | 2.8 | ITD | 18/22 (82) | 9/9 (100) | ITD | 7/7 (100) | 7/7 (100) | ||||||
| 23 | 75 | 3 (0.3) | 0.62 | ITD | 19/22 (86) | 3/3 (100) | ITD | 4/4 (100) | NG | ||||||
| 24 | 93 | 2 (0.1) | 0.82 | ITD | 3/3 (100) | 1/1 (100) | ITD | 3/3 (100) | 4/4 (100) | ||||||
BM indicates bone marrow; NA, not available; NG, no growth; ITD, FLT3/ITD-positive; WT, FLT3 wild type
Patients lacking FLT3/ITD detection in CD34+/CD33- progenitors
CFU-GM colonies derived from CD34+/CD33- progenitors were tested to define the extent of the mutation's early progenitor involvement. Of the 19 patients identified as having FLT3/ITD involvement of CD34+/CD33- progenitors, FLT3/ITD was detected in CFU-GM colonies of 15 of 19 patients. The remaining 4 patient specimens either lacked CFU-GM growth (n = 1) or had fewer than 10 colonies available for analysis (n = 3), which may have precluded detection of the mutation. Of the total 18 patients with evaluable CFU-GM colonies, FLT3/ITD involvement ranged from 0% to 100% of colonies analyzed per patient sample with a median of 19% of colonies affected per patient sample. In 7 patients, greater than 50% of colonies tested were FLT3/ITD positive (Table 1).
Similar criteria were used for evaluation of the CD34+/CD33+ population. Unlike the heterogeneity seen previously, FLT3/ITD was uniformly detected in this more mature precursor population. Twenty-one of 24 patient specimens had FACS-purified CD34+/CD33+ cells for analysis, and all 21 samples were FLT3/ITD positive. The remaining 3 of 24 patients who lacked freshly sorted cells for analysis had CFU-GM colonies available and were deemed FLT3/ITD positive based on their analysis. In general, CD34+/CD33+ progenitors had limited CFU-GM growth whereby 18 of 24 patient samples either had no growth (n = 4) or growth fewer than 10 CFU-GM colonies (n = 14) which precluded complete assessment of this population by CFC assay. Of the total 20 patients with evaluable CFU-GM, FLT3/ITD involvement ranged from 0% to 100% of colonies analyzed per patient sample (median, 31.5%) (Table 1).
FLT3/ITD analysis of BFU-E colonies and sorted lymphocytes
BFU-E colonies and lymphocytes were assessed to determine whether FLT3/ITD mutations affect multipotent or bipotent progenitors. Nineteen of 24 patients in our study had CD34+/CD33--derived BFU-E colonies available for analysis (median, 9 BFU-E colonies per patient sample; range, 1-21 colonies). Five of the 19 evaluable patients were found to have FLT3/ITD-positive CD34+/CD33--derived BFU-E colonies. Of these 5, the median percentage of FLT3/ITD involvement of CD34+/CD33- BFU-E colonies was 100% (range, 5%-100%). Presence of FLT3/ITD in erythroid progenitors correlated with FLT3/ITD detection in CD34+/CD33- freshly sorted cells and resultant CFU-GM colonies in every case. This finding suggested that, at least in some patients, FLT3/ITD evolved in a bipotent hematopoietic progenitor. Because only 9 of 19 patients had sufficient (10 or more) BFU-E colonies for analysis, BFU-E involvement cannot be ruled out in other patient samples (Table 1). Eleven of 24 patients had CD34+/CD33+-derived BFU-E colonies available for analysis (median, 2 BFU-E colonies per patient sample; range, 1-7 BFU-E colonies). FLT3/ITD was detected in 4 of 11 patient samples; all 4 had previously been identified as having CD34+/CD33--derived BFU-E involvement (Table 1).
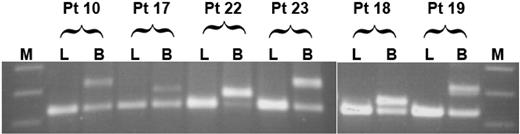
We further inquired whether FLT3/ITD may have evolved in a multipotent progenitor cell. We reasoned that, if this were the case, then one would expect the mutation to be present in lymphocytes as well as myeloid blasts. To test this possibility, we analyzed lymphocytes from a subset of 12 of 19 study patients that had demonstrated FLT3/ITD involvement of CD34+/CD33- progenitors, including 4 of 5 patient specimens that had demonstrated bipotent FLT3/ITD involvement. FACS-sorted lymphocytes were subjected to FLT3/ITD analysis by PCR; lymphocyte fractions from all patients were negative for FLT3/ITD, arguing that, in our patient population, FLT3/ITD involvement appears restricted to the myeloid lineage but may arise in a progenitor cell with bipotent potential (Figure 1).
FLT3/ITD detection in a CD34+/CD33- progenitor population correlates with clinical outcome
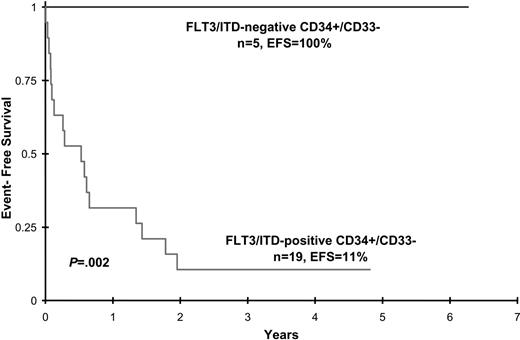
Disease characteristics and clinical outcome data were analyzed in patients with FLT3/ITD involvement of CD34+/CD33- progenitors and compared with that of FLT3/ITD-positive patients with AML who lacked detection of the mutation in this early precursor population. Patient age, sex, diagnostic WBC count, diagnostic bone marrow blast percentage, and CD34+/CD33- cell count did not differ between the 2 groups, although the latter attribute approached statistical significance (Table 2). Postremission therapy (chemotherapy versus hematopoietic cell transplantation) was also comparable (Table 2). All 5 patients without CD34+/CD33- involvement achieved CR at end of induction and remain in long-term remission (median, 3 years; range 2.2-6 years after induction). In contrast, of the 19 patients with FLT3/ITD involvement of CD34+/CD33- progenitors, 5 (26%) died of toxicity during induction therapy and 4 (21%) had no response to treatment. The remaining 10 patients initially achieved CR (n = 9) or PR (n = 1), but 8 of 10 subsequently relapsed. Median time to relapse from remission was 250.5 days (range, 92-616 days). The 2 patients who remain alive and disease free (patients 9 and 23) received allogeneic stem cell transplants. Actuarial EFS at 4 years from study entry for patients with and without FLT3/ITD detection in CD34+/CD33- precursors is 11% ± 14% versus 100% ± 0%, respectively (P = .002) (Figure 2). Corresponding OS for patients with and without FLT3/ITD detection is 18% ± 19% versus 100% ± 0%, respectively, (P = .007).
Clinical characteristics of patients with and without detection of the FLT3/ITD mutation in CD34+/CD33- progenitor cells
. | Without FLT3/ITD . | With FLT3/ITD . | P . |
|---|---|---|---|
| Total, n (%) | 5 (21) | 19 (79) | |
| Median age, y (range) | 16.3 (8.9-19.8) | 14.3 (3.0-18.7) | .57 |
| Sex | |||
| Male, n (%) | 2 (40) | 12 (63) | .615 |
| Female, n (%) | 3 (60) | 7 (37) | |
| Median WBC count, × 109/L (range) | 64.1 (5.6-250.3) | 42.1 (1.6-860) | .943 |
| Median marrow blast, % (range) | 81 (36-96) | 74 (44-93) | .776 |
| Median CD34+/CD33- cell count, × 104 (range) | 0.2 (0.05-32) | 3.3 (0.08-150) | .075 |
| Allelic ratio | |||
| No more than 0.4, n (%) | 4 (67) | 3 (16) | .015 |
| More than 0.4, n (%) | 1 (33) | 16 (84) | |
| Postinduction therapy | |||
| Allo BMT, n (%) | 2 (40) | 3 (30) | .99 |
| Chemo, n (%) | 3 (60) | 7 (70) | |
| Induction outcome | |||
| CR/PR, n (%) | 5/0 (100) | 9/1 (53) | .118 |
| Fail, n (%) | 0 (0) | 4 (21) | .544 |
| Death, n (%) | 0 (0) | 5 (26) | .544 |
. | Without FLT3/ITD . | With FLT3/ITD . | P . |
|---|---|---|---|
| Total, n (%) | 5 (21) | 19 (79) | |
| Median age, y (range) | 16.3 (8.9-19.8) | 14.3 (3.0-18.7) | .57 |
| Sex | |||
| Male, n (%) | 2 (40) | 12 (63) | .615 |
| Female, n (%) | 3 (60) | 7 (37) | |
| Median WBC count, × 109/L (range) | 64.1 (5.6-250.3) | 42.1 (1.6-860) | .943 |
| Median marrow blast, % (range) | 81 (36-96) | 74 (44-93) | .776 |
| Median CD34+/CD33- cell count, × 104 (range) | 0.2 (0.05-32) | 3.3 (0.08-150) | .075 |
| Allelic ratio | |||
| No more than 0.4, n (%) | 4 (67) | 3 (16) | .015 |
| More than 0.4, n (%) | 1 (33) | 16 (84) | |
| Postinduction therapy | |||
| Allo BMT, n (%) | 2 (40) | 3 (30) | .99 |
| Chemo, n (%) | 3 (60) | 7 (70) | |
| Induction outcome | |||
| CR/PR, n (%) | 5/0 (100) | 9/1 (53) | .118 |
| Fail, n (%) | 0 (0) | 4 (21) | .544 |
| Death, n (%) | 0 (0) | 5 (26) | .544 |
Allo indicates allogeneic hematopoietic cell transplantation; BMT, bone marrow transplantation; chemo, chemotherapy
FLT3/ITD detection in a CD34+/CD33- progenitor population correlates with high FLT3/ITD allelic ratio
We previously demonstrated the clinical significance of FLT3/ITD allelic ratio (ITD-AR)10 and defined an ITD-AR of greater than 0.4 as the clinically significant ITD-AR threshold that defines relapse risk in pediatric patients positive for FLT3/ITD.29 Of the 24 patients in our study, 17 had ITD-ARs greater than 0.4. There was a significant association between FLT3/ITD detection in CD34+/CD33- progenitor cells and high ITD-AR, as 16 (84%) of 19 patient samples with early progenitor involvement had ITD-ARs greater than 0.4 compared with 1 (20%) of 5 patients who lacked evidence of the mutation in CD34+/CD33- precursors (P = .015) (Table 2).
Discussion
Our study suggests that pediatric patients with FLT3/ITD-positive AML have heterogeneous disease involvement in CD34+/CD33- progenitors, whereas more mature CD34+/CD33+ progenitors are universally affected. Detection of FLT3/ITD in both CFU-GM and BFU-E colonies in a subset of patients implies that the mutation arises, in at least some instances, in a bipotent myeloid progenitor. Moreover, detection of the mutation in CD34+/CD33- progenitors may correlate with disease resistance. These data suggest that the absolute presence of FLT3/ITD may be an unreliable predictor of clinical response. Instead, efforts to define the degree to which the mutation affects immature hematopoietic precursors may improve accuracy of clinical prognostication and ultimately ensure that efficacious therapeutic regimens are used.
Extent of lineage involvement of FLT3/ITD. Flow-sorted lymphocytes (L) and myeloblasts (B) from 12 patients positive for FLT3/ITD were tested for the presence of FLT3/ITD mutations; results from 6 patients are displayed. The first 4 patients (patients 10, 17, 22, and 23) had FLT3/ITD involvement of both CD34+/CD33--derived CFU-GM and BFU-E colonies. Patients 18 and 19 had FLT3/ITD involvement of CD34+/CD33- CFU-GM colonies only. M indicates molecular weight marker.
Extent of lineage involvement of FLT3/ITD. Flow-sorted lymphocytes (L) and myeloblasts (B) from 12 patients positive for FLT3/ITD were tested for the presence of FLT3/ITD mutations; results from 6 patients are displayed. The first 4 patients (patients 10, 17, 22, and 23) had FLT3/ITD involvement of both CD34+/CD33--derived CFU-GM and BFU-E colonies. Patients 18 and 19 had FLT3/ITD involvement of CD34+/CD33- CFU-GM colonies only. M indicates molecular weight marker.
Although clinical outcome in AML has improved considerably over the past decades, the risk of relapse remains high.21 Disease-free survival may improve if the hematopoietic cell targeted for leukemic transformation is better defined, because recurrent/refractory disease may reflect an inability of current therapies to eradicate the leukemic cell responsible for ongoing proliferation of the leukemic clone.22 The origin of this cell, defined as the leukemic stem cell (LSC), remains in question. Because the LSC and a normal hematopoietic stem cell have phenotypic and functional similarities, it is possible that the LSC arises from the latter.22,23 However, pediatric AML is a heterogeneous disease that appears to be myeloid restricted in some patients, raising the possibility that an inciting mutational event arises in a stem cell but requires additional mutational events in more mature progenitors for frank leukemic transformation to occur.1,2,24 Alternatively, disease may evolve from a committed myeloid progenitor, as is thought to be the case with acute promyelocytic leukemia (APL).3,25,26 Ultimately, the maturational stage at which frank leukemic transformation occurs may dictate clinical response. For example, APL, a disease that is limited to a mature precursor population, has excellent clinical outcome.3,26,27 Moreover, studies in monosomy 7 (-7) AML demonstrate that response to induction chemotherapy correlates with extent of early progenitor cell involvement, with patients demonstrating the cytogenetic abnormality in less-mature CD34+/CD33- hematopoietic progenitors having worse response to induction therapy than patients in which -7 is limited to a more differentiated population.3 Our data further support this concept, suggesting that FLT3/ITD detection is heterogeneous in CD34+/CD33- myeloid progenitors and that disease involvement of this precursor population may correlate with poor clinical outcome.
Event-free survival for patients with and without FLT3/ITD detection in CD34+/CD33- progenitor cells.
Event-free survival for patients with and without FLT3/ITD detection in CD34+/CD33- progenitor cells.
Our findings clearly show dominance of a CD34+/CD33- FLT3/ITD-positive clone in 19 of 24 patient specimens analyzed. However, for the remaining 5 patients in which CD34+/CD33- cells were not found to harbor the mutation, our studies do not rule out the possibility of rare CD34+/CD33- FLT3/ITD-positive cells that do not undergo uncontrolled proliferation and attain clonal dominance until subsequent maturation into CD34+/CD33+ precursors. In addition, these studies do not determine whether FLT3/ITD-negative CD34+/CD33- cells isolated from these 5 patient specimens are leukemic in nature. We were able to demonstrate in one case that the progenitors analyzed indeed harbored a leukemia-associated fusion transcript (data not shown). Comprehensive studies that demonstrate evolution of the FLT3 mutation in FLT3/ITD-negative progenitors containing other leukemia-associated abnormalities would validate our findings and support the hypothesis that maturation-related cooperating mutations contribute to myeloid leukemogenesis.
FLT3/ITD detection in CD34+/CD33- progenitors was also correlated with allelic ratio, because previous studies have demonstrated that patients with high ITD-AR are at significantly greater risk of relapse.7,8,10,29 Here, we demonstrate that there is a significant association between FLT3/ITD detection in CD34+/CD33- progenitors and high ITD-AR. This raises the possibility that in patients with FLT3/ITD-positive AML, allelic imbalance may be a secondary process and that the acquisition of FLT3/ITD in an early precursor population may increase the probability that this subsequent event occurs.
Once our findings are extended and validated in other patient populations and with other disease markers, we may have a clinical paradigm that will aid treatment allocation and clinical prognostication. Specifically, patients with disease limited to CD34+/CD33+ progenitors have a better chance of responding to conventional chemotherapy and may be spared the morbidity associated with myeloablative regimens. CD33-targeted therapy with gemtuzumab ozogamicin (GO) would be an attractive approach for these patients whereby, similar to patients with APL,28 therapy with GO may effectively eradicate disease. In contrast, the majority of patients that demonstrate disease involvement of less mature CD34+/CD33- progenitors might be expected to do poorly with conventional therapy and not respond to GO, requiring alternative treatment approaches to target this less-mature leukemic progenitor. Traditionally, patients with high-risk disease have been referred for hematopoietic cell transplantation. Indeed, in our study, the only 2 patients with CD34+/CD33- disease involvement who remain alive and relapse free received allogeneic stem cell transplants, suggesting that a myeloablative approach may eradicate immature leukemic cells responsible for proliferation of the leukemic clone. Ultimately, by making a paradigm shift from treatment strategies defined by the presence of a specific cytogenetic or molecular abnormality to approaches that consider the extent to which the abnormality affects immature hematopoietic progenitors, we might improve clinical outcomes and decrease morbidity associated with unnecessary and/or ineffectual treatment.
Our study provides insight into leukemogenesis, suggesting that differential involvement of requisite molecular mutations affects clinical outcome. Additional prospective studies are ongoing to determine whether direct analysis of sorted CD34+/CD33- cells from AML diagnostic samples aids clinical prognostication and assignment to efficacious therapeutic regimens.
Prepublished online as Blood First Edition Paper, June 29, 2006; DOI 10.1182/blood-2006-04-012260.
Supported by the National Institutes of Health (grants 5T32CA009351-27 [J.A.P. and I.D.B.], R01 CA092316 [I.D.B.], U10 CA98543 [I.D.B.], U24 CA114766 [I.D.B.], R21 CA102624 [S.M.], and R01 CA114563 [S.M.]) and by the Leukemia and Lymphoma Society (Specialized Center of Research [SCOR] 7040-03 [I.D.B.]).
J.A.P. designed and performed research, analyzed the data, and wrote the paper; T.A.A. and R.B.G. analyzed the data and wrote the paper; W.G.W. was principal investigator for Children's Cancer Group (CCG) protocol 2891; B.J.L. was principal investigator for CCG protocols 2941 and 2961; D.A.S. and J.P.R. designed the research; I.D.B. designed the research, analyzed the data, and wrote the paper; and S.M. designed and performed the research, analyzed the data, and wrote the paper.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dave Flowers, Carolyn Brashem-Stein, Cristina Galer, and Kristin Miller for technical assistance and Mariko Kawabori and the COG AML Reference Laboratory for providing diagnostic specimens.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal