Abstract
Waldenström macroglobulinemia (WM) is a B-cell malignancy characterized by the ability of the B-cell clone to differentiate into plasma cells. Although the clinical syndrome and the pathologic characteristics are well defined, little is known about its biology and controversy still exists regarding its cell of origin. In this gene-expression study, we compared the transcription profiles of WM with those of other malignant B cells including (chronic lymphocytic leukemia [CLL] and multiple myeloma [MM]) as well as normal cells (peripheral-blood B cells and bone marrow plasma cells). We found that WM has a homogenous gene expression regardless of 6q deletion status and clusters with CLL and normal B cells on unsupervised clustering with very similar expression profiles. Only a small gene set has expression profiles unique to WM compared to CLL and MM. The most significantly up-regulated gene is IL6 and the most significantly associated pathway for this set of genes is MAPK signaling. Thus, IL6 and its downstream signaling may be of biologic importance in WM. Further elucidation of the role of IL-6 in WM is warranted as this may offer a potential therapeutic avenue.
Introduction
Waldenström's macroglobulinemia (WM) is a clinicopathologically distinct B-cell malignancy characterized by intramedullary monoclonal expansion of predominantly small B lymphocytes with variable plasmacytoid differentiation in the bone marrow (BM), associated with serum IgM paraprotein. Histologically, this represents BM involvement by lymphoplasmacytic lymphoma.1
Although the clinical disease spectrum has been well established, little is known about its biology. The cell of origin is thought to be an unusual memory B cell that has undergone somatic hypermutation in the absence of antigenic selection with failure in the class switch recombination.2,3 Genetically, unlike many other B-cell malignancies, translocations involving the IgH locus on chromosome 14q32 are rare in WM.4,5 Karyotypic analysis is hampered by a low yield of abnormal metaphases. The most common genetic abnormality is deletion of the long arm of chromosome 6.5,6 However, this cytogenetic abnormality is not unique to WM and its biologic and clinical significance in WM has yet to be elucidated.
In this study, we undertook a gene-expression profiling study of WM using CD138+ and CD19+ selected malignant BM cells and compared their transcription program with the related B-cell malignancies chronic lymphocytic leukemia (CLL) and multiple myeloma (MM) as well as the normal B-cell and plasma-cell (PC) counterparts. The selection of cells using both CD138 and CD19 should encompass the full spectrum of malignant cells, so that we can define a molecular signature for WM, determine its relation to other normal and malignant B-cell counterparts, and identify biologically relevant and important pathways, new disease markers, and potential therapeutic targets in an unbiased fashion. We found that the transcription profile of WM is very similar to CLL and normal B cells but distinct from MM and normal PCs. Only a small gene set is unique to WM. Pathway analysis of genes unique to WM provide further insights into the biology of this disease.
Materials and methods
Samples
Twenty-three WM, 8 CLL, 101 MM, 24 smoldering myeloma (SMM), 22 monoclonal gammopathy of unknown significance (MGUS; including 1 IgM MGUS), 15 normal PC, and 7 normal B-cell samples were included in this study. Blood and BM samples were procured after informed consent was obtained in accordance with the Declaration of Helsinki. The study was approved by the Institutional Review Board of the Mayo Clinic. The samples were enriched for cell populations of interest using immunomagnetic beads (AutoMACS; Miltenyi-Biotec, Auburn, CA). For WM and the case of IgM MGUS, CD19+ and CD138+ (concurrent but not sequential) selected BM cells were used. For MM, SMM, MGUS, and normal PCs, CD138+ selected BM cells were used. For CLL and normal B cells, CD19+ selected peripheral-blood cells were used. The CLL, WM, and PC neoplasm samples were consecutive samples with quantitatively and qualitatively adequate RNA for gene-expression study. The common cytogenetic abnormalities and categories were present (6q deletion for WM, IgH translocations, 13 deletion and hyperdiploidy for MM, and mutated and unmutated IgVH for CLL) in these patients at frequencies similar to published data (data not shown).
Gene-expression profiling
RNA was extracted from the enriched cells. Gene-expression analysis was performed using the Affymetrix U133A chip for WM, MM, SMM, MGUS, and normal PCs and the U133A and U133B chips for CLL and normal B cells (Affymetrix, Santa Clara, CA). RNA isolation, purification, and microarray hybridization have been previously reported.7 Gene-expression intensity values were log transformed, normalized to the median, and analyzed using GeneSpring 7 (Agilent Technologies, Palo Alto, CA).
Gene-expression analysis
For all analyses, the universal gene list was all genes on the U133A chip minus immunoglobulin genes (total, 22 104 genes). To define the relation between WM, CLL, PC neoplasm, normal PCs, and B cells, we first generated a list of genes whose expression varied significantly across individual samples by Welch ANOVA using variance computed by applying the Cross-Gene Error Model (CGEM) based on deviation from 1 available within GeneSpring. This overcomes the lack of replicates and variance associated with the individual samples and is similar in principle to variance filtering. The generated gene list was then used for unsupervised clustering of the CLL, WM, PC neoplasm, normal PC, and B-cell samples using a hierarchical agglomerative algorithm. To detect possible heterogeneity and identify subtypes within WM, all genes were used for unsupervised clustering. The Pearson correlation coefficient and centroid linkage were used as similarity and linkage methods, respectively.
To identify genes with unique gene-expression profiles for WM, CLL, and MM, we first identified genes with significantly different expression between WM, MM, and CLL by ANOVA with Benjamini and Hochberg multiple testing corrections. Genes whose P value is less than .05 and that pass a false discovery rate (FDR) of 5% were selected for further filtering by fold change (2-fold or higher different expression between WM and others). A similar sequence was applied to MM and CLL. Finally, a Venn diagram incorporating the 3 filtered gene sets (1 each for WM, MM, and CLL) was used to identify genes unique to each disease. The specificity of the WM signature was further validated by leave-one-out cross-validation (LOOCV) using 2 algorithms: the k-nearest neighbor (KNN) and the support vector machines (SVMs). In the KNN method, a sample is classified based on a majority vote of the classes of the k neighbor that are closest to it in terms of Euclidean distance. In the SVM method, a sample is classified based on its position relative to an optimal linear decision boundary constructed on a transformed feature space of the microarray data. For both methods, the predictor genes are those with unique profiles in WM compared to CLL or MM (significant by ANOVA after multiple testing corrections and greater than 2-fold difference in expression) as outlined. For the KNN method, optimal settings are number of neighbors of 15 and P value ratio of 0.1. For SVM, radial kernal function was used.
Inspection of some of these disease-specific gene lists revealed genes that could represent contaminating cell populations acquired during selection; pre-B cells (expressing NPTT [tdt], MME [CD10], VPREB1), and monocyte/macrophages (expressing CD36, CD163, CD14, APOE). Subsequently, we generated lists of genes whose expression may be the result of these contaminating cell populations by finding genes whose expression profiles were correlated with tdt (for pre-B cells) and CD36 (for monocytes) using a Pearson correlation coefficient of 0.7 as the cutoff value.
Because the initial unsupervised clustering of samples may be affected by the presence of these contaminating cells, we extracted previously published CLL signature (unique to CLL when compared to different B-cell types and B-cell malignancies although not including WM or MM).8 Unique MM signature has not been previously defined. As a compromise we extracted genes differentially expressed between MM and normal PCs previously published9 as a surrogate for an MM signature. Interestingly, the 2 gene sets do not overlap. Using both gene signatures, we clustered WM with CLL and MM samples to confirm the similarity between WM and CLL but not MM.
Gene ontology and pathway/network analysis was performed using a web-based software, MetaCore (GeneGo, St Joseph, MI). The software contains an interactive, manually annotated database derived from literature publications on proteins and small molecules that allows for representation of biologic functionality and integration of functional, molecular, or clinical information. Several algorithms to enable both the construction and analysis of gene networks were integrated as previously described.10 The output P values reflect scoring, prioritization, and statistical significance of networks according to the relevance of input data.
Results
Gene-expression profile of WM is related to CLL and normal B cells
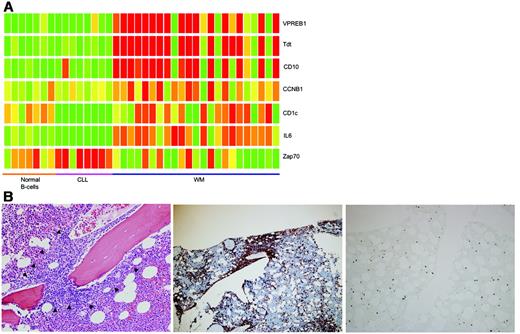
On unsupervised clustering, WM clustered together with normal B cells and CLL samples (first branching of the dendogram, Figure 1A). Furthermore, inspection of the heatmap suggested very similar profiles between these conditions. Of interest, the 1 case of IgM MGUS in the data set clustered with WM instead of MGUS or normal PCs. WM samples were split into 2 groups with CLL and IgM MGUS clustered in between. The separation of WM into 2 clusters had no relation to 6q deletion status because both clusters had similar numbers of patients with 6q deletion (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The expression profile of the smaller WM cluster (which clustered with IgM MGUS, WM cluster 1) was closer to normal B cells than the larger WM cluster (WM cluster 2; Figure 1A). In addition, 2 WM samples clustered next to normal PCs and 1 sample clustered with MM and MGUS (WM cluster 3). In general, the percentage of plasma cells in the bead-purified cells used for the gene-expression profile (GEP) increased from WM cluster 1 to WM cluster 3, suggesting the clustering of WM into these groups may reflect a difference in degree of plasmacytic differentiation of the tumor cells (Table 1).
Percentage of PCs and BM involvement by tumor in WM samples and their correlation with contamination signature and clustering
Samples . | PCs in purified samples, % . | BM involvement, % . | Contamination signature . | Clustering . |
|---|---|---|---|---|
| WM8 | 25 | 20-30 | Nil | WM 1 |
| WM15 | 16 | 50-60 | Nil | WM 1 |
| WM22 | NA | 60-70 | Nil | WM 1 |
| WM25 | 8 | 80 | Nil | WM 1 |
| WM2 | 45 | 10 | Monocytes | WM 1 |
| WM10 | 26 | 80-90 | Nil | WM 1 |
| WM24 | 70 | 10-20 | Nil | WM 2 |
| WM17 | 38 | 80 | Nil | WM 2 |
| WM3 | 18 | 30 | Monocytes | WM 2 |
| WM12 | 26 | 20 | Nil | WM 2 |
| WM5 | 90 | 60-70 | Nil | WM 2 |
| WM19 | NA | NA | Nil | WM 2 |
| WM20 | NA | NA | Monocytes | WM 2 |
| WM11 | 28 | 30 | Monocytes | WM 2 |
| WM4 | 36 | 20-25 | Pre-B | WM 2 |
| WM18 | NA | NA | Pre-B | WM 2 |
| WM7 | NA | 30 | Pre-B | WM 2 |
| WM13 | 23 | 20 | Pre-B | WM 2 |
| WM16 | 14 | 20-30 | Pre-B | WM 2 |
| WM9 | 22 | 10 | Pre-B | WM 2 |
| WM23 | 95 | 30 | Monocytes | WM 3 |
| WM26 | 94 | 10 | Monocytes | WM 3 |
| WM14 | 49 | <5 | Monocytes | WM 3 |
Samples . | PCs in purified samples, % . | BM involvement, % . | Contamination signature . | Clustering . |
|---|---|---|---|---|
| WM8 | 25 | 20-30 | Nil | WM 1 |
| WM15 | 16 | 50-60 | Nil | WM 1 |
| WM22 | NA | 60-70 | Nil | WM 1 |
| WM25 | 8 | 80 | Nil | WM 1 |
| WM2 | 45 | 10 | Monocytes | WM 1 |
| WM10 | 26 | 80-90 | Nil | WM 1 |
| WM24 | 70 | 10-20 | Nil | WM 2 |
| WM17 | 38 | 80 | Nil | WM 2 |
| WM3 | 18 | 30 | Monocytes | WM 2 |
| WM12 | 26 | 20 | Nil | WM 2 |
| WM5 | 90 | 60-70 | Nil | WM 2 |
| WM19 | NA | NA | Nil | WM 2 |
| WM20 | NA | NA | Monocytes | WM 2 |
| WM11 | 28 | 30 | Monocytes | WM 2 |
| WM4 | 36 | 20-25 | Pre-B | WM 2 |
| WM18 | NA | NA | Pre-B | WM 2 |
| WM7 | NA | 30 | Pre-B | WM 2 |
| WM13 | 23 | 20 | Pre-B | WM 2 |
| WM16 | 14 | 20-30 | Pre-B | WM 2 |
| WM9 | 22 | 10 | Pre-B | WM 2 |
| WM23 | 95 | 30 | Monocytes | WM 3 |
| WM26 | 94 | 10 | Monocytes | WM 3 |
| WM14 | 49 | <5 | Monocytes | WM 3 |
NA indicates not available
Normal PCs and most of the MGUS samples clustered together. A significant number of genes overexpressed by these samples are genes expressed in monocytes/macrophages (CD163, CD36, CD14, APOE, VCAM1). These genes were also overexpressed in some WM cases (Figure 1A). This probably represents genes expressed by contaminating monocytes/macrophages resulting from immunomagnetic bead selection of BM cells. This could be due to nonspecific binding of beads to Fc receptors present on the surface of monocytes/macrophages or phagocytosis of these beads by the monocytes/macrophages. This was most apparent when the percentage of cells selected for was low in the BM (in this case CD138+ PCs in normal and MGUS BM or WM samples with fewer malignant cells). This problem is less apparent if the tissue source of selection is peripheral blood (normal B cells and CLL samples) because monocytes are very minor populations of peripheral-blood cells. As corroborating evidence, the WM cases with this monocyte contamination signature had the lowest degree of BM tumor involvement (Table 1). In view of the possible impact of these contaminating cells and their gene expression on interpretation of results, we created list of genes that are highly correlated with these pre-B-cell and monocytes/macrophage genes, so that they can be subtracted from subsequent analysis of genes with expression profiles unique to the individual tumor types. There were also cases of MGUS that clustered with MM samples (Figure 1A). This is consistent with the well-known similarity in global gene expression between MGUS and MM.9
The GEP of WM is closer to CLL than MM. (A) Using 2162 gene probes that are variably expressed across the WM, CLL, and MM samples, unsupervised clustering of these samples together with normal B cells, PCs, MGUS, and SMM was performed. Selected genes that clustered together and were overexpressed in different sample clusters are highlighted. Restricting our analysis to only WM, CLL, and MM samples, unsupervised clustering was performed using published (B) CLL and (C) MM signatures. WM samples clustered predominantly with CLL samples and also exhibited closer approximation of the GEP using both signatures to CLL. Some genes of interests are highlighted. In both heatmaps, the colored bar at the bottom indicates the tumor type: blue, CLL; yellow, WM; and red, MM. The scale of the gene-expression data are similar to that in panel A.
The GEP of WM is closer to CLL than MM. (A) Using 2162 gene probes that are variably expressed across the WM, CLL, and MM samples, unsupervised clustering of these samples together with normal B cells, PCs, MGUS, and SMM was performed. Selected genes that clustered together and were overexpressed in different sample clusters are highlighted. Restricting our analysis to only WM, CLL, and MM samples, unsupervised clustering was performed using published (B) CLL and (C) MM signatures. WM samples clustered predominantly with CLL samples and also exhibited closer approximation of the GEP using both signatures to CLL. Some genes of interests are highlighted. In both heatmaps, the colored bar at the bottom indicates the tumor type: blue, CLL; yellow, WM; and red, MM. The scale of the gene-expression data are similar to that in panel A.
The 2-dimensional clustering also revealed different clusters of genes that were overexpressed in the different tumor/tissue types. The expression profiles of normal B cells and WM and CLL samples were very similar (Figure 1A). These samples overexpressed a cluster of genes that included B-cell markers such as CD22, CD19, CD79b, and CD11b, potential therapeutic targets such as CD52, prognostic markers (ZAP70), a set of genes normally expressed in pre-B cells (MME [CD10]), VPREB1, NPTT [tdt]). and genes of potential biologic relevance (IL6) (for a full list of genes in this cluster refer to Table S1). The appropriate expression of B-cell markers provided some internal validation to the gene-expression data. We decided to look closer at the expression of IL6, ZAP70, MME, VPREB1, and NPTT in CLL and WM (Figure 2A). IL6 expression was high (> 2-fold higher than CLL and MM) in most WM compared to CLL and normal B cells. ZAP70 expression was high in 7 of the 8 CLL samples and a subset of patients with WM. MME, VPREB1, and NPTT were overexpressed in the same WM samples and most likely represent contaminating BM pre-B cells resulting from the CD19 selection process. Interestingly, the expression of ZAP70 in WM was highest in the samples with the presumed pre-B-cell contamination. In view of previous studies showing ZAP70 expression in normal B cells11,12 and lack of ZAP70 expression in WM,13 the ZAP70 expression in the subset of WM was most likely because of the contaminating pre-B cells.
A subset of WM appeared to overexpress genes involved in cell cycle and proliferation (MAD2L1, CCNB1, CCNB2, TOP2A, NEK2, CENPE, CDC2). Interestingly these corresponded to the WM samples with the highest expression of ZAP70, but they did not constitute all the samples with the contaminating “pre-B-cell signature” (Figures 1A and 2A). Because ZAP-70 is overexpressed in “activated” B cells,11 it is possible that these samples are contaminated with more activated and proliferating pre-B cells as compared to other WM samples.
To validate that these genes were expressed by contaminating cells, staining for tdt by immunohistochemistry was performed by an expert hematopathologist for 5 samples with the contamination signature and 5 without and showed that tdt was positive only in the nonmalignant pre-B cells (hematogones) in the 5 samples with the contamination signature and tdt staining was absent in the 5 samples without the contamination signature. A representative example is shown in Figure 2B. This strongly suggested that overexpression of tdt and the strongly correlated set of proliferation genes including ZAP-70 was due to the presence of contaminating pre-B cells.
Because our unsupervised clustering may be affected by these contaminating cells, we sought to verify the observation that the expression profile of WM was similar to CLL but different from MM by using published CLL and MM signatures (see “Materials and methods”). It was clear that using either signature, WM was clustered together with CLL and not MM. Furthermore, the expression profiles of genes constituting the 2 signatures were more similar between WM and CLL than MM (Figure 1B-C).
WM has a homogeneous gene-expression profile
Initial attempts to identify genes with variable expression across the 23 WM samples using ANOVA yielded a very small list of genes (< 50). Therefore, we performed unsupervised clustering of WM samples using all genes. This analysis suggested a relatively homogeneous expression profile among the WM samples except for 4 samples that underexpressed a subset of genes (Figure S1). On closer inspection, these cases had the highest PC percentage and low BM involvement, and the underexpression of these genes was also seen in normal PCs and MGUS but not in CLL or B cells (data not shown). These 4 cases were not IgM myeloma because the patients had no bone disease and were negative for t(11;14)14 ; instead, they had lymphadenopathy or splenomegaly and in 1 case also a 6q deletion, shown by fluorescence in situ hybridization (FISH), which were all hallmarks of WM. There were no obvious differences in the expression profiles between samples with the 6q deletion and those without the deletion because they clustered together on unsupervised clustering and no genes were differentially expressed between 6q-deleted and nondeleted cases.
Contributions of genes from contaminating cell populations. (A) Closer inspection of some of the genes within the gene cluster overexpressed in normal B cells, CLL, and WM revealed some interesting differences. In particular, the concurrent overexpression of NPTT (tdt), MME (CD10), and VPREB1 in the WM samples suggested likely contamination with pre-B cells. In addition, ZAP70 and CCNB1 (as a representative gene from the proliferation cluster overexpressed in a subset of patients with WM) overexpression is tightly associated with samples overexpressing NPTT (tdt), MME (CD10), and VPREB1. (B) To verify that tdt expression was originating from contaminating pre-B cells, immunostaining for tdt was performed on BM biopsies from patients with WM with and without the pre-B-cell contamination signature. This figure is representative for samples with the contamination signature. Malignant cells (CD20+) formed intramedullary clusters (marked by arrows; left panel, hematoxylin and eosin staining; middle panel, CD20 staining). In contrast, nuclear tdt staining was seen in scattered interstitial cells that represented pre-B cells (right panel). In samples without the contamination signature, no tdt+ cells were seen. All images were acquired using a 40×/0.8 numeric aperture objective lens. The microscope used is a Zeiss Axioskop (Carl Zeiss Microimaging, Thornwood, NY). Images were captured by the Olympus DP70 CCD camera using DP controller image capture software (Olympus, Center Valley, PA).
Contributions of genes from contaminating cell populations. (A) Closer inspection of some of the genes within the gene cluster overexpressed in normal B cells, CLL, and WM revealed some interesting differences. In particular, the concurrent overexpression of NPTT (tdt), MME (CD10), and VPREB1 in the WM samples suggested likely contamination with pre-B cells. In addition, ZAP70 and CCNB1 (as a representative gene from the proliferation cluster overexpressed in a subset of patients with WM) overexpression is tightly associated with samples overexpressing NPTT (tdt), MME (CD10), and VPREB1. (B) To verify that tdt expression was originating from contaminating pre-B cells, immunostaining for tdt was performed on BM biopsies from patients with WM with and without the pre-B-cell contamination signature. This figure is representative for samples with the contamination signature. Malignant cells (CD20+) formed intramedullary clusters (marked by arrows; left panel, hematoxylin and eosin staining; middle panel, CD20 staining). In contrast, nuclear tdt staining was seen in scattered interstitial cells that represented pre-B cells (right panel). In samples without the contamination signature, no tdt+ cells were seen. All images were acquired using a 40×/0.8 numeric aperture objective lens. The microscope used is a Zeiss Axioskop (Carl Zeiss Microimaging, Thornwood, NY). Images were captured by the Olympus DP70 CCD camera using DP controller image capture software (Olympus, Center Valley, PA).
Analysis of expression of CD markers and genes involved in cell-cycle regulation
In an attempt to identify possible diagnostic markers, we next analyzed the expression of cluster of differentiation (CD) markers among the samples. The expression of CD markers in WM was similar to that in CLL and B cells and different from that in MM and normal PCs (Figure S2A). The expression of B-cell markers such as CD19, CD20, CD22, and CD23 and the common leukocyte antigen (CD45) in normal B cells, CLL, and WM but not MM, the expression of CD5 in CLL but not normal B cells, WM, MM, or normal PCs, and the expression of PC markers such as CD38 and CD138 in MM and normal PCs but not in normal B-cells, CLL, or WM provided internal validation of the gene-expression results as these were known immunophenotypic markers for the respective cell types (Figure S2B). The low but higher expression of CD45 and CD19 in normal PCs compared to MM and expression of CD56 in MM but not normal PCs are consistent with the difference in the expression of these markers between normal and malignant plasma cells.15,16 In terms of potential new disease markers, CD1C was highly expressed in almost all cases of WM but was not expressed in CLL or MM; however, strong expression was also seen in normal B cells (Figures 2A and S2B). CD200 was strongly expressed in CLL but not in WM or MM, and expression was weaker in normal B cells. Of therapeutic interest, CD52 was expressed in CLL and WM but not MM, whereas CD117 (c-kit) was expressed in MM but not WM or CLL. CD20 appears to be expressed strongly in normal B cells, CLL, and WM, with weak expression in MM (Figure S2B). Consistent with published data,17 the expression of CD20 was found predominantly in patients with t(11;14).
Because deregulation of the cell cycle is common in B-cell malignancies and these deregulated pathways are potential therapeutic targets,18 we investigated and compared the gene expression of genes involved in these processes between CLL, WM, and MM (Figure S3). The expression of cell-cycle genes was very similar between CLL and normal B cells, whereas there were more differences between MM and normal PCs. WM seemed to have an intermediate pattern of expression for these genes. In terms of the D-type cyclins, CLL and WM only expressed CCND3 whereas MM expressed all 3 cyclin D genes. Of note, the CDKI genes were predominantly up-regulated in the malignant conditions compared to their normal cellular counterpart. In MM, p14, p15, p18, and p21 were up-regulated and only p27 and p57 were down-regulated compared to normal PCs. In WM, p15, p18, and p57 were up-regulated compared to normal B cells (Figure S3). This suggests that on the whole, CDKIs are up-regulated in these B-cell malignancies and we speculate that these are secondary responses to check proliferation of tumor cells. The expression of CCNB1 and CDC2 is usually an indication of proliferation. These were not expressed in CLL consistent with its known low proliferative index. The expression of these genes was significantly higher in WM and correlated with expression of DNTT, probably representing pre-B-cell contamination (Figure 2A).
Genes with distinct expression profiles in WM, CLL, and MM are involved in different pathways relevant to their biology
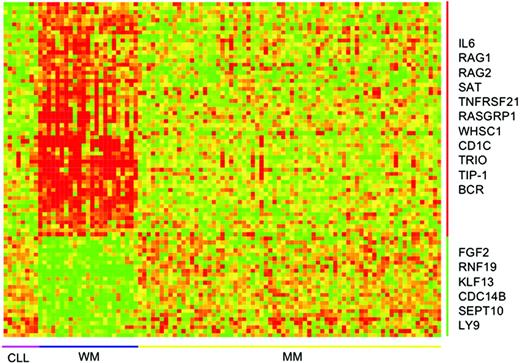
Seventy-three genes had a distinct expression profile in WM compared to CLL and MM (Figure 3; Table 2). The specificity of this WM signature was further tested by LOOCV. Using both KNN and SVM, all the WM cases were correctly predicted. Of all the non-WM cases, only 1 case was not predicted by KNN (did not pass the P value ratio) and wrongly predicted by SVM. Forty-eight of these were up-regulated and 25 down-regulated in WM. Interestingly, the most significantly up-regulated gene in WM is IL6. The aforementioned CD1C was also among the top 10 most significantly up-regulated genes.
Genes with unique gene-expression profile in WM
Probe . | P . | Gene names . | Fold change relative to MM . | Fold change relative to CLL . | Descriptions . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up-regulated in WM compared with CLL and MM | ||||||||||
| 205207_at | < .001 | IL6 | 4.39 | 6.41 | Interleukin 6 (interferon-β2) | |||||
| 204081_at | < .001 | NRGN | 3.23 | 5.38 | Neurogranin (protein kinase C substrate, RC3) | |||||
| 201310_s_at | < .001 | P311 | 4.9 | 6.29 | P311 protein | |||||
| 209626_s_at | < .001 | OSBPL3 | 3.63 | 2.77 | Oxysterol binding protein-like 3 | |||||
| 205987_at | < .001 | CD1C | 24.4 | 17.99 | CD1C antigen, c polypeptide | |||||
| 210640_s_at | < .001 | GPR30 | 9.75 | 8.93 | G protein-coupled receptor 30 | |||||
| 205240_at | < .001 | HSU54999 | 2.28 | 3.66 | LGN protein | |||||
| 211829_s_at | < .001 | GPR30 | 6.08 | 8.93 | G protein–coupled receptor 30 | |||||
| 202497_x_at | < .001 | SLC2A3 | 7.55 | 4.39 | Solute carrier family 2 (facilitated glucose transporter), member 3 | |||||
| 215464_s_at | < .001 | TIP-1 | 4.1 | 4.85 | Tax interaction protein 1 | |||||
| 209053_s_at | .004 | WHSC1 | 3.12 | 2.94 | Wolf-Hirschhorn syndrome candidate 1 | |||||
| Down-regulated in WM compared with CLL and MM | ||||||||||
| 222154_s_at | < .001 | DKFZP564 A2416 | 6.33 | 6.64 | DKFZP564A2416 protein | |||||
| 219878_s_at | < .001 | KLF13 | 5.99 | 6.04 | Kruppel-like factor 13 | |||||
| 221163_s_at | < .001 | WBSCR14 | 3.39 | 4.58 | Williams-Beuren syndrome critical region gene 14 | |||||
| 216869_at | < .001 | PDE1C | 2.42 | 3.21 | PDE1C3 splice variant; 3′,5′ cyclic nucleotide phosphodiesterase | |||||
| 218182_s_at | < .001 | CLDN1 | 2.05 | 3.55 | Claudin 1 | |||||
| 219630_at | < .001 | DD96 | 2.16 | 3.22 | Epithelial protein up-regulated in carcinoma, membrane-associated protein 17 | |||||
| 206736_x_at | < .001 | CHRNA4 | 3.09 | 3.75 | Cholinergic receptor, nicotinic, α-polypeptide 4 | |||||
| 206994_at | < .001 | CST4 | 3.24 | 2.97 | Cystatin S | |||||
| 215967_s_at | < .001 | LY9 | 3.27 | 2.93 | Lymphocyte antigen 9 | |||||
| 207553_at | < .001 | OPRK1 | 3.88 | 3.09 | Opioid receptor, κ1 | |||||
Probe . | P . | Gene names . | Fold change relative to MM . | Fold change relative to CLL . | Descriptions . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up-regulated in WM compared with CLL and MM | ||||||||||
| 205207_at | < .001 | IL6 | 4.39 | 6.41 | Interleukin 6 (interferon-β2) | |||||
| 204081_at | < .001 | NRGN | 3.23 | 5.38 | Neurogranin (protein kinase C substrate, RC3) | |||||
| 201310_s_at | < .001 | P311 | 4.9 | 6.29 | P311 protein | |||||
| 209626_s_at | < .001 | OSBPL3 | 3.63 | 2.77 | Oxysterol binding protein-like 3 | |||||
| 205987_at | < .001 | CD1C | 24.4 | 17.99 | CD1C antigen, c polypeptide | |||||
| 210640_s_at | < .001 | GPR30 | 9.75 | 8.93 | G protein-coupled receptor 30 | |||||
| 205240_at | < .001 | HSU54999 | 2.28 | 3.66 | LGN protein | |||||
| 211829_s_at | < .001 | GPR30 | 6.08 | 8.93 | G protein–coupled receptor 30 | |||||
| 202497_x_at | < .001 | SLC2A3 | 7.55 | 4.39 | Solute carrier family 2 (facilitated glucose transporter), member 3 | |||||
| 215464_s_at | < .001 | TIP-1 | 4.1 | 4.85 | Tax interaction protein 1 | |||||
| 209053_s_at | .004 | WHSC1 | 3.12 | 2.94 | Wolf-Hirschhorn syndrome candidate 1 | |||||
| Down-regulated in WM compared with CLL and MM | ||||||||||
| 222154_s_at | < .001 | DKFZP564 A2416 | 6.33 | 6.64 | DKFZP564A2416 protein | |||||
| 219878_s_at | < .001 | KLF13 | 5.99 | 6.04 | Kruppel-like factor 13 | |||||
| 221163_s_at | < .001 | WBSCR14 | 3.39 | 4.58 | Williams-Beuren syndrome critical region gene 14 | |||||
| 216869_at | < .001 | PDE1C | 2.42 | 3.21 | PDE1C3 splice variant; 3′,5′ cyclic nucleotide phosphodiesterase | |||||
| 218182_s_at | < .001 | CLDN1 | 2.05 | 3.55 | Claudin 1 | |||||
| 219630_at | < .001 | DD96 | 2.16 | 3.22 | Epithelial protein up-regulated in carcinoma, membrane-associated protein 17 | |||||
| 206736_x_at | < .001 | CHRNA4 | 3.09 | 3.75 | Cholinergic receptor, nicotinic, α-polypeptide 4 | |||||
| 206994_at | < .001 | CST4 | 3.24 | 2.97 | Cystatin S | |||||
| 215967_s_at | < .001 | LY9 | 3.27 | 2.93 | Lymphocyte antigen 9 | |||||
| 207553_at | < .001 | OPRK1 | 3.88 | 3.09 | Opioid receptor, κ1 | |||||
The top 10 up- and down-regulated genes plus other genes of interest are shown
A total of 396 genes had a distinct expression profile in CLL compared to WM and MM; 174 of these genes were up-regulated and 222 down-regulated in CLL. As expected, up-regulated genes included BCL2 and ZAP70. Among the down-regulated genes were cell-cycle-related genes (CDC2, CDC20, NEK2), and several CDKIs (CDKN1C, CDKN1A, CDKN2C).
A total of 1247 genes had a distinct expression pattern in MM compared to CLL and WM; 577 of these were up-regulated and 670 down-regulated in MM. Many of the up-regulated genes are involved in signaling pathways known to be important in MM such as WNT signaling (DKK1, FRZB, WNT10B, WNT5A, FZD6, WNT6), IL6R, IGF1R, MET (HGF receptor), and HGF. Among the down-regulated genes were several known to be important in early B-cell receptor signaling and differentiation that tend to be down-regulated with terminal differentiation to PCs (PAX5, CD19, VAV). The top 10 up- and down-regulated genes and other selected genes of interest in WM, CLL, and MM are appended in Tables 2, 3, and 4, respectively (the full gene lists are presented in Table S1).
Genes with unique expression profile in CLL
Probe . | Gene names . | Fold change relative to MM . | Fold change relative to WM . | Description . | ||||
|---|---|---|---|---|---|---|---|---|
| Up-regulated in CLL compared with WM and MM | ||||||||
| 218704_at | FLJ20315 | 3.34 | 3.15 | Hypothetic protein FLJ20315 | ||||
| 221010_s_at | SIRT5 | 3.44 | 2.86 | Sirtuin silent mating type information regulation 2 homolog 5 | ||||
| 203072_at | MYO1E | 7.97 | 7.77 | Myosin 1E | ||||
| 204155_s_at | KIAA0999 | 3.51 | 2.83 | ESTs | ||||
| 204446_s_at | ALOX5 | 9.13 | 8.97 | Arachidonate 5-lipoxygenase | ||||
| 208858_s_at | KIAA0747 | 4.95 | 4.93 | KIAA0747 protein | ||||
| 214366_s_at | ALOX5 | 7.66 | 4.36 | Arachidonate 5-lipoxygenase | ||||
| 208269_s_at | ADAM28 | 38.87 | 5.47 | A disintegrin and metalloproteinase domain 28 | ||||
| 205389_s_at | ANK1 | 10.23 | 4.74 | Aankyrin 1, erythrocytic | ||||
| 209670_at | TRAC | 13.03 | 7.38 | T-cell receptor α-chain (VDJC); human T-cell receptor active α-chain mRNA from JM-cell line, complete clusters of differentiation | ||||
| 203685_at | BCL2 | 3.96 | 3.01 | B-cell CLL/lymphoma 2 | ||||
| 214032_at | ZAP70 | 11.79 | 6.71 | ζ-chain (TCR)–associated protein kinase (70 kDa) | ||||
| Down-regulated in CLL compared with WM and MM | ||||||||
| 222315_at | ESTs | 28.09 | 40 | ESTs | ||||
| 202768_at | FOSB | 82.6 | 107.76 | FBJ murine osteosarcoma viral oncogene homolog B | ||||
| 202708_s_at | H2BFQ | 4.88 | 3.07 | H2B histone family, member Q | ||||
| 201830_s_at | NET1 | 19.4 | 23.7 | Neuroepithelial-cell transforming gene 1 | ||||
| 201694_s_at | EGR1 | 8.62 | 6.41 | Early growth response 1 | ||||
| 205780_at | BIK | 9.61 | 16.8 | BCL2-interacting killer (apoptosis-inducing) | ||||
| 202733_at | P4HA2 | 3.32 | 3.1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), α polypeptide II | ||||
| 211143_x_at | NR4A1 | 3.95 | 3.19 | Nuclear receptor subfamily 4, group A, member 1 | ||||
| 202095_s_at | BIRC5 | 4.78 | 6.41 | Baculoviral IAP repeat-containing 5 (survivin) | ||||
| 209911_x_at | H2BFB | 4.88 | 2.94 | H2B histone family, member B | ||||
| 204493_at | BID | 9.26 | 6.21 | BH3 interacting domain death agonist | ||||
| 213182_x_at | CDKN1C | 4.33 | 6.94 | Vyclin-dependent kinase inhibitor 1C (p57, Kip2) | ||||
| 200670_at | XBP1 | 13.73 | 7.81 | X-box binding protein 1 | ||||
| 203362_s_at | MAD2L1 | 3.34 | 3.53 | MAD2 mitotic arrest deficient-like 1 (yeast) | ||||
| 206665_s_at | BCL2L1 | 8.2 | 4.67 | BCL2-like 1 | ||||
| 209642_at | BUB1 | 3.05 | 3.1 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | ||||
| 202284_s_at | CDKN1A | 15.27 | 9 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | ||||
| 202870_s_at | CDC20 | 4.44 | 7.58 | CDC20-cell division cycle 20 homolog (Saccharomyces cerevisiae) | ||||
| 210559_s_at | CDC2 | 3.36 | 4.08 | Homo sapiens mRNA for CDC2 δ T, complete clusters of differentiation | ||||
| 211792_s_at | CDKN2C | 3.25 | 3.05 | Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | ||||
| 204641_at | NEK2 | 3.51 | 5.6 | NIMA (never in mitosis gene a)-related kinase 2 | ||||
Probe . | Gene names . | Fold change relative to MM . | Fold change relative to WM . | Description . | ||||
|---|---|---|---|---|---|---|---|---|
| Up-regulated in CLL compared with WM and MM | ||||||||
| 218704_at | FLJ20315 | 3.34 | 3.15 | Hypothetic protein FLJ20315 | ||||
| 221010_s_at | SIRT5 | 3.44 | 2.86 | Sirtuin silent mating type information regulation 2 homolog 5 | ||||
| 203072_at | MYO1E | 7.97 | 7.77 | Myosin 1E | ||||
| 204155_s_at | KIAA0999 | 3.51 | 2.83 | ESTs | ||||
| 204446_s_at | ALOX5 | 9.13 | 8.97 | Arachidonate 5-lipoxygenase | ||||
| 208858_s_at | KIAA0747 | 4.95 | 4.93 | KIAA0747 protein | ||||
| 214366_s_at | ALOX5 | 7.66 | 4.36 | Arachidonate 5-lipoxygenase | ||||
| 208269_s_at | ADAM28 | 38.87 | 5.47 | A disintegrin and metalloproteinase domain 28 | ||||
| 205389_s_at | ANK1 | 10.23 | 4.74 | Aankyrin 1, erythrocytic | ||||
| 209670_at | TRAC | 13.03 | 7.38 | T-cell receptor α-chain (VDJC); human T-cell receptor active α-chain mRNA from JM-cell line, complete clusters of differentiation | ||||
| 203685_at | BCL2 | 3.96 | 3.01 | B-cell CLL/lymphoma 2 | ||||
| 214032_at | ZAP70 | 11.79 | 6.71 | ζ-chain (TCR)–associated protein kinase (70 kDa) | ||||
| Down-regulated in CLL compared with WM and MM | ||||||||
| 222315_at | ESTs | 28.09 | 40 | ESTs | ||||
| 202768_at | FOSB | 82.6 | 107.76 | FBJ murine osteosarcoma viral oncogene homolog B | ||||
| 202708_s_at | H2BFQ | 4.88 | 3.07 | H2B histone family, member Q | ||||
| 201830_s_at | NET1 | 19.4 | 23.7 | Neuroepithelial-cell transforming gene 1 | ||||
| 201694_s_at | EGR1 | 8.62 | 6.41 | Early growth response 1 | ||||
| 205780_at | BIK | 9.61 | 16.8 | BCL2-interacting killer (apoptosis-inducing) | ||||
| 202733_at | P4HA2 | 3.32 | 3.1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), α polypeptide II | ||||
| 211143_x_at | NR4A1 | 3.95 | 3.19 | Nuclear receptor subfamily 4, group A, member 1 | ||||
| 202095_s_at | BIRC5 | 4.78 | 6.41 | Baculoviral IAP repeat-containing 5 (survivin) | ||||
| 209911_x_at | H2BFB | 4.88 | 2.94 | H2B histone family, member B | ||||
| 204493_at | BID | 9.26 | 6.21 | BH3 interacting domain death agonist | ||||
| 213182_x_at | CDKN1C | 4.33 | 6.94 | Vyclin-dependent kinase inhibitor 1C (p57, Kip2) | ||||
| 200670_at | XBP1 | 13.73 | 7.81 | X-box binding protein 1 | ||||
| 203362_s_at | MAD2L1 | 3.34 | 3.53 | MAD2 mitotic arrest deficient-like 1 (yeast) | ||||
| 206665_s_at | BCL2L1 | 8.2 | 4.67 | BCL2-like 1 | ||||
| 209642_at | BUB1 | 3.05 | 3.1 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | ||||
| 202284_s_at | CDKN1A | 15.27 | 9 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | ||||
| 202870_s_at | CDC20 | 4.44 | 7.58 | CDC20-cell division cycle 20 homolog (Saccharomyces cerevisiae) | ||||
| 210559_s_at | CDC2 | 3.36 | 4.08 | Homo sapiens mRNA for CDC2 δ T, complete clusters of differentiation | ||||
| 211792_s_at | CDKN2C | 3.25 | 3.05 | Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | ||||
| 204641_at | NEK2 | 3.51 | 5.6 | NIMA (never in mitosis gene a)-related kinase 2 | ||||
The top 10 up- and down-regulated genes plus other genes of interest are shown. For all probes, P < .001
Genes with unique expression profile in MM
Probe . | P . | Gene name . | Fold change relative to WM . | Fold change relative to CLL . | Description . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up-regulated in MM compared to CLL and WM | ||||||||||
| 212253_x_at | < .001 | BPAG1 | 3.52 | 5.62 | Bullous pemphigoid antigen 1 (230/240 kDa) | |||||
| 201064_s_at | < .001 | PABPC4 | 2.36 | 4.22 | Poly(A) binding protein, cytoplasmic 4 (inducible form) | |||||
| 209968_s_at | < .001 | NCAM1 | 2.3 | 4.1 | Neural cell adhesion molecule 1 | |||||
| 204271_s_at | < .001 | EDNRB | 17.6 | 12.5 | Endothelin receptor type B | |||||
| 205309_at | < .001 | ASML3B | 3.41 | 4.67 | Acid sphingomyelinase-like phosphodiesterase | |||||
| 204602_at | < .001 | DKK1 | 9.9 | 7.41 | Dickkopf homolog 1 (Xenopus laevis) | |||||
| 202973_x_at | < .001 | KIAA0914 | 5.56 | 6.53 | KIAA0914 gene product | |||||
| 203697_at | < .001 | Frizzled-related protein | 13.61 | 17.6 | FRZB; frizzled protein homolog | |||||
| 202170_s_at | < .001 | AASDHPPT | 2.11 | 2.58 | Aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | |||||
| 215059_at | < .001 | Homo sapiens mRNA: cDNA DKFZp564G112 (from clone DKFZp564G112) | 6.76 | 10.76 | Homo sapiens mRNA; cDNA DKFZp564G112 (from clone DKFZp564G112) | |||||
| 210755_at | < .001 | HGF | 4.18 | 3.36 | Hepatocyte growth factor (hepapoietin A; scatter factor) | |||||
| 209347_s_at | < .001 | MAF | 2.43 | 2.78 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | |||||
| 205071_x_at | < .001 | XRCC4 | 5.26 | 6.58 | X-ray repair complementing defective repair in Chinese hamster cells 4 | |||||
| 206213_at | < .001 | WNT10B | 2.72 | 4.72 | Wingless-type MMTV integration site family, member 10B | |||||
| 213693_s_at | < .001 | MUC1 | 2.22 | 2.61 | Mucin 1, transmembrane | |||||
| 213425_at | < .001 | WNT5A | 3.33 | 4.55 | Wingless-type MMTV integration site family, member 5A | |||||
| 203510_at | < .001 | MET | 4.27 | 4.9 | met proto-oncogene (hepatocyte growth factor receptor) | |||||
| 212097_at | < .001 | CAV1 | 17.45 | 18.7 | Caveolin 1, caveolae protein, 22kD | |||||
| 205945_at | < .001 | IL6R | 2.48 | 3.41 | Interleukin 6 receptor | |||||
| 203987_at | < .001 | FZD6 | 2.87 | 4.97 | Frizzled homolog 6 (Drosophila) | |||||
| 221609_s_at | < .001 | WNT6 | 2.26 | 2.73 | Wingless-type MMTV integration site family, member 6 | |||||
| 203628_at | .018 | IGF1R | 2.33 | 2.85 | Insulin-like growth factor 1 receptor | |||||
| Down-regulated in MM compared to CLL and WM | ||||||||||
| 209269_s_at | < .001 | SYK | 30.17 | 26.91 | Spleen tyrosine kinase | |||||
| 34210_at | < .001 | CD52 | 94.71 | 180.4 | CAMPATH-1 (human); mRNA sequence | |||||
| 215537_x_at | < .001 | DDAH2 | 5.04 | 7.25 | Dimethylarginine dimethylaminohydrolase 2 | |||||
| 204661_at | < .001 | CD52 | 62.91 | 115.2 | CDW52 antigen (CAMPATH-1 antigen) | |||||
| 201721_s_at | < .001 | LAPTM5 | 9.51 | 16.39 | Lysosomal-associated multispanning membrane protein-5 | |||||
| 203037_s_at | < .001 | KIAA0429 | 10.66 | KIAA0429 gene product | ||||||
| 216237_s_at | < .001 | MCM5 | 5.2 | 8.1 | MCM5 minichromosome maintenance deficient 5, cell division cycle 46 (Saccharomyces cerevisiae) | |||||
| 201954_at | < .001 | ARPC1B | 2.08 | 3.51 | Actin-related protein 2/3 complex, subunit 1 B (41 kDa) | |||||
| 41220_at | < .001 | MSF | 3.8 | 7.59 | MLL septin-like fusion | |||||
| 200934_at | < .001 | DEK | 21.98 | 28.82 | DEK oncogene (DNA binding) | |||||
| 221969_at | < .001 | PAX5 | 6.58 | 12.04 | ESTs, Weakly similar to S57447 HPBRII-7 protein (Homo sapiens) | |||||
| 206398_s_at | < .001 | CD19 | 23.57 | 45.66 | CD19 antigen | |||||
| 205536_at | < .001 | VAV2 | 2.76 | 4.82 | vav 2 oncogene | |||||
| 218806_s_at | < .001 | VAV3 | 2.86 | 5.21 | vav 3 oncogene | |||||
| 202414_at | < .001 | ERCC5 | 2.03 | 3.01 | Excision repair cross-complementing rodent repair deficiency, complementation group 5 (Xeroderma pigmentosum) | |||||
| 204054_at | < .001 | PTEN | 2.05 | 2.64 | Phosphatase and tensin homolog | |||||
| 212888_at | < .001 | DICER1 | 2.32 | 2.88 | Homo sapiens clone 23938 mRNA sequence | |||||
| 209193_at | < .001 | PIM1 | 3.18 | 3.06 | pim-1 oncogene | |||||
| 201700_at | < .001 | CCND3 | 3.07 | 3.78 | Cyclin D3 | |||||
| 205647_at | < .001 | RAD52 | 2.45 | 4.22 | RAD52 homolog (S cerevisiae) | |||||
| 216299_s_at | .002 | XRCC3 | 3.1 | 3.17 | X-ray repair complementing defective repair in Chinese hamster cells 3 | |||||
Probe . | P . | Gene name . | Fold change relative to WM . | Fold change relative to CLL . | Description . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up-regulated in MM compared to CLL and WM | ||||||||||
| 212253_x_at | < .001 | BPAG1 | 3.52 | 5.62 | Bullous pemphigoid antigen 1 (230/240 kDa) | |||||
| 201064_s_at | < .001 | PABPC4 | 2.36 | 4.22 | Poly(A) binding protein, cytoplasmic 4 (inducible form) | |||||
| 209968_s_at | < .001 | NCAM1 | 2.3 | 4.1 | Neural cell adhesion molecule 1 | |||||
| 204271_s_at | < .001 | EDNRB | 17.6 | 12.5 | Endothelin receptor type B | |||||
| 205309_at | < .001 | ASML3B | 3.41 | 4.67 | Acid sphingomyelinase-like phosphodiesterase | |||||
| 204602_at | < .001 | DKK1 | 9.9 | 7.41 | Dickkopf homolog 1 (Xenopus laevis) | |||||
| 202973_x_at | < .001 | KIAA0914 | 5.56 | 6.53 | KIAA0914 gene product | |||||
| 203697_at | < .001 | Frizzled-related protein | 13.61 | 17.6 | FRZB; frizzled protein homolog | |||||
| 202170_s_at | < .001 | AASDHPPT | 2.11 | 2.58 | Aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | |||||
| 215059_at | < .001 | Homo sapiens mRNA: cDNA DKFZp564G112 (from clone DKFZp564G112) | 6.76 | 10.76 | Homo sapiens mRNA; cDNA DKFZp564G112 (from clone DKFZp564G112) | |||||
| 210755_at | < .001 | HGF | 4.18 | 3.36 | Hepatocyte growth factor (hepapoietin A; scatter factor) | |||||
| 209347_s_at | < .001 | MAF | 2.43 | 2.78 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | |||||
| 205071_x_at | < .001 | XRCC4 | 5.26 | 6.58 | X-ray repair complementing defective repair in Chinese hamster cells 4 | |||||
| 206213_at | < .001 | WNT10B | 2.72 | 4.72 | Wingless-type MMTV integration site family, member 10B | |||||
| 213693_s_at | < .001 | MUC1 | 2.22 | 2.61 | Mucin 1, transmembrane | |||||
| 213425_at | < .001 | WNT5A | 3.33 | 4.55 | Wingless-type MMTV integration site family, member 5A | |||||
| 203510_at | < .001 | MET | 4.27 | 4.9 | met proto-oncogene (hepatocyte growth factor receptor) | |||||
| 212097_at | < .001 | CAV1 | 17.45 | 18.7 | Caveolin 1, caveolae protein, 22kD | |||||
| 205945_at | < .001 | IL6R | 2.48 | 3.41 | Interleukin 6 receptor | |||||
| 203987_at | < .001 | FZD6 | 2.87 | 4.97 | Frizzled homolog 6 (Drosophila) | |||||
| 221609_s_at | < .001 | WNT6 | 2.26 | 2.73 | Wingless-type MMTV integration site family, member 6 | |||||
| 203628_at | .018 | IGF1R | 2.33 | 2.85 | Insulin-like growth factor 1 receptor | |||||
| Down-regulated in MM compared to CLL and WM | ||||||||||
| 209269_s_at | < .001 | SYK | 30.17 | 26.91 | Spleen tyrosine kinase | |||||
| 34210_at | < .001 | CD52 | 94.71 | 180.4 | CAMPATH-1 (human); mRNA sequence | |||||
| 215537_x_at | < .001 | DDAH2 | 5.04 | 7.25 | Dimethylarginine dimethylaminohydrolase 2 | |||||
| 204661_at | < .001 | CD52 | 62.91 | 115.2 | CDW52 antigen (CAMPATH-1 antigen) | |||||
| 201721_s_at | < .001 | LAPTM5 | 9.51 | 16.39 | Lysosomal-associated multispanning membrane protein-5 | |||||
| 203037_s_at | < .001 | KIAA0429 | 10.66 | KIAA0429 gene product | ||||||
| 216237_s_at | < .001 | MCM5 | 5.2 | 8.1 | MCM5 minichromosome maintenance deficient 5, cell division cycle 46 (Saccharomyces cerevisiae) | |||||
| 201954_at | < .001 | ARPC1B | 2.08 | 3.51 | Actin-related protein 2/3 complex, subunit 1 B (41 kDa) | |||||
| 41220_at | < .001 | MSF | 3.8 | 7.59 | MLL septin-like fusion | |||||
| 200934_at | < .001 | DEK | 21.98 | 28.82 | DEK oncogene (DNA binding) | |||||
| 221969_at | < .001 | PAX5 | 6.58 | 12.04 | ESTs, Weakly similar to S57447 HPBRII-7 protein (Homo sapiens) | |||||
| 206398_s_at | < .001 | CD19 | 23.57 | 45.66 | CD19 antigen | |||||
| 205536_at | < .001 | VAV2 | 2.76 | 4.82 | vav 2 oncogene | |||||
| 218806_s_at | < .001 | VAV3 | 2.86 | 5.21 | vav 3 oncogene | |||||
| 202414_at | < .001 | ERCC5 | 2.03 | 3.01 | Excision repair cross-complementing rodent repair deficiency, complementation group 5 (Xeroderma pigmentosum) | |||||
| 204054_at | < .001 | PTEN | 2.05 | 2.64 | Phosphatase and tensin homolog | |||||
| 212888_at | < .001 | DICER1 | 2.32 | 2.88 | Homo sapiens clone 23938 mRNA sequence | |||||
| 209193_at | < .001 | PIM1 | 3.18 | 3.06 | pim-1 oncogene | |||||
| 201700_at | < .001 | CCND3 | 3.07 | 3.78 | Cyclin D3 | |||||
| 205647_at | < .001 | RAD52 | 2.45 | 4.22 | RAD52 homolog (S cerevisiae) | |||||
| 216299_s_at | .002 | XRCC3 | 3.1 | 3.17 | X-ray repair complementing defective repair in Chinese hamster cells 3 | |||||
The top 10 up- and down-regulated genes plus other genes of interest are shown
When these disease-specific genes were analyzed for relevant processes and pathways using MetaCore, interesting differences were obtained. The gene ontology (GO) processes most associated with the genes unique to MM were involved in signal transduction and intracellular signaling, in particular cell-surface receptor-linked signaling, whereas the most relevant pathways include AKT, IGF-1R, and WNT signaling as well as prostacyclin synthesis, angiopoietin signaling, and integrin-mediated cell adhesion. For CLL, the relevant processes were immune response, apoptosis, and cell-cycle regulation and the most relevant pathways were involved in apoptosis regulation (data not shown). Due to the small list of genes unique to WM, no significant pathways related to the set of genes were detected. However, the most relevant GO process is in activation of MAPK activity. Of note, the MAPK cascade is one of the signaling pathways activated by IL-6.19
Gene-expression signature unique to WM. Seventy-three genes, 48 up-regulated and 25 down-regulated, constitute a gene-expression signature unique to WM. Here the samples are ordered according to diagnosis and genes according to fold difference in expression between WM and CLL and MM. Some interesting genes are highlighted. For a more complete list, see Table S1. The scale of the gene-expression data is similar to that in Figure 1A.
Gene-expression signature unique to WM. Seventy-three genes, 48 up-regulated and 25 down-regulated, constitute a gene-expression signature unique to WM. Here the samples are ordered according to diagnosis and genes according to fold difference in expression between WM and CLL and MM. Some interesting genes are highlighted. For a more complete list, see Table S1. The scale of the gene-expression data is similar to that in Figure 1A.
Discussion
To our knowledge, this is the first gene-expression profiling study in WM and its contrast to other B-cell malignancies. In this study, we attempted to define a molecular signature of WM in the context of normal cellular counterparts (B cells and PCs) and malignant cells in related B-cell malignancies (CLL and MM).
WM has a homogeneous transcription profile, clusters with CLL and normal B cells on unsupervised clustering, and clearly has a similar expression profile to CLL. In contrast, the expression profile of WM is very different from MM and normal PCs. Previous studies have shown that CLL, regardless of immunoglobulin heavy-chain (IgH) mutation status, has a homogeneous expression profile8,20 very similar to peripheral-blood resting B cells.21,22 In another study comparing CLL to different tonsillar B-cell populations, the expression profile of CLL is most closely related to memory B cells,8 which also constitutes a significant population of the peripheral-blood B-cell pool.23 The similarity between the expression profile of WM and CLL is perhaps not surprising given the known biologic and clinical characteristics of these diseases. Both are indolent tumors characterized by low proliferation. Unlike other B-cell malignancies, chromosomal translocations involving the IgH locus are relatively uncommon in both CLL and WM.24,25 These IgH translocations are thought to occur either during immunoglobulin VDJ recombination in maturing B cells or during immunoglobulin somatic hypermutation and isotype switching in mature B cells within the germinal center (GC).26 They are therefore common in B-cell lymphomas, which are usually derived from GC B cells.27 This observation is consistent with the notion that both CLL and WM are derived from memory B cells where these mechanisms have been inactivated. Despite these similarities, differences such as differential expression of immunophenotypic markers and demographics in IgVH mutation status (almost all WM cases are IgV mutated3,28 compared to 50%-70% of CLLs29,30 ) suggest that they may be derived from different memory B-cell populations.23
It is possible that the expression profile of WM is closer to normal B cells and CLL than MM and normal PCs due to the additional CD19+ selection (normal B cells and CLL are CD19+ selected) as compared to only CD138+ selection in MM and normal PCs. There are several arguments against this: (1) the bulk of CD19+ cells should represent the cell of interest (malignant WM and CLL cells and normal B cells); (2) the tissue source for CD19 selection is different (BM for WM and peripheral blood for normal B cells and CLL) and hence the predominant nonmalignant B-cell population selected should be different (pre-B cells for WM and mature B cells for CLL); and (3) WM but not CLL samples are additionally CD138 selected. Furthermore, our strategy of using both CD138 and CD19 to select for malignant WM cells has the added advantage of allowing the analysis of the entire malignant population. Therefore, the respective expression signature should, in general, reflect tumor phenotype rather than contaminating cells or other byproducts resulting from CD19 selection. The similarity between WM and CLL and not MM is further confirmed using independently derived CLL and MM signatures.
Detailed analyses of gene expression for CD markers and genes involved in cell-cycle regulation also show similarities between WM and CLL and normal B cells but not MM and normal PCs. In total, our analysis suggests that WM is defined by a B cell-like signature and in terms of gene transcription signature is closer to CLL than MM. The clustering of the IgM MGUS case together with WM and the similar expression of genes with expression profile unique to WM (IL-6 and CD1c; data not shown) suggest a shared phenotype between IgM MGUS and WM. This is consistent with current notion that a subset of IgM MGUS represents the precursor state of WM.31
Defining genes with unique expression in a disease may provide insight into its biology. This would be particularly useful in WM where little is known about its biology. Despite the purity of more than 90% in most samples after immunomagnetic bead-positive selection, presence of contaminating normal cells may still contribute to the overall gene-expression profile. In our data set, we noted signatures of contaminating monocyte/macrophages and pre-B cells, particularly in the samples with lower number of target cells (eg, MGUS, normal samples, and WM with lower BM involvement). To ensure that all the genes with unique expression profiles in each of the malignant conditions are relevant to the malignant cells, we subtracted genes that constitute the signature of contaminating cell types. Only a small set of genes have a unique expression profile in WM. Among the up-regulated genes, IL6 is the most significant. It has been demonstrated that IL-6 levels are elevated in WM 32,33 and that IL-6 is required for plasmacytic differentiation of the clonal B cells in WM.34 However, remarkably little is published regarding the potential role of IL6 in WM biology given its prominent role as a growth and survival factor in MM.35,36 Our data regarding IL6 expression in WM is validated by 2 abstract presentations that used gene-expression approaches. In the first study, the GEP identified deregulated elements in the IL-6 signaling cascade,37 whereas in the second study, using a similar analysis approach as ours, they found that IL6 expression is higher in WM B cells than normal B cells.38 Its potential relevance to WM biology is highlighted by the fact that the GO process most relevant to the WM unique genes in our study is activation of MAPK, which is involved in IL-6 signaling.19 Furthermore, because our gene-expression analysis is performed on highly purified malignant cells and not total BM cells (ie, including stromal cells), the differential expression of IL-6 in WM cells is more supportive of an autocrine source for IL-6 in WM compared to MM where the source of IL-6 is predominantly from the stromal cells. The functional and biologic importance of IL-6 in WM should be investigated because this may represent an important therapeutic avenue.
As for CLL and MM, the differentially expressed genes suggest that apoptosis regulation and receptor-mediated signal transduction are critical processes central to CLL and MM biology, respectively. These results are consistent with the known biochemical and signaling pathways found in the malignant B cells from patients with MM and CLL. It is well known that MM growth and survival are mediated through various cytokines secreted by stromal cells in the BM milieu on interaction with MM cells.35,36 Similarly, the fundamental role of antiapoptosis mechanisms is well established in CLL.39 More importantly, this comparative analysis shows that different pathways are deregulated in these tumors.
In conclusion, our GEP study suggests that WM samples have a homogenous expression profile very similar to CLL and normal peripheral blood B cells. A small set of genes is distinctly expressed in WM, with IL6 the most significantly up-regulated gene. These genes are most significantly associated with MAPK signaling. This, in turn, suggests that IL-6 and its signaling network may be of biologic significance in WM.
Prepublished online as Blood First Edition Paper, June 27, 2006; DOI 10.1182/blood-2006-02-005488.
Supported in part by grants R01 CA83724-01, Specialized Program of Research Excellence (SPORE) P50 CA100707-01, and P01 CA62242 from the National Cancer Institute; the International Waldenström's Macroglobulinemia Foundation; and the Fund to Cure Myeloma. N.K. is supported in part by National Institutes of Health/National Cancer Institute grant CA 95241 and philanthropic support from the Donaldson Charitable Trust, Mr E. Spencer, and the Donner families. W.J.C. is funded by an International Fellowship from the Agency for Science, Technology and Research (A*STAR), Singapore.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Ellen Remstein, MD (Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN) for help with performing and interpreting IHC staining for tdt.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal