To identify the mechanism of loss of heterozygosity (LOH) and potential modifier gene(s), we investigated the molecular basis of somatic NF1 inactivation in myeloid malignancies from 10 children with neurofibromatosis type 1. Loci across a minimal 50-Mb region of primarily the long arm of chromosome 17 showed LOH in 8 cases, whereas a less than 9-Mb region of loci flanking NF1 had LOH in the remaining 2 cases. Two complementary techniques, quantitative polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH), were used to determine whether the copy number at loci that showed LOH was 1 or 2 (ie, deleted or isodisomic). The 2 cases with LOH limited to less than 9 Mb were intrachromosomal deletions. Among the 8 leukemias with 50-Mb LOH segments, 4 had partial uniparental isodisomy and 4 had interstitial uniparental isodisomy. These isodisomic cases showed clustering of the centromeric and telomeric LOH breakpoints. This suggests that the cases with interstitial uniparental isodisomy arose in a leukemia-initiating cell by double-homologous recombination events at intervals of preferred mitotic recombination. Homozygous inactivation of NF1 favored outgrowth of the leukemia-initiating cell. Our studies demonstrate that LOH analyses of loci distributed along the chromosomal length along with copy-number analysis can reveal novel mechanisms of LOH that may potentially identify regions harboring “cryptic” tumor suppressor or modifier genes whose inactivation contributes to tumorigenesis.
Introduction
Tumor suppressor gene (TSG) inactivation commonly occurs by sequential somatic inactivation of both alleles or, in individuals who inherit a germline mutation, by a somatic mutation in the single normal homolog. In both groups of patients, somatic inactivation is frequently associated with loss of heterozygosity (LOH) at the TSG locus and at flanking loci.1,2 Defining the minimal chromosomal region with LOH in a collection of tumors has been a successful strategy for mapping and cloning TSGs. LOH can occur by multiple mechanisms as demonstrated by the extensive analyses of retinoblastomas, which identified RB1 intragenic deletions, segmental chromosomal deletions, loss of the entire chromosome, or mitotic recombination.2-4 While hundreds of TSGs have been identified, LOH studies typically focus on the TSG and/or closely flanking loci. For most TSGs, remarkably little is known about the extent and underlying mechanism(s) of LOH in tumor tissues.
In this study, we sought to examine the extent and mechanism of LOH in myeloid leukemias from children affected with neurofibromatosis 1 (NF1). The gene responsible for this disorder, NF1 at chromosome band 17q11.2, encodes neurofibromin, a GTPase-activating protein that negatively regulates the biochemical activation of p21ras (Ras) family members (reviewed in Boguski and McCormick5 and Donovan et al6 ). Germline NF1 mutations cause NF1, a dominant familial cancer syndrome that affects about 1 in 4000 people. Clinical features of NF1 include neurocutaneous abnormalities, learning disabilities, and a predisposition to specific benign and malignant tumors (reviewed in Friedman and Riccardi7 ). Children with NF1 are at a markedly increased risk of developing myeloid malignancies, particularly juvenile myelomonocytic leukemia (JMML).8 JMML is an aggressive myeloproliferative disease (MPD) characterized by monocytosis, thrombocytopenia, splenomegaly, and by malignant infiltration of the skin, lymph nodes, lungs, liver, and other organs (reviewed in Arico et al9 and Emanuel et al10 ). Together, the biochemical activity of neurofibromin and the dominant cancer predisposition seen in affected persons suggests that NF1 functions as a TSG. Indeed, LOH at the NF1 locus occurs in JMML and in other NF1-associated cancers (reviewed in Side and Shannon11 ). Similarly, tumorigenesis in heterozygous Nf1 mutant mice is associated with loss of the wild-type Nf1 allele.12,13 Consistent with the Knudson model of familial cancer genes, tumors from individuals with familial NF1 invariably show loss of the allele inherited from the unaffected parent (reviewed in Side and Shannon11 ). Somatic intragenic NF1 mutations have been identified in primary neoplasms, (reviewed in Stephens14 ), providing compelling evidence that functional inactivation of NF1 is central to tumorigenesis. Deregulated Ras signaling has been reported in tumors from NF1 patients and Nf1 mutant mice. These data are consistent with the idea that the tumor suppressor function of NF1 is related to the ability of neurofibromin to negatively regulate Ras output (reviewed in Side and Shannon,11 Cichowski and Jacks,15 and Dasgupta and Gutmann16 ).
In the course of investigating the extent and mechanism of LOH at the NF1 locus in myeloid malignancies, we unexpectedly identified interstitial isodisomy for a large segment of chromosome 17 as a frequent underlying genetic mechanism. Remarkably, the LOH breakpoints clustered within centromeric and telomeric marker intervals in these leukemias. To our knowledge, this is the first report of interstitial isodisomy as a frequent mechanism of somatic TSG inactivation. These data have implications for uncovering novel TSGs and for understanding pathogenic mechanisms that contribute to the development of hematopoietic malignancies as well as solid tumors.
Patients, materials, and methods
Patients
Clinical descriptions, LOH, and mutation analyses of the NF1 gene have been reported for most of the patients.17-19 Selected demographic and laboratory data are summarized in Table 1. Additional patient characteristics from previous reports are in Table S1, available on the Blood website (see the Supplemental Materials link at the top of the online article). Study procedures involving human subjects were approved by the University of California at San Francisco (UCSF) Committee for Human Research. Informed consent was provided according to the Declaration of Helsinki.
NF1 gene dosage in bone marrows of children with NF1 and malignant myeloid disorders
. | . | . | . | Parental origin . | . | . | . | |
|---|---|---|---|---|---|---|---|---|
| Patient no. . | Sex . | Age at onset, mo . | Diagnosis . | NF1 mutation . | LOH at NF1 locus . | NF1 gene dosage value*† . | Predicted NF1 gene copy no.‡ . | |
| 1 | M | 9 | MPS | Paternal | Maternal | 0.99 | Disomy | |
| 2 | M | 10 | AML | Paternal | Maternal | 0.93 | Disomy | |
| 3 | M | 24 | Monosomy 7 | Unknown | Maternal | 0.96 | Disomy | |
| 4 | M | 14 | JMML | Maternal | Paternal | 0.86 | Disomy | |
| 5 | F | 30 | JMML | Maternal | Paternal | 0.94 | Disomy | |
| 6 | M | 10 | JMML | Maternal | Paternal | 0.87 | Disomy | |
| 7 | M | 5 | Monosomy 7 | Maternal | Paternal | 0.90 | Disomy | |
| 8 | F | 18 | MPS | Paternal | Maternal* | 0.85 | Disomy | |
| 9 | M | 60 | JMML | Maternal | Paternal | 0.57 | Monosomy | |
| 10 | M | 19 | Monosomy 7 | De novo | Maternal* | 0.48 | Monosomy | |
. | . | . | . | Parental origin . | . | . | . | |
|---|---|---|---|---|---|---|---|---|
| Patient no. . | Sex . | Age at onset, mo . | Diagnosis . | NF1 mutation . | LOH at NF1 locus . | NF1 gene dosage value*† . | Predicted NF1 gene copy no.‡ . | |
| 1 | M | 9 | MPS | Paternal | Maternal | 0.99 | Disomy | |
| 2 | M | 10 | AML | Paternal | Maternal | 0.93 | Disomy | |
| 3 | M | 24 | Monosomy 7 | Unknown | Maternal | 0.96 | Disomy | |
| 4 | M | 14 | JMML | Maternal | Paternal | 0.86 | Disomy | |
| 5 | F | 30 | JMML | Maternal | Paternal | 0.94 | Disomy | |
| 6 | M | 10 | JMML | Maternal | Paternal | 0.87 | Disomy | |
| 7 | M | 5 | Monosomy 7 | Maternal | Paternal | 0.90 | Disomy | |
| 8 | F | 18 | MPS | Paternal | Maternal* | 0.85 | Disomy | |
| 9 | M | 60 | JMML | Maternal | Paternal | 0.57 | Monosomy | |
| 10 | M | 19 | Monosomy 7 | De novo | Maternal* | 0.48 | Monosomy | |
Clinical descriptions, LOH, and NF1 mutation analyses have been reported previously for most of the patients.17-19
Data from this study employing quantitative PCR at NF1 gene segment
Measured in unfractionated bone marrow cells except in patients 3 and 9, for whom leukemic cells in peripheral blood were used
Predicted gene copy number based on NF1 gene dosage values. Validation of the assay on nontumor DNA demonstrated that NF1 disomy gave dosage values of 0.98 ± 0.08 SD, while monosomy gave values of 0.45 ± 0.04 SD. A range of 2 SD was used to approximate a 95% confidence interval. Therefore, values from 0.82 to 1.14 predicted disomy, and values from 0.37 to 0.53 predicted monosomy
NF1 gene dosage assay
This assay measures the copy number of NF1 exon 32 by quantitating polymerase chain reaction (PCR) amplicons relative to those of a competitively amplified disomic control locus. Validation of the assay on nontumor DNA demonstrated that NF1 disomy gave dosage values of 0.98 ± 0.08 SD, while monosomy gave values of 0.45 ± 0.04 SD (SD indicates 1 standard deviation). A range involving 2 SD was used to predict gene copy number (Table 1, ‡ footnote). Briefly, the assay is a quantitative, competitive PCR adapted from the method of Celi et al.20 Assay conditions are available upon request.
FISH
Metaphase chromosome preparations of immortalized lymphoblastoid cells from patient 1 were prepared and hybridized as described previously.21 The bacteriophage P1 probe P1-12 contains approximately 55 kb of sequence from NF1 intron 27b.21 Bacterial artificial chromosome (BAC) clone 1000G21 (17q25) was identified by hybridization of D17S928 amplicons to filter arrays of the RPCI-11 human male BAC library, segment 4 (Roswell Park Cancer Institute, Buffalo, NY). Hybridization signals were detected using a commercial system (Vector Laboratories, Burlingame, CA). Chromosomes were banded using Hoechst 33258-actinomycin D staining, counterstained with propidium iodide, and signals visualized by fluorescence microscopy using a dual-band pass filter (Omega, Brattleboro, VT). Images in Figure 2A-B were visualized using a Zeiss Axioskop 2 plus microscope (Carl Zeiss, Thornwood, NY) equipped with a 100×/1.2 numeric aperture (NA) oil objective. Images were acquired using Spot software version 3.5.9 (Diagnostic Instruments, Sterling Heights, MI) and were processed using Adobe Photoshop software (Adobe Systems, San Jose, CA).
Cryopreserved bone marrow samples were thawed and cultured at 1 × 106 cells/mL for 24 hours (90% RPMI 1640/10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 mM HEPES) at 37°C in 95% air/5% CO2. Following incubation, the cells were exposed to hypotonic KCl (0.75 M for 8 minutes at 37°C), fixed in absolute methanol-glacial acetic acid (3:1), and air-dried on slides. NF1 probes were P1 bacteriophage clone P1-9, which spans approximately 65 kb of the NF1 gene, including exons 2-11, and clone P1-12.21 Centromere-specific probes for chromosomes 7 and 17 (CEP7-Spectrum Green and CEP17-Spectrum Green; Abbott Molecular, Abbott Park, IL), and the P1-derived artificial chromosome (PAC) clone P263P1 (Genome Systems, St Louis, MO), were hybridized as controls. P263P1 was isolated by screening the PAC library using primers for D5S479, and contains an insert of 70 kb derived from 5q31. Labeled probes were prepared by nick-translation using Bio-11-dUTP (Enzo Diagnostics, New York, NY) or digoxigenin-11-dUTP (Boehringer Mannheim, Indianapolis, IN). Interphase fluorescence in situ hybridization (FISH) was performed as described previously.22 Hybridization of probes labeled with either biotin or digoxigenin was detected with fluorescein-conjugated avidin (Vector Laboratories) and rhodamine-conjugated antidigoxigenin antibodies (Boehringer Mannheim), respectively. Nuclei were counterstained with DAPI. The slides were randomized and examined by 2 observers in a blinded fashion, with 500 cells scored by each observer for each probe. We established control values by hydridizing the probes to cryopreserved bone marrow cells from patients in remission (C1, AML-M4) or with myeloid leukemias that retained heterozygosity at NF1 (C2-C5 in Table S2). The cutoff value was set as the mean ± 3 SD.
The distribution of hybridization signals per nucleus for the CEP17 probe was determined in bone marrow cells from healthy control individuals (n = 10; Table S2).
Mapping the LOH region
Polymorphic loci were genotyped by PCR. LOH at the NF1 locus was evaluated by PCR analysis of at least 1 informative intragenic site including exon 5, intron 27B AluI/AluII, and intron 38. LOH was determined by comparing the genotype of the patient's tumor DNA to that of peripheral blood DNA of the patient's parents. For patients 1, 4, 5, 9, and 10, normal tissue or an Epstein-Barr virus (EBV)-transformed cell line was available to confirm a constitutional genotype with biparental inheritance of NF1 alleles.18,23 Segregation of alleles from parents to child for multiple informative loci on autosomes other than 17 was consistent with parentage as stated for each case (data not shown). Physical distances between chromosome 17 loci are based on the May 2004 assembly of the human genome (http://genome.ucsc.edu).
Results
Delineation of a large region of isodisomy in an NF1-associated MPD
Previous molecular analysis of bone marrow cells from children with NF1 revealed LOH at NF1 in CD34+ cells in 3 informative cases, whereas lymphoblasts immortalized by EBV retained heterozygosity in 2 of these patients.18 The remaining child with LOH at NF1 in EBV-transformed lymphoblasts was a 9-month-old boy with an unusual MPD and loss of the maternal NF1 allele (Table 1, patient 1).18 The retained paternal allele carried a de novo R1276X mutation (Table S1) that encoded a truncated protein lacking the GTPase-activating protein (GAP) domain.19 We performed extended LOH analyses of chromosome 17, which demonstrated loss of maternally derived alleles from D17S975 at 17q11.2 to D17S1830 at 17q25.3, a segment of 50.3 Mb (Figure 1). Although a deletion of this size is readily detected by cytogenetic techniques, the bone marrow and lymphoblastoid cells of patient 1 had a normal 46, XY karyotype. These data suggested that the 50.3-Mb LOH region was not deleted, but present on both chromosome 17 homologs. To address this possibility, the lymphoblastoid cells were analyzed by FISH with an NF1 intron 27b probe.21 Hybridization signals were detected on both chromosome 17 homologs (Figure 2A). Together, these data suggest that a 50.3-Mb interstitial interval of the maternal chromosome had been replaced with a homologous paternal DNA segment, resulting in homozygosity for the mutant R1276X NF1 allele (Figure 1). FISH with a BAC harboring the D17S928 locus confirmed that the heterozygous 17qter segment had not translocated elsewhere in the genome of the leukemic clone (Figure 2B). Taken together, these data confirmed interstitial isodisomy.
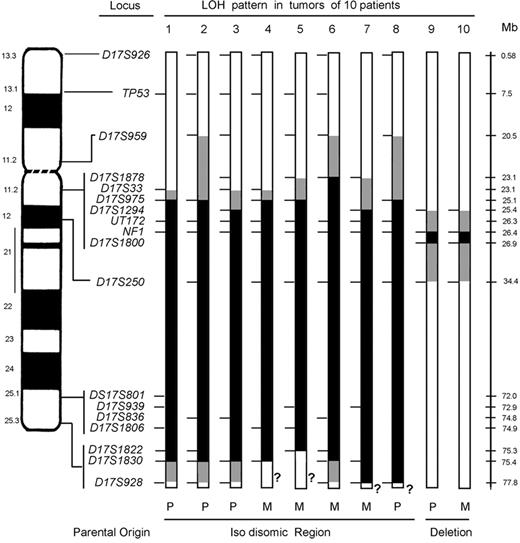
LOH at chromosome 17 loci in NF1-associated myeloid malignancies. Ideogram and schematic of chromosome 17 showing the loci that were screened for LOH. For each tumor, a bar shows the single chromosome 17 that underwent LOH with informative loci (tick marks), segments showing biparental inheritance (□), segments that underwent LOH (▪), segments where a recombination event occurred ( ), and qter segments that lacked informative loci (?). Below the schematic, the parental origin is given for the isodisomic and deleted regions for each tumor. For patients 1, 2, 3, and 8, the maternal homolog is shown with the region of paternal isodisomy indicated in black. For patients 4 through 7, the paternal homolog is shown with the region of maternal isodisomy indicated in black. LOH in patients 9 and 10 occurred by intrachromosomal deletion indicated in black. Additional chromosome 17 loci that were tested, but not informative, are not shown. Physical distances (rounded to the nearest tenth of a megabase) are based on the May 2004 assembly of the human genome (http://genome.uscs.edu), in which the length of chromosome 17 is given as 78 774 742 bp.
), and qter segments that lacked informative loci (?). Below the schematic, the parental origin is given for the isodisomic and deleted regions for each tumor. For patients 1, 2, 3, and 8, the maternal homolog is shown with the region of paternal isodisomy indicated in black. For patients 4 through 7, the paternal homolog is shown with the region of maternal isodisomy indicated in black. LOH in patients 9 and 10 occurred by intrachromosomal deletion indicated in black. Additional chromosome 17 loci that were tested, but not informative, are not shown. Physical distances (rounded to the nearest tenth of a megabase) are based on the May 2004 assembly of the human genome (http://genome.uscs.edu), in which the length of chromosome 17 is given as 78 774 742 bp.
LOH at chromosome 17 loci in NF1-associated myeloid malignancies. Ideogram and schematic of chromosome 17 showing the loci that were screened for LOH. For each tumor, a bar shows the single chromosome 17 that underwent LOH with informative loci (tick marks), segments showing biparental inheritance (□), segments that underwent LOH (▪), segments where a recombination event occurred ( ), and qter segments that lacked informative loci (?). Below the schematic, the parental origin is given for the isodisomic and deleted regions for each tumor. For patients 1, 2, 3, and 8, the maternal homolog is shown with the region of paternal isodisomy indicated in black. For patients 4 through 7, the paternal homolog is shown with the region of maternal isodisomy indicated in black. LOH in patients 9 and 10 occurred by intrachromosomal deletion indicated in black. Additional chromosome 17 loci that were tested, but not informative, are not shown. Physical distances (rounded to the nearest tenth of a megabase) are based on the May 2004 assembly of the human genome (http://genome.uscs.edu), in which the length of chromosome 17 is given as 78 774 742 bp.
), and qter segments that lacked informative loci (?). Below the schematic, the parental origin is given for the isodisomic and deleted regions for each tumor. For patients 1, 2, 3, and 8, the maternal homolog is shown with the region of paternal isodisomy indicated in black. For patients 4 through 7, the paternal homolog is shown with the region of maternal isodisomy indicated in black. LOH in patients 9 and 10 occurred by intrachromosomal deletion indicated in black. Additional chromosome 17 loci that were tested, but not informative, are not shown. Physical distances (rounded to the nearest tenth of a megabase) are based on the May 2004 assembly of the human genome (http://genome.uscs.edu), in which the length of chromosome 17 is given as 78 774 742 bp.
Uniparental isodisomy is a frequent mechanism of LOH in NF1-associated leukemias
The unexpected findings in patient 1 prompted us to use a quantitative NF1 gene dosage PCR assay to assess NF1 copy numbers in 9 additional NF1-associated leukemia specimens with LOH at NF1.17-19 Bone marrow DNA from patient 1 (Table 1) and from the normal tissues of his parents (data not shown) gave NF1 gene dosage values ranging from 0.91 to 1.08, which are consistent with disomy. Surprisingly, the NF1 dosage values for 7 of the 9 remaining NF1-associated myeloid malignancies were also consistent with disomy, whereas 2 cases had values consistent with monosomy (Table 1). Cryopreserved bone marrow specimens were available from 5 of these patients for copy number confirmation. FISH analyses using a chromosome 17 centromere-specific probe (Cep17) and 2 NF1 probes (P1-9 and P1-12) provided physical confirmation that the leukemias of all 4 cases in which the dosage assay predicted disomy contained 2 NF1 alleles (Figure 2D; patients 3-6 in Table S2). By contrast, the bone marrow of patient 10 demonstrated monosomy for NF1 with 2 signals in 67% and 1 signal in 29% of cells (Figure 2D and Table S2), which was also consistent with the gene dosage assay (Table 1). To confirm that the cells being examined were from the malignant clone with LOH at NF1, a chromosome 7-specific probe (Cep7) was hybridized to bone marrow cells of patient 3, who had monosomy 7 and was disomic at NF1 as measured by both gene dosage and FISH (Table 1, Figure 2D, Table S2). As expected, dual-color FISH revealed monosomy 7 in cells that also had 2 structural NF1 alleles (Figure 2C-D and Table S2). Together these studies demonstrate that LOH at NF1 in myeloid malignancies is preferentially associated with isodisomy of a chromosomal segment carrying the mutant NF1 allele.
FISH analysis of NF1-associated myeloid malignancies. (A-B) Metaphase spreads of EBV-transformed cells from patient 1 that were hybridized with NF1 probe P1-12 (A) and BAC clone 1000G21, which contains the D17S928 locus at 17q25 (B). Each of 20 metaphase cells examined showed signals on both chromosome 17 homologs, consistent with disomy. The chromosome 17 homologs were identified by Hoechst-actinomycin D staining, which reveals a Q-banding-like pattern. (C) Dual-color FISH performed by cohybridizing a digoxigenin-labeled probe P1-12 (rhodamine signal) and an α-satellite probe specific for the centromere of chromosome 7 (CEP7, SpectrumGreen), which showed monosomy 7 and NF1 disomy in bone marrow cells from patient 3. Interphase nuclei were counter-stained with DAPI, and the slides mounted with PDD antifade solution. Images were visualized using a Zeiss Axioplan microscope equipped with a 63×/1.25 NA oil Plan Neofluar lens, Optivar setting 1.6. Images were acquired using a Photometrics cooled charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) and NIH Image software (National Institutes of Health, Bethesda, MD), and were processed using Adobe Photoshop. (D) Graphic summary of interphase FISH analyses of myeloid leukemia cells with NF1 (red triangle) and control probes. Probe 263P1 is a 70-kb PAC clone containing D5S479 (chromosome band 5q31; yellow squares). CEP17 is a centromere-specific probe for chromosome 17 (green circles), and CEP7 is a centromere-specific probe for chromosome 7 (blue diamond). C1 is a cryopreserved bone marrow sample from a patient with AML-M4 in complete remission. Control samples C2 through C5 are cryopreserved bone marrow samples from 4 children with myeloid leukemias that retained heterozygosity at the NF1 locus. The mean distribution of signals for the chromosome 17 centromere-specific probe was determined by the interphase analysis of bone marrow cells from 10 healthy individuals. This graph is a summary of data given in Table S2.
FISH analysis of NF1-associated myeloid malignancies. (A-B) Metaphase spreads of EBV-transformed cells from patient 1 that were hybridized with NF1 probe P1-12 (A) and BAC clone 1000G21, which contains the D17S928 locus at 17q25 (B). Each of 20 metaphase cells examined showed signals on both chromosome 17 homologs, consistent with disomy. The chromosome 17 homologs were identified by Hoechst-actinomycin D staining, which reveals a Q-banding-like pattern. (C) Dual-color FISH performed by cohybridizing a digoxigenin-labeled probe P1-12 (rhodamine signal) and an α-satellite probe specific for the centromere of chromosome 7 (CEP7, SpectrumGreen), which showed monosomy 7 and NF1 disomy in bone marrow cells from patient 3. Interphase nuclei were counter-stained with DAPI, and the slides mounted with PDD antifade solution. Images were visualized using a Zeiss Axioplan microscope equipped with a 63×/1.25 NA oil Plan Neofluar lens, Optivar setting 1.6. Images were acquired using a Photometrics cooled charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) and NIH Image software (National Institutes of Health, Bethesda, MD), and were processed using Adobe Photoshop. (D) Graphic summary of interphase FISH analyses of myeloid leukemia cells with NF1 (red triangle) and control probes. Probe 263P1 is a 70-kb PAC clone containing D5S479 (chromosome band 5q31; yellow squares). CEP17 is a centromere-specific probe for chromosome 17 (green circles), and CEP7 is a centromere-specific probe for chromosome 7 (blue diamond). C1 is a cryopreserved bone marrow sample from a patient with AML-M4 in complete remission. Control samples C2 through C5 are cryopreserved bone marrow samples from 4 children with myeloid leukemias that retained heterozygosity at the NF1 locus. The mean distribution of signals for the chromosome 17 centromere-specific probe was determined by the interphase analysis of bone marrow cells from 10 healthy individuals. This graph is a summary of data given in Table S2.
Clustering and parental origin of chromosome 17 LOH breakpoints
Each of the 8 leukemias with isodisomy at NF1 showed a large segment of LOH that minimally ranged from 50 to 52.7 Mb (Figure 1). Among these cases, both the proximal and distal LOH breakpoints were clustered. In all 8, the centromeric breakpoints mapped to a maximum interval of 4.9 Mb (6% of the chromosome 17 length of 78.77 Mb) between D17S959 and D17S1294. In some leukemias, additional informative markers narrowed the breakpoint interval, as in patient 5 with a 2-Mb interval between D17S1878 and D17S975. With the exception of case 6, the minimum common breakpoint region is 2 Mb and is delineated by D17S33 and D17S975. The distal breakpoints in patients 1, 2, 3, and 6 were clustered between D17S1830 and D17S928, an interval of 2.4 Mb (Figure 1). Lack of informativeness at D17S928 and other 17qter loci tested precluded determining whether the large LOH segments in the tumors of patients 4, 5, 7, and 8 were isodisomic.
The bone marrows of patients 9 and 10 had LOH at loci flanking the NF1 region. The leukemias of these 2 patients showed structural deletions that included the NF1 locus as determined by gene dosage (Table 1) and FISH data (Figure 2D; Table S2). The centromeric breakpoints mapped to a 1.2-Mb interval between D17S1294 and NF1 intron 38 and the telomeric breakpoints were in a 7.5-Mb interval between D17S1800 and D17S250. These data are consistent with an interstitial deletion ranging from 268 kb (NF1 ex38 to D17S1800) to 9 Mb (D17S1294 to D17S250).
The bone marrows of patients 1, 2, 3, and 8 lost chromosome 17 maternal alleles and were isodisomic for a paternally derived DNA segment, whereas the cells of patients 4 through 7 lost paternal alleles and were isodisomic for a maternally derived interval of comparable length (Figure 1). In each patient with familial NF1, the isodisomic segment was derived from the parent with NF1 (Tables 1, S1). The bone marrow of patient 10, who had de novo NF1, showed loss of the maternal NF1 allele (Table 1; Figure 1B). These data infer that patient 10 carried a germline mutation of the paternal NF1 allele and underwent somatic deletion of the normal maternal NF1 allele, which is consistent with the reported parental predisposition for NF1 germline mutations.24,25
Discussion
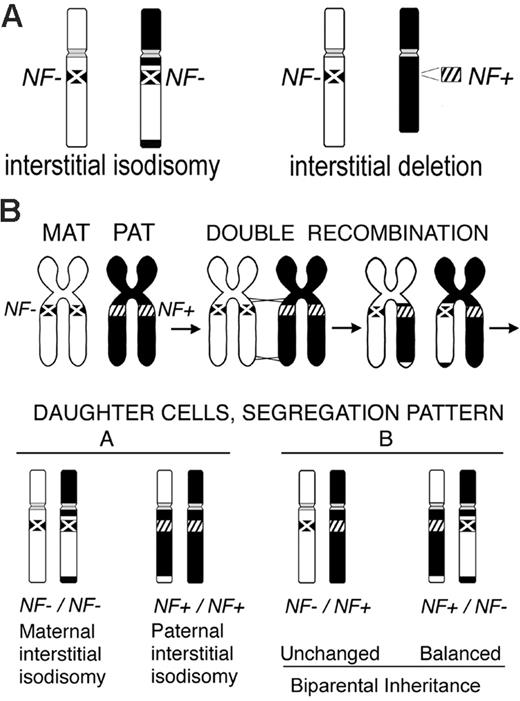
Our analyses of myeloid malignancies from children with NF1 uncovered 2 distinct mechanisms underlying inactivation of the normal NF1 allele: interstitial isodisomy and interstitial deletion (Figure 3A). The somatic interstitial deletions involving NF1 are of interest as they may occur by a mechanism similar to that identified in constitutional and somatic mosaic NF1 microdeletions in normal (nontumor) tissues. In these cases, the deletions are 1.2 to 1.4 Mb and occur by nonallelic homologous recombination between pairs of high-identity, low copy number repeat (LCR) elements that flank the NF1 gene (reviewed in Stephens26 ). Microdeletions mediated by germline or somatic recombination between different LCR pairs27,28 both involve the entire NF1 gene and the D17S1800 locus, which are deleted in the tumors of patients 9 and 10. A mechanism of LCR-mediated recombination could be tested by precise mapping of the deletion breakpoints by single nucleotide polymorphism (SNP) mapping in the myeloid malignancies. Alternatively, the length of these somatic microdeletions in myeloid malignancies may be constrained if a 50% reduction in the expression of a critical flanking gene(s) inhibited outgrowth of the leukemic clone.
While segmental or interstitial uniparental isodisomy has been reported in constitutional rearrangements (reviewed in Vianna-Morgante29 ), we were surprised to find that interstitial isodisomy for 50 to 52.7 Mb of chromosome 17 is a common mechanism underlying LOH in NF1-associated myeloid malignancies. The leukemias of each of the 4 patients (1, 2, 3, and 6 in Figure 1) in whom D17S928 was informative had interstitial isodisomy. The remaining 4 tumors (4, 5, 7, and 8 in Figure 1) may also be interstitial, but D17S928 or other regional markers were not informative. We propose that interstitial isodisomy results from a double mitotic recombination event between chromatids of the 2 chromosome 17 homologs during the S/G2 phase of the cell cycle of a leukemia-initiating cell (Figure 3B). Depending upon the segregation pattern during mitosis, the daughter cells would have biparental inheritance at all chromosome 17 loci, or alternatively, would show interstitial isodisomy (Figure 3B). Only 1 of 4 possible daughter cells would have interstitial isodisomy along with homozygous inactivation of NF1, the latter of which is presumably essential for leukemic outgrowth.30,31 The low frequency of double-mitotic recombination events, estimated at about 10-10 in normal lymphocytes,32 infers that NF1 inactivation confers a strong proliferative advantage in the leukemia-initiating cell. A possible mechanism may involve nonallelic mitotic homologous recombination between LCRs or Alu elements, which has been implicated in recurring translocations, isochromosomes, deletions, and amplifications in tumor tissues.33-35 Other mechanisms that could give rise to 17q interstitial isodisomy are less likely. For example, 2 sequential single recombination events in different precursor cells are also possible, but would imply that each independent event conferred a proliferative advantage. A gene conversion-like event of a 50-Mb segment would be unprecedented as estimated conversion tracts in humans are typically less than 2 kb.36,37 The clustered breakpoint intervals of the isodisomic segments suggest that mitotic recombination may be favored in these regions.
LOH in NF1-associated myeloid malignancies and proposed mechanism of interstitial isodisomy. (A) The schematic depicts the 2 different patterns of LOH observed in the tumors. The inactivated NF1 allele (NF-) is marked with an X on the chromosome, while the normal NF1 allele (NF+) is indicated by diagonal hashmarks (////). The interstitial isodisomic and deleted regions can be of maternal or paternal origin. (B) Proposed mechanism for double mitotic recombination during the S/G2 phase of the cell cycle leading to interstitial uniparental isodisomy in a leukemic-initiating cell. The 4 possible daughter cells are depicted, along with their NF1 genotypes and disomy patterns. Although this example depicts a cell with maternal interstitial isodisomy and NF1 inactivation, paternal interstitial isodisomy was also observed in our study (Figure 1).
LOH in NF1-associated myeloid malignancies and proposed mechanism of interstitial isodisomy. (A) The schematic depicts the 2 different patterns of LOH observed in the tumors. The inactivated NF1 allele (NF-) is marked with an X on the chromosome, while the normal NF1 allele (NF+) is indicated by diagonal hashmarks (////). The interstitial isodisomic and deleted regions can be of maternal or paternal origin. (B) Proposed mechanism for double mitotic recombination during the S/G2 phase of the cell cycle leading to interstitial uniparental isodisomy in a leukemic-initiating cell. The 4 possible daughter cells are depicted, along with their NF1 genotypes and disomy patterns. Although this example depicts a cell with maternal interstitial isodisomy and NF1 inactivation, paternal interstitial isodisomy was also observed in our study (Figure 1).
Our data are intriguing in light of recent reports showing JAK2 point mutations in most patients with polycythemia vera (PV) and in some cases of essential thrombocythemia (ET) and chronic idiopathic myelofibrosis (CIMF).38-41 An unexpected and intriguing result of these studies was the finding of biallelic mutations in approximately 30% of the PV specimens. The underlying genetic mechanism in these cases was a mitotic recombination that led to loss of the normal JAK2 allele and resulted in isodisomy for a segment of the short arm of chromosome 9 estimated to span approximately 40 cM.42 The most telomeric marker studied showed LOH in many cases, which is consistent with a single recombination event. We did not prove that the isodisomic regions were interstitial in patients 4, 5, 7, and 8 due to a lack of informative polymorphic markers near 17qter (Figure 1), and it is therefore possible that isodisomy resulted from a single recombination event in 1 or more of our cases. Similarly, it is possible that studies with additional markers near 9pter would uncover double-mitotic recombination events in some PV samples. Whereas biallelic NF1 inactivation deregulates Ras signaling in response to hematopoietic growth factors, it is less obvious why loss of the normal JAK2 allele and isodisomy of the mutant homolog would confer a growth advantage beyond that of a dominant heterozygous mutation. Since JAK2 molecules that are recruited to activated growth factor receptors transphosphorylate each other, it is possible that the normal protein has a dominant interfering activity that impairs the ability of mutant JAK2 to deregulate downstream effectors. Consistent with this idea, James et al38 found that coexpressing wild-type and mutant JAK2 proteins restored erythropoietin-dependence in the Ba/F3 pro-B-cell line.
A broad implication of our work and of the recent studies of JAK2 mutations in MPD is that segmental uniparental isodisomy may be a frequent but unrecognized mechanism in human cancers. Although isodisomy of a 25-cM interval was first described in children with Down syndrome who developed acute lymphoblastic leukemia by Rogan et al,43 this genetic mechanism has received limited attention in hematologic malignancies until recently. The availability of automated allelotyping and the use of SNP and high-density arrays have been developed for high-resolution analysis of allelic losses and gains in tumors.44-46 Interestingly, a number of investigators are now identifying regions of isodisomy in acute myeloid leukemia.47-49 Our data extend these studies by showing that inactivation of a known myeloid TSG is frequently associated with acquired uniparental disomy. Importantly, DNA segments that are associated with partial or interstitial isodisomy will appear normal when examined by conventional cytogenetic analysis, FISH, or comparative genomic hybridization, making these approaches of limited use for cancers where LOH results in isodisomy. Together, LOH and copy-number analyses provide the opportunity to define new genetic mechanisms of somatic mutation, mitotic recombination sites, putative modifying or imprinted genes, and/or correlations between tumor genotype and neoplastic transformation. In addition, mono- or biallelic expression from a locus (loci), other than the TSG itself, could affect the efficacy of putative therapeutic agents.
Prepublished online as Blood First Edition Paper, May 11, 2006; DOI 10.1182/blood-2005-11-011486.
Supported by U.S. Army Medical Research and Materiel Command grants DAMD17-97-1-7344 and DAMD17-03-1-0203 (K.S.), and National Institutes of Health grants PO1 CA40046 (M.M.L., K.M.S.), R01 CA72614 (K.M.S.), and K24 CA80916 (P.D.E.).
K.S. designed research, generated primary data and performed analyses in her laboratory, and wrote the paper; M.W. performed genotyping experiments in K.S.'s lab; K.A.L. generated and analyzed primary cytogenetic data; K.M. developed gene dosage assay and performed experiments in K.S.'s lab; P.D.E. contributed well-characterized patient samples; M.M.L.B. helped to design research, generated primary data on interphase FISH, and performed analysis in her laboratory; K.M.S. helped to design research, identified LOH in NF1-associated leukemias, and assisted in writing the paper.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Virginia P. Sybert and Eric Sievers for referring patient 1, Melvin H. Freedman for bone marrow samples, Michael Dorschner for BAC1000G21, and Elizabeth M. Davis and Rafael Espinosa III for technical assistance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal