Abstract
Hemolytic uremic syndrome (HUS) is a thrombotic microangiopathy with manifestations of hemolytic anemia, thrombocytopenia, and renal impairment. Genetic studies have shown that mutations in complement regulatory proteins predispose to non–Shiga toxin–associated HUS (non-Stx–HUS). We undertook genetic analysis on membrane cofactor protein (MCP), complement factor H (CFH), and factor I (IF) in 156 patients with non-Stx–HUS. Fourteen, 11, and 5 new mutational events were found in MCP, CFH, and IF, respectively. Mutation frequencies were 12.8%, 30.1%, and 4.5% for MCP, CFH, and IF, respectively. MCP mutations resulted in either reduced protein expression or impaired C3b binding capability. MCP-mutated patients had a better prognosis than CFH-mutated and nonmutated patients. In MCP-mutated patients, plasma treatment did not impact the outcome significantly: remission was achieved in around 90% of both plasma-treated and plasma-untreated acute episodes. Kidney transplantation outcome was favorable in patients with MCP mutations, whereas the outcome was poor in patients with CFH and IF mutations due to disease recurrence. This study documents that the presentation, the response to therapy, and the outcome of the disease are influenced by the genotype. Hopefully this will translate into improved management and therapy of patients and will provide the way to design tailored treatments.
Introduction
Hemolytic uremic syndrome (HUS) is a rare disease with manifestations of microangiopathic hemolytic anemia, thrombocytopenia, and renal impairment.1 In most cases HUS is triggered by Shiga toxin (Stx)–producing Escherichia coli (Stx-HUS)2 and manifests with watery or bloody diarrhea. Approximately half of the patients require dialysis during the acute episode, but renal function recovers in most of them.1,3
Non–Shiga toxin–associated HUS (non-Stx–HUS) is rare and comprises a heterogeneous group of patients in whom an infection by Stx-producing bacteria can be excluded as a cause of disease. It may be sporadic or familial. The clinical outcome is unfavorable with up to 50% of cases progressing to end-stage renal failure (ESRF) and 25% dying during the acute phase.4,5
Clustering of affected individuals within families suggested a genetic predisposition to the disease. Both autosomal dominant and recessive transmission have been reported,6 with precipitating events such as pregnancy, viruslike disease, or sepsis identified in some cases.7,8 Persistent and remarkably depressed levels of the third component (C3) of the complement system have been documented in some patients with non-Stx–HUS,9-11 suggesting the presence of an inherited defect causing hyperactivation of the complement cascade. Indeed, recent genetic studies have shown that mutations in genes encoding complement regulatory proteins12-26 predispose to the development of non-Stx–HUS.
More than 50 different mutations in complement factor H (CFH), a plasma protein that inhibits the activation of the alternative pathway of complement, have been described in non-Stx–HUS.12-17,23,24 The majority of them are heterozygous and cause either single–amino acid substitutions or premature translation interruption within the protein C-terminus, where binding sites for C3b/3d and heparin have been mapped.27
More recently, 4 mutations in membrane cofactor protein (MCP), a surface-bound complement regulator that degrades both C3b and C4b on host cells, have been reported in patients with non-Stx–HUS.19,20,26 Finally, 6 mutations in the gene encoding factor I (IF), a circulating serine protease that inactivates cell-bound C3b to iC3b, have been reported in patients with sporadic18,25 and familial26 non-Stx–HUS.
In this study MCP, CFH, and IF genetic analyses were undertaken in a large number of non-Stx–HUS patients referred to our International Registry of HUS/thrombotic thrombocytopenic purpura (TTP) with the following aims: (1) to establish the frequencies of mutations in the 3 genes and the relative penetrance of the syndrome; and (2) to analyze the clinical phenotypes of patients with MCP, CFH, or IF mutations to establish correlates between diverse genetic changes and clinical presentation, response to therapy, and outcome.
Patients, materials, and methods
Patients
One hundred fifty-six patients with a diagnosis of non-Stx–HUS were recruited through the database of the International Registry of Recurrent and Familial HUS/TTP. HUS was diagnosed in all cases reported to have one or more episodes of microangiopathic hemolytic anemia and thrombocytopenia defined on the basis of hematocrit (Ht) less than .3 (30%), hemoglobin (Hb) level less than 100 g/L (10 g/dL), serum lactate dehydrogenase (LDH) level greater than 460 U/L, undetectable haptoglobin level, fragmented erythrocytes in the peripheral blood smear, and platelet count less than 150 × 109/L (150 000/μL), associated with acute renal failure. Patients with Stx-HUS, defined as the presence of Shiga toxin in the stools (by the Vero cell assay) and/or of serum antibodies against Shiga toxin (by enzyme-linked immunosorbent assay [ELISA]) and/or LPS (O157, O26, O103, O111, and O145, by ELISA), were excluded. Familial non-Stx–HUS was diagnosed when 2 or more members of the same family were affected by the disease at least 6 months apart and exposure to a common triggering infectious agent was excluded. Sporadic non-Stx–HUS was diagnosed when one or more episodes of the disease manifested in a subject with no familial history of the disease. No patient included in the study had HIV-associated HUS. All patients were white (70% from Italy, 12% from other European countries, 12% from the United States), with the exception of the previously published12 Bedouin family comprising 10 affected subjects (6%). Healthy controls, matched for sex and geographic origin (100 from Europe, 20 from the United States), were also recruited. All participants provided informed written consent. The Institutional Review Board of the Mario Negri Institute approved the protocol.
Single-strand conformation polymorphism, denaturing HPLC, and sequencing
Genomic DNA was extracted from peripheral blood leukocytes (Nucleon BACC2 kit; Amersham, Little Chalfont, United Kingdom). The coding sequence and the intronic flanking regions of MCP and CFH were screened by polymerase chain reaction–single-strand conformation polymorphism method (PCR-SSCP). The experimental conditions were optimized for each exon by testing different PCR amplicons and different electrophoretic conditions. DNA samples from subjects with known mutations or polymorphisms were run in parallel to check for variations in sensitivity and specificity. IF was screened by denaturing high-performance liquid chromatography (DHPLC; WAVE DNA Fragment Analysis System, model MD4000plus; Transgenomics, Cedex, France) following the manufacturer's instructions and previously published methods.28 Each analysis was performed at 3 different denaturing temperatures.
Primers were synthesized by Sigma Genosys LTD (Sigma-Aldrich House, Haverhill, United Kingdom). The sequences of primers for CFH and IF screening have been already published,13,18,25 and the primers for MCP screening are shown in Table 1. For the first 5 exons, primers were constructed to avoid coamplification of MCP-like genes. PCR reactions, gel electrophoresis in nondenaturing conditions, and staining were performed as previously described.13 DNA from subjects showing aberrant bands were sequenced using a CEQ 8000 XL sequencer (Beckman Coulter, Berkeley, CA).
Sequences of primers used for MCP genetic screening
Exon . | Forward primer . | Reverse primer . |
|---|---|---|
| I | 5′-ctgtcctgcagcactggatg-3′ | 5′-cacggcctgctgtgagc-3′ |
| II | 5′-acttcatcttcatgttcctattctcttatc-3′ | 5′-acaagaagaaaatcatcatcaccg-3′ |
| III | 5′-aattatattcccacccattcaaaaga-3′ | 5′-ttcccttatttcctctaaggagca-3′ |
| IV | 5′-ccaccccctcaaactactgtagtg-3′ | 5′-agaaacctctttgggatctttgtta-3′ |
| V | 5′-tgtcttaatcttttacatttcctttcctct-3′ | 5′-cacatacacctgctttgtttatctgt-3′ |
| VI | 5′-cttgtctctgttcacactggaaattact-3′ | 5′-cagcaacaacaataacaaaccaaga-3′ |
| VII + VIII | 5′-cccaagtggttgatcttctaacatt-3′ | 5′-agcaggaaattactaaacctgaggc-3′ |
| IX | 5′-ttgataaggccctggtgaattt-3′ | 5′-cctgcacgctgtgcaca-3′ |
| X | 5′-ccctatgagtttaaaggattttaagctt-3′ | 5′-cctatgtttgggcacctcataa-3′ |
| XI | 5′-ggagatccatgtgttcaacatctt-3′ | 5′-tcggtttaaccaatttacaagctg-3′ |
| XII | 5′-ttgaccactgaaatgtaaccaaca-3′ | 5′-tgaagctgcacaaaagcatgt-3′ |
| XIII | 5′-tcgtttctttttggtttgaagtca-3′ | 5′-gccaatatctctttgctcaggttat-3′ |
| XIV | 5′-tcattttctgaataggcttctggaat-3′ | 5′-gtcaaaagatgaactggcaaacc-3′ |
Exon . | Forward primer . | Reverse primer . |
|---|---|---|
| I | 5′-ctgtcctgcagcactggatg-3′ | 5′-cacggcctgctgtgagc-3′ |
| II | 5′-acttcatcttcatgttcctattctcttatc-3′ | 5′-acaagaagaaaatcatcatcaccg-3′ |
| III | 5′-aattatattcccacccattcaaaaga-3′ | 5′-ttcccttatttcctctaaggagca-3′ |
| IV | 5′-ccaccccctcaaactactgtagtg-3′ | 5′-agaaacctctttgggatctttgtta-3′ |
| V | 5′-tgtcttaatcttttacatttcctttcctct-3′ | 5′-cacatacacctgctttgtttatctgt-3′ |
| VI | 5′-cttgtctctgttcacactggaaattact-3′ | 5′-cagcaacaacaataacaaaccaaga-3′ |
| VII + VIII | 5′-cccaagtggttgatcttctaacatt-3′ | 5′-agcaggaaattactaaacctgaggc-3′ |
| IX | 5′-ttgataaggccctggtgaattt-3′ | 5′-cctgcacgctgtgcaca-3′ |
| X | 5′-ccctatgagtttaaaggattttaagctt-3′ | 5′-cctatgtttgggcacctcataa-3′ |
| XI | 5′-ggagatccatgtgttcaacatctt-3′ | 5′-tcggtttaaccaatttacaagctg-3′ |
| XII | 5′-ttgaccactgaaatgtaaccaaca-3′ | 5′-tgaagctgcacaaaagcatgt-3′ |
| XIII | 5′-tcgtttctttttggtttgaagtca-3′ | 5′-gccaatatctctttgctcaggttat-3′ |
| XIV | 5′-tcattttctgaataggcttctggaat-3′ | 5′-gtcaaaagatgaactggcaaacc-3′ |
Microsatellite polymorphism genotyping and linkage analysis
Microsatellites D1S2735, D1S2796, and D1S2692 flanking MCP were studied in family no. 099 and in subject S222 212. PCR, gel electrophoresis in denaturing conditions, and staining were performed as previously described.13 Haplotypes were reconstructed including 2 single-nucleotide polymorphisms (SNPs; rs4844390, rs1111850; NCBI, Bethesda, MD).
mRNA extraction, cDNA synthesis, and analysis
The mRNA was extracted from peripheral blood mononuclear cells (PBMCs; standard protocol) of family no. 099 members. Reverse transcription–PCR (RT-PCR) was performed using a forward primer constructed on exon I (5′-GCGAGTGTCCCTTTCCTTCCT-3′) and a reverse primer on exon III (5′-AAAGTGCATCTGATAACCAAACTGG-3′) of MCP. The amplicons were sequenced either directly or after cloning in E coli.
Expression and functional studies on MCP mutants
Expression of MCP on PBMCs was evaluated by fluorescence-activated cell sorter (FACS) as described.19 PBMCs were incubated with a fluorescein isothiocyanate (FITC)–conjugated mouse anti–human monoclonal antibody (20 μL/106 PBMCs, clone E4.3 recognizing an epitope within short consensus repeat 1 [SCR1] of MCP; BD Biosciences Pharmingen, San Diego, CA) and analyzed by FACSort (BD Biosciences, Mountain View, CA). Samples incubated with FITC-conjugated mouse IgG1 were used as negative controls.
For mutagenesis and expression experiments, the MCP isoform BC1 was used as a template and transient transfections were performed in Chinese hamster ovary (CHO) cells, as previously reported.20 MCP expression in CHO cells was analyzed by FACS and Western blotting.20 MCP was quantified in cell lysates by ELISA.20 For functional assessments, C3b and C4b binding capability was evaluated by ELISA, as described.20
To model the IVS2 –2A>G mutation, we constructed a pET-MCP minigene. MCP genomic DNA (2843 bp) from unaffected individuals was amplified from intron 1 (IVS1 –62) to intron 4 (IVS4 +70) using primers introducing NotI restriction sites (Figure 1A). The PCR product was subcloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA). The IVS2 –2A>G mutant was produced by the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutant and wild-type DNA fragments were cloned into the exon trap cloning vector pET (MoBiTec, Goettingen, Germany). Transfections were performed into 293T cells and RNA was extracted 40 hours later. After RT-PCR, the cDNA products were separated on 1.5% agarose gel and subcloned into pCR2.1-TOPO for sequencing.
Statistical analysis
Differences in clinical and genetic data in patients with MCP or CFH mutations or with no mutations were analyzed by the chi-square test. The cumulative fraction of patients free of events (defined as the combination of the occurrence of chronic renal insufficiency or initiation of dialysis or death, whichever occurred first after the onset of HUS) was estimated by Kaplan-Meier analysis. Differences between groups were calculated by the log-rank test. The differences were considered statistically significant at P values of less than .05.
Results
MCP mutation screening
Results of mutation screening of 156 non-Stx–HUS patients (familial HUS, n = 58; sporadic HUS, n = 98) are summarized in Table 2 and in Figures 2 and 3. Fourteen independent mutational events in overall 20 white patients were found in MCP. Three patients (within one family) are compound heterozygotes, 2 patients from one family carry a homozygous mutation, the others are heterozygotes. Of note, the IVS1 –1G>C mutation has been counted once, as a founder effect was established by microsatellite analysis for family no. 099 and patient S222 212, who was of Sardinian origin (Figure 3). None of the mutations were found in any of 120 healthy white controls.
MCP gene mutations in non-Stx–HUS patients from our registry
Exon/intron, subject/family code . | Mutation . | SCR . | Effect . | Subgroups . | Inheritance . | Unaffected carriers . | Origin of the mutation . |
|---|---|---|---|---|---|---|---|
| Int 1 | See Figure 3 | ||||||
| F166 099 | IVS1-1G > C | NA | C1Stop | Familial | Homozygote | 4/5 | |
| F167 099 | IVS1-1G > C | NA | C1Stop | Familial | Homozygote | 4/5 | |
| F168 099 | IVS1-1G > C | NA | C1Stop | Familial | Heterozygote | 4/5 | |
| S222 212 | IVS1-1G > C | NA | C1Stop | Sporadic | Heterozygote | ND | |
| Ex II | |||||||
| S202 048 | 218C > T | 1 | R25Stop | Sporadic | Heterozygote | ND | Unknown |
| S019 085 | 218C > T | 1 | R25Stop | Sporadic | Heterozygote | ND | Unknown |
| S044 146 | 218C > T | 1 | R25Stop | Sporadic | Heterozygote | ND | Unknown |
| F106 024* | 218C > T | 1 | R25Stop | Familial | Heterozygote | 1/3 | Paternal |
| F108 024* | 218C > T | 1 | R25Stop | Familial | Heterozygote | 1/3 | Paternal |
| D9 024* | 218C > T | 1 | R25Stop | Familial | Heterozygote | 1/3 | Paternal |
| F106 024 | 147G > A | 1 | C1Y | Familial | Heterozygote | 1/3 | Maternal |
| F108 024 | 147G > A | 1 | C1Y | Familial | Heterozygote | 1/3 | Maternal |
| D9 024 | 147G > A | 1 | C1Y | Familial | Heterozygote | 1/3 | Maternal |
| S203 188 | 147G > A | 1 | C1Y | Sporadic | Heterozygote | ND | Unknown |
| S204 202 | 235T > C + (236-241)delA | 1 + 2 | 39 aa change + L72Stop | Sporadic | Heterozygote | 1/2 | Unknown |
| Ex III | |||||||
| S207 199 | 338T > C | 2 | C65R | Sporadic | Heterozygote | 2/3 | Paternal |
| Int 2 | |||||||
| S199 192† | IVS2-2A > G | 2 | (62-95)del + G96I + Y97I + Y98T + L99Stop | Sporadic | Heterozygote | ND | Unknown |
| Ex VI | |||||||
| F164 088‡ | 843-844delAC | 4 | T233fsStop236 | Familial | Heterozygote | 2/3 | Paternal |
| F165 088‡ | 843-844delAC | 4 | T233fsStop236 | Familial | Heterozygote | 2/3 | Paternal |
| S045 169 | 858-872del15bp + 875C > T | 4 | 238-242del + D243N + P244S | Sporadic | Heterozygote | ND | Unknown |
| F169 130* | 768T > G | 4 | F208C | Familial | Heterozygote | 5/19 | Maternal |
| F170 130* | 768T > G | 4 | F208C | Familial | Heterozygote | 5/19 | Maternal |
| Ex XI | |||||||
| S050 172 | 1056C > T | NA | A304V | Sporadic | Heterozygote | ND | Unknown |
Exon/intron, subject/family code . | Mutation . | SCR . | Effect . | Subgroups . | Inheritance . | Unaffected carriers . | Origin of the mutation . |
|---|---|---|---|---|---|---|---|
| Int 1 | See Figure 3 | ||||||
| F166 099 | IVS1-1G > C | NA | C1Stop | Familial | Homozygote | 4/5 | |
| F167 099 | IVS1-1G > C | NA | C1Stop | Familial | Homozygote | 4/5 | |
| F168 099 | IVS1-1G > C | NA | C1Stop | Familial | Heterozygote | 4/5 | |
| S222 212 | IVS1-1G > C | NA | C1Stop | Sporadic | Heterozygote | ND | |
| Ex II | |||||||
| S202 048 | 218C > T | 1 | R25Stop | Sporadic | Heterozygote | ND | Unknown |
| S019 085 | 218C > T | 1 | R25Stop | Sporadic | Heterozygote | ND | Unknown |
| S044 146 | 218C > T | 1 | R25Stop | Sporadic | Heterozygote | ND | Unknown |
| F106 024* | 218C > T | 1 | R25Stop | Familial | Heterozygote | 1/3 | Paternal |
| F108 024* | 218C > T | 1 | R25Stop | Familial | Heterozygote | 1/3 | Paternal |
| D9 024* | 218C > T | 1 | R25Stop | Familial | Heterozygote | 1/3 | Paternal |
| F106 024 | 147G > A | 1 | C1Y | Familial | Heterozygote | 1/3 | Maternal |
| F108 024 | 147G > A | 1 | C1Y | Familial | Heterozygote | 1/3 | Maternal |
| D9 024 | 147G > A | 1 | C1Y | Familial | Heterozygote | 1/3 | Maternal |
| S203 188 | 147G > A | 1 | C1Y | Sporadic | Heterozygote | ND | Unknown |
| S204 202 | 235T > C + (236-241)delA | 1 + 2 | 39 aa change + L72Stop | Sporadic | Heterozygote | 1/2 | Unknown |
| Ex III | |||||||
| S207 199 | 338T > C | 2 | C65R | Sporadic | Heterozygote | 2/3 | Paternal |
| Int 2 | |||||||
| S199 192† | IVS2-2A > G | 2 | (62-95)del + G96I + Y97I + Y98T + L99Stop | Sporadic | Heterozygote | ND | Unknown |
| Ex VI | |||||||
| F164 088‡ | 843-844delAC | 4 | T233fsStop236 | Familial | Heterozygote | 2/3 | Paternal |
| F165 088‡ | 843-844delAC | 4 | T233fsStop236 | Familial | Heterozygote | 2/3 | Paternal |
| S045 169 | 858-872del15bp + 875C > T | 4 | 238-242del + D243N + P244S | Sporadic | Heterozygote | ND | Unknown |
| F169 130* | 768T > G | 4 | F208C | Familial | Heterozygote | 5/19 | Maternal |
| F170 130* | 768T > G | 4 | F208C | Familial | Heterozygote | 5/19 | Maternal |
| Ex XI | |||||||
| S050 172 | 1056C > T | NA | A304V | Sporadic | Heterozygote | ND | Unknown |
NA indicates not applicable; ND, not done; aa, amino acid.
Patients carrying both MCP and CFH mutations.
Patient carrying both MCP and IF mutations.
See Noris et al.19
Modeling of IVS2 –2A>G mutation in MCP. (A) The pET (MoBiTec) exon traps cloning vectors containing wild-type or mutated genomic DNA from intron 1 (IVS1 –62) to intron 4 (IVS4 +70) of MCP. The vectors were transfected into 293T cells and the products were analyzed by RT-PCR and sequencing. Arrows 2 and 3 are the primers used for RT-PCR. Arrow 4 is the primer used for sequencing of cDNA product. MCP2, MCP3, and MCP4 indicate exons II, III, and IV of MCP. (B) Ethidium bromide–stained 1.5% agarose gel of RT-PCR products from splicing assays of transfected 293T cells. The wild-type minigene generated a product of 624 bp containing MCP exons II, III, and IV, whereas the IVS2 –2A>G mutant minigene produced a product of 521 bp lacking exon III. The wild-type sequence inserted in the reverse orientation gave a product (246 bp) that did not contain any spliced product. (C) Sequencing of cloned RT-PCR product. The RT-PCR products were subcloned into pCR2.1-TOPO and sequenced. The IVS2–2A>G mutation results in skipping of exon III. This alteration predicts a 34–amino acid loss (62-95del) in SCR2 followed by 3 amino acid changes (G96I +Y97I + Y98T) and a premature stop at L99.
Modeling of IVS2 –2A>G mutation in MCP. (A) The pET (MoBiTec) exon traps cloning vectors containing wild-type or mutated genomic DNA from intron 1 (IVS1 –62) to intron 4 (IVS4 +70) of MCP. The vectors were transfected into 293T cells and the products were analyzed by RT-PCR and sequencing. Arrows 2 and 3 are the primers used for RT-PCR. Arrow 4 is the primer used for sequencing of cDNA product. MCP2, MCP3, and MCP4 indicate exons II, III, and IV of MCP. (B) Ethidium bromide–stained 1.5% agarose gel of RT-PCR products from splicing assays of transfected 293T cells. The wild-type minigene generated a product of 624 bp containing MCP exons II, III, and IV, whereas the IVS2 –2A>G mutant minigene produced a product of 521 bp lacking exon III. The wild-type sequence inserted in the reverse orientation gave a product (246 bp) that did not contain any spliced product. (C) Sequencing of cloned RT-PCR product. The RT-PCR products were subcloned into pCR2.1-TOPO and sequenced. The IVS2–2A>G mutation results in skipping of exon III. This alteration predicts a 34–amino acid loss (62-95del) in SCR2 followed by 3 amino acid changes (G96I +Y97I + Y98T) and a premature stop at L99.
Thirteen of 14 independent mutational events (93%) cluster in the 4 SCRs at the amino-terminal region of MCP, thus confirming previously reported data on the importance of this region for complement regulation.29 Five mutations determine the introduction of a premature stop codon resulting in truncated proteins within the 4 SCRs.
The frequency of MCP mutations in our population of HUS patients is 12.8% (20/156). Mutation frequency in familial forms of HUS is 19% if we consider only one patient from each family and 10.1% in sporadic forms. Analyses of available relatives revealed a penetrance of 54%.
Functional and expression studies on MCP mutations
The IVS1 –1G>C mutation appeared to cause the loss of the splice acceptor site of intron 1, as documented by splice-site score analysis (http://www.itba.mi.cnr.it/oriel/). RT-PCR amplification of PBMC mRNA of patient F166 099, who is homozygous for this mutation, indicated that the mutation causes an aberrant splicing 2 bp downstream so that the first 2 bp of exon II are not transcribed. Cloning and sequencing of patients' cDNA confirmed the aberrant splicing in 5 of 5 clones. This abnormality is predicted to cause the introduction of a premature stop codon that blocks translation at the very beginning of the protein (C1stop). Data that MCP protein expression on patient PBMCs (by FACS) is severely reduced compared with control PBMCs (Figure 4A) would support the interpretation.
PBMCs from patients F106 024 and F108 024, who carry the combined heterozygous 147G>A (C1Y) and 218C>T (R25Stop) mutations, showed almost no MCP staining by FACS (Figure 4A). The R25Stop causes loss of the entire transmembrane domain of MCP so that the mutant protein is not expressed on PBMCs. PBMCs from these patients' mother, who carries only the heterozygous C1Y mutation, had 50% reduction in MCP median fluorescence intensity compared with PBMCs from healthy controls (Figure 4A) and from the patients' sister without mutations (not shown). Similarly, PBMCs from patient S207 199, carrying the heterozygous C65R mutation, showed reduced MCP expression (Figure 4A). Western blot analysis of the C1Y and C65R mutants expressed in CHO cells (Figure 4B) showed no mature form and a small amount of an aberrant one (C1Y only).20 These data suggest that the proteins do not get expressed on the cell surface. This possibility was further substantiated by FACS data of transfected CHO cells (not shown), which demonstrated no surface expression.
Summary of MCP mutations in non-Stx–HUS patients from our registry. The corresponding number of mutational events is indicated in parentheses. SCR indicates short consensus repeat; STP, serine-threonine-proline–rich domain; TM, transmembrane domain; and CT, cytoplasmic tail.
Summary of MCP mutations in non-Stx–HUS patients from our registry. The corresponding number of mutational events is indicated in parentheses. SCR indicates short consensus repeat; STP, serine-threonine-proline–rich domain; TM, transmembrane domain; and CT, cytoplasmic tail.
Patient S045 169, with the heterozygous 238-242del+ D243N+P244S mutation, had reduced MCP expression on PBMCs (Figure 4A). The D243N+P244S mutant was expressed in CHO cells and Western blot analysis revealed that the predominant form expressed was the precursor (Figure 4B).
Due to the unavailability of patient material, the IVS2 –2A>G mutation was modeled using a pET-MCP minigene exon trap vector. When transfected into 293T cells, the wild-type minigene generated an RNA product containing MCP exons II, III, and IV, whereas the mutated minigene generated a product lacking exon III (Figure 1B-C). Splice-site score analysis confirmed that the mutation causes the loss of the splicing acceptor site and that no alternative high-score splice site is present in intron 2, exon III, and intron 3. The exon skipping is predicted to cause the loss of the first 34 amino acids in SCR2 of MCP followed by 3–amino acid change and protein interruption at L99 (Table 2).
Both F208C and A304V mutants, expressed in CHO cells, showed a normal phenotype on Western blot. F208C had severely decreased C3b (15% ± 2%) and decreased C4b (71% ± 6%) binding capabilities compared with wild-type protein, whereas C3b and C4b binding by A304V mutant was 84% ± 6% and 82% ± 6%, respectively (mean ± SEM of 6 separate experiments; see also Figure 4B-C). We could not obtain PBMCs from carriers of the 2 mutations.
Haplotype analysis on markers flanking MCP gene of family no. 099 and of patient S222 212, both of Sardinian origin, showing that a common allele carrying the mutation is present. The mutation is present in homozygosity in 2 affected siblings (F166 099 and F167 099) in family no. 099 from nonconsanguineous parents, both carrying the mutation in heterozygosity. Of note, the 2 siblings developed HUS very early in life (before 4 years of age) whereas their father (F168 099) developed HUS in adulthood and their mother is still healthy. Three other healthy family members carry the mutation in heterozygosity (marked with ·). Circles indicate females; squares, males; filled symbols, affected individuals; and open symbols, unaffected individuals.
Haplotype analysis on markers flanking MCP gene of family no. 099 and of patient S222 212, both of Sardinian origin, showing that a common allele carrying the mutation is present. The mutation is present in homozygosity in 2 affected siblings (F166 099 and F167 099) in family no. 099 from nonconsanguineous parents, both carrying the mutation in heterozygosity. Of note, the 2 siblings developed HUS very early in life (before 4 years of age) whereas their father (F168 099) developed HUS in adulthood and their mother is still healthy. Three other healthy family members carry the mutation in heterozygosity (marked with ·). Circles indicate females; squares, males; filled symbols, affected individuals; and open symbols, unaffected individuals.
CFH mutation screening
The complete CFH sequence was analyzed in 66 patients (analysis of the other 90 patients was previously published12,13 ) with non-Stx–HUS. Eleven independent new mutational events were found in 12 patients, as summarized in Table 3 and in Figure 5. All mutated patients are white, and all of them have sporadic non-Stx–HUS with the exception of 2 siblings from family no. 210 (Table 3).
New CFH gene mutations in non-Stx–HUS patients from our registry
Exon, subject/family code . | Mutation . | SCR . | Effect . | Subgroups . | Inheritance . | CFH serum levels, mg/L* . | Unaffected carriers . |
|---|---|---|---|---|---|---|---|
| XVIII | |||||||
| S176 137 | 2742G > T | 15 | S8901 | Sporadic | Heterozygote | 788 | ND |
| S177 161 | 2742G > T | 15 | S8901 | Sporadic | Heterozygote | 748 | ND |
| S178 181 | 2759del15bp | 15 | 896-900del5aa | Sporadic | Heterozygote | 285 | ND |
| S181 160 | 2770T > A | 15 | Y899Stop | Sporadic | Homozygote | 196 | ND |
| XIX | |||||||
| S182 155 | 2981A > G | 16 | 1970V | Sporadic | Heterozygote | 550 | ND |
| XX | |||||||
| S185 151 | (3103-3105)delG† | 17 | 1014Stop | Sporadic | Heterozygote | 414 | ND |
| XXIII | |||||||
| S187 194 | dup (3546-3581) | 20 | Ins12aa | Sporadic | Heterozygote | 407 | ND |
| F192 210 | 3663T > C | 20 | V1197A | Familial | Heterozygote | 665 | 2/2 |
| D193 210 | 3663T > C | 20 | V1197A | Familial | Heterozygote | ND | 2/2 |
| S196 177 | 3701C > T | 20 | R1210C | Sporadic | Heterozygote | 793 | ND |
| S187 090 | 3701C > T | 20 | R1210C | Sporadic | Heterozygote | 511 | ND |
| S198 206 | 3701C > T | 20 | R1210C | Sporadic | Heterozygote | 688 | ND |
Exon, subject/family code . | Mutation . | SCR . | Effect . | Subgroups . | Inheritance . | CFH serum levels, mg/L* . | Unaffected carriers . |
|---|---|---|---|---|---|---|---|
| XVIII | |||||||
| S176 137 | 2742G > T | 15 | S8901 | Sporadic | Heterozygote | 788 | ND |
| S177 161 | 2742G > T | 15 | S8901 | Sporadic | Heterozygote | 748 | ND |
| S178 181 | 2759del15bp | 15 | 896-900del5aa | Sporadic | Heterozygote | 285 | ND |
| S181 160 | 2770T > A | 15 | Y899Stop | Sporadic | Homozygote | 196 | ND |
| XIX | |||||||
| S182 155 | 2981A > G | 16 | 1970V | Sporadic | Heterozygote | 550 | ND |
| XX | |||||||
| S185 151 | (3103-3105)delG† | 17 | 1014Stop | Sporadic | Heterozygote | 414 | ND |
| XXIII | |||||||
| S187 194 | dup (3546-3581) | 20 | Ins12aa | Sporadic | Heterozygote | 407 | ND |
| F192 210 | 3663T > C | 20 | V1197A | Familial | Heterozygote | 665 | 2/2 |
| D193 210 | 3663T > C | 20 | V1197A | Familial | Heterozygote | ND | 2/2 |
| S196 177 | 3701C > T | 20 | R1210C | Sporadic | Heterozygote | 793 | ND |
| S187 090 | 3701C > T | 20 | R1210C | Sporadic | Heterozygote | 511 | ND |
| S198 206 | 3701C > T | 20 | R1210C | Sporadic | Heterozygote | 688 | ND |
ND indicates not done; italics indicate below-normal CFH serum levels.
CFH serum levels were measured by radial immunodiffusion (RID) assay (normal range, 350-750 mgNDL).
Deletion of 1 of the 3 consecutive guanines (3103-3105).
Expression and functional studies on MCP mutants. (A) Flow cytometry analysis of MCP (CD46) expression in PBMCs from 5 mutation carriers (C1X ho: patient F166 099, homozygous for the IVS1 –1G>C mutation; C1Y he: unaffected healthy carrier of family no. 024, heterozygous for the 147G>A mutation, mother of patients F106 and F108; R25X/C1Y: patient F106 24, compound heterozygous for the 218C>T and 147G>A mutations; C65R he: patient S207 199, heterozygous for the 338T>C mutation; 238-242del* he: patient S045 169, heterozygous for the 858-872del 15bp+875C>T mutation) and from healthy controls (Control, n = 6). Percentages of CD46+ cells and median fluorescence intensity (MFI) are presented (control: mean ± SD). PBMCs separated by density gradient centrifugation were incubated with an FITC-conjugated mouse anti–human CD46 monoclonal antibody (mAb) or with FITC-mouse IgG1 (isotype control, empty curve) and analyzed by FACSort. 238-242del* indicates deletion of 238-242 amino acids+D243N+P244S; ho, homozygous; and he, heterozygous. No PBMCs could be obtained from patients carrying the 39–amino acid change + L72Stop, the 62-95del+G96I+Y97I+Y98I+L99Stop, or the F208C or A304V mutations. (B) Western blot of CHO cell lysates probed with a rabbit polyclonal Ab to MCP. Lane 6 shows the phenotype of wild-type MCP as expressed by transfected CHO cells.29 Lanes 1 to 5 are the MCP mutations identified in HUS patients. The precursor form is predominant for the C1Y, C65R, and D243N+P244S mutants, indicating an altered folding with minimal processing to the mature form so that the proteins do not get expressed on the cell surface. Both the C1Y and C65R mutants give a faint signal on Western blot, likely due to degradation of unstable precursor protein. The F208C and the A304V mutations show a normal phenotype on Western blot. Lane 7 is a CHO cell not expressing MCP. (C) C3b (left) and C4b (right) binding activity of MCP derived from lysates of CHO cells. F208C and A304V indicate CHO cells expressing these mutants; MCP pos, CHO cells transfected with wild-type MCP; and MCP neg, CHO cells not expressing MCP. An ELISA format was used for ligand binding in which C3b or C4b were coated onto wells of a microtiter plate. Binding assay was performed using diluted CHO extracts (5 × 106 to 25 × 106 MCP molecules as quantified in ELISA). Data are from 1 representative experiment of 6.
Expression and functional studies on MCP mutants. (A) Flow cytometry analysis of MCP (CD46) expression in PBMCs from 5 mutation carriers (C1X ho: patient F166 099, homozygous for the IVS1 –1G>C mutation; C1Y he: unaffected healthy carrier of family no. 024, heterozygous for the 147G>A mutation, mother of patients F106 and F108; R25X/C1Y: patient F106 24, compound heterozygous for the 218C>T and 147G>A mutations; C65R he: patient S207 199, heterozygous for the 338T>C mutation; 238-242del* he: patient S045 169, heterozygous for the 858-872del 15bp+875C>T mutation) and from healthy controls (Control, n = 6). Percentages of CD46+ cells and median fluorescence intensity (MFI) are presented (control: mean ± SD). PBMCs separated by density gradient centrifugation were incubated with an FITC-conjugated mouse anti–human CD46 monoclonal antibody (mAb) or with FITC-mouse IgG1 (isotype control, empty curve) and analyzed by FACSort. 238-242del* indicates deletion of 238-242 amino acids+D243N+P244S; ho, homozygous; and he, heterozygous. No PBMCs could be obtained from patients carrying the 39–amino acid change + L72Stop, the 62-95del+G96I+Y97I+Y98I+L99Stop, or the F208C or A304V mutations. (B) Western blot of CHO cell lysates probed with a rabbit polyclonal Ab to MCP. Lane 6 shows the phenotype of wild-type MCP as expressed by transfected CHO cells.29 Lanes 1 to 5 are the MCP mutations identified in HUS patients. The precursor form is predominant for the C1Y, C65R, and D243N+P244S mutants, indicating an altered folding with minimal processing to the mature form so that the proteins do not get expressed on the cell surface. Both the C1Y and C65R mutants give a faint signal on Western blot, likely due to degradation of unstable precursor protein. The F208C and the A304V mutations show a normal phenotype on Western blot. Lane 7 is a CHO cell not expressing MCP. (C) C3b (left) and C4b (right) binding activity of MCP derived from lysates of CHO cells. F208C and A304V indicate CHO cells expressing these mutants; MCP pos, CHO cells transfected with wild-type MCP; and MCP neg, CHO cells not expressing MCP. An ELISA format was used for ligand binding in which C3b or C4b were coated onto wells of a microtiter plate. Binding assay was performed using diluted CHO extracts (5 × 106 to 25 × 106 MCP molecules as quantified in ELISA). Data are from 1 representative experiment of 6.
All mutations are heterozygous, with the exception of the homozygous 2770T>A transversion in exon XVIII causing introduction of a premature stop codon in SCR15 (Y899Stop) in patient S181 160 from consanguineous parents. CFH serum levels, as measured by radial immunodiffusion (RID),9 were lower than normal in 2 CFH-mutated patients (Table 3).
Figure 5 summarizes all the CFH mutations found in patients from our Registry including the ones reported here and those previously published by our group.12,13 A total of 47 patients were mutated in CFH (Caprioli et al12,13 and present data). All are heterozygous mutations, with the exception of patient S181 160 and of family no. 029, showing recessive transmission.
Sixty-one percent of the overall independent mutational events (17/28) cluster in SCR20 (Figure 5). Moreover, 9 other mutational events are located in SCR15 (n = 4), SCR16 (n = 3), SCR17 (n = 1), and SCR19 (n = 1), thus confirming the importance of the C-terminus of CFH12-17 to the pathogenesis of HUS.
Five mutations determine the introduction of a premature stop codon, resulting in truncated proteins at SCR8 (n = 1), SCR15 (n = 1), SCR17 (n = 1), and SCR20 (n = 2), whereas the others are missense mutations (Table 4). None of the mutations described were found in any of 120 healthy white controls.
IF gene mutations in non-Stx–HUS patients from our registry
Exon/intron, subject/family code . | Mutation . | Effect . | Subgroups . | Inheritance . | IF serum levels, %* . |
|---|---|---|---|---|---|
| Ex V | |||||
| S211 117 | 719C > G | A240G | Sporadic | Heterozygote | 98 |
| Ex IX | |||||
| F215 010 | 949C > T | R317W | Familial | Heterozygote | ND |
| F216 010 | 949C > T | R317W | Familial | Heterozygote | 98 |
| Ex XII | |||||
| S199 192† | (1446-1450)delTTCAC | L484V + Q485G + W486Stop | Sporadic | Heterozygote | 77 |
| Int 12 | |||||
| S214 150 | 1534 + 5G > T | Splice score decrease from 93 to 86 | Sporadic | Heterozygote | 103 |
| Ex XIII | |||||
| F118 034 | 1555G > A | D519N | Familial | Heterozygote | ND |
| F119 034 | 1555G > A | D519N | Familial | Heterozygote | ND |
Exon/intron, subject/family code . | Mutation . | Effect . | Subgroups . | Inheritance . | IF serum levels, %* . |
|---|---|---|---|---|---|
| Ex V | |||||
| S211 117 | 719C > G | A240G | Sporadic | Heterozygote | 98 |
| Ex IX | |||||
| F215 010 | 949C > T | R317W | Familial | Heterozygote | ND |
| F216 010 | 949C > T | R317W | Familial | Heterozygote | 98 |
| Ex XII | |||||
| S199 192† | (1446-1450)delTTCAC | L484V + Q485G + W486Stop | Sporadic | Heterozygote | 77 |
| Int 12 | |||||
| S214 150 | 1534 + 5G > T | Splice score decrease from 93 to 86 | Sporadic | Heterozygote | 103 |
| Ex XIII | |||||
| F118 034 | 1555G > A | D519N | Familial | Heterozygote | ND |
| F119 034 | 1555G > A | D519N | Familial | Heterozygote | ND |
ND indicates not done.
IF serum levels were measured by ELISA assay as described by Fremeaux-Bacchi et al18 (normal range, 70%-130%).
Patient carrying both MCP and IF mutations.
The frequency of CFH mutations in our population of patients is 30.1% (38% in familial forms, if we consider only one patient from each family, and 20% in sporadic forms). The penetrance of the disease in mutation carriers is 59%. Of note, the 5 patients from family no. 024 (n = 3) and no. 130 (n = 2) (Table 2, *) carry mutations in both MCP and CFH.
Summary of CFH mutations in non-Stx–HUS patients from our registry. The corresponding number of mutational events is indicated in parentheses.
Summary of CFH mutations in non-Stx–HUS patients from our registry. The corresponding number of mutational events is indicated in parentheses.
IF mutation screening
Five independent mutational events in 7 white patients (3 with sporadic HUS and 4 with the familial form; Table 4) were found in IF. All mutations are heterozygous. Four mutations cluster in the serine protease domain of IF30 : 2 are missense mutations; 1 is a 5-bp deletion that causes a frameshift and the introduction of a premature stop codon; and 1 affects the donor site of intron 12, causing a reduction of splice score from 93 to 86. The fifth mutation causes an amino acid change in the low-density lipoprotein receptor (LDLR) module30 of IF (Table 4). The frequency of IF mutations in our population of HUS patients is 4.5% (7/156). Of note, patient S199 192 (Tables 2 and 4†) carries mutations both in MCP and IF. Serum IF levels, as measured by ELISA,18 were within normal range in all patients (Tables 4, 5).
Patient characteristics
Characteristic . | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P1 . | P2 . | P3 . |
|---|---|---|---|---|---|---|---|
| Disease onset | |||||||
| Childhood, younger than 18 y | 52 (86) | 30 (42) | 10 (14) | 3 (6) | .22 | .43 | > .999 |
| Adulthood, 18 y or older | 34 (86) | 12 (42) | 4 (14) | 3 (6) | .22 | .43 | > .999 |
| Triggers | |||||||
| Pregnancy | 6 (66) | 1 (26) | 0 (12) | 2 (5) | .39 | .28 | .49 |
| Drugs | 4 (66) | 1 (26) | 0 (12) | None | .67 | .38 | .49 |
| Flulike, gastroenteritis, other infections | 46 (66) | 18 (26) | 12 (12) | 3 (5) | .96 | .03 | .03 |
| No triggers | 1 (66) | 1 (26) | 0 (12) | None | NA | NA | NA |
| Other triggers | 10 (66) | 5 (26) | 0 (12) | None | .63 | .15 | .10 |
| Recurrences | 30 (84) | 15 (42) | 9 (14) | 2 (6) | > .999 | .04 | .06 |
| Biochemical evaluation | |||||||
| Reduced C3 serum levels, 83 mg/dL or less* | 22 (71) | 16 (31) | 4 (12) | 3 (5) | .047 | .87 | .28 |
| Reduced C4 serum levels, 15 mg/dL or less* | 6 (69) | 1 (29) | 0 (12) | 0 (4) | .36 | .29 | .51 |
| Reduced CFH serum levels, 350 mg/L or less† | 1 (73) | 5 (34) | 0 (12) | 0 (5) | .005 | .68 | .16 |
| Reduced IF serum levels, below 70%‡ | 0 (57) | 0 (23) | 0 (10) | 0 (3) | NA | NA | NA |
Characteristic . | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P1 . | P2 . | P3 . |
|---|---|---|---|---|---|---|---|
| Disease onset | |||||||
| Childhood, younger than 18 y | 52 (86) | 30 (42) | 10 (14) | 3 (6) | .22 | .43 | > .999 |
| Adulthood, 18 y or older | 34 (86) | 12 (42) | 4 (14) | 3 (6) | .22 | .43 | > .999 |
| Triggers | |||||||
| Pregnancy | 6 (66) | 1 (26) | 0 (12) | 2 (5) | .39 | .28 | .49 |
| Drugs | 4 (66) | 1 (26) | 0 (12) | None | .67 | .38 | .49 |
| Flulike, gastroenteritis, other infections | 46 (66) | 18 (26) | 12 (12) | 3 (5) | .96 | .03 | .03 |
| No triggers | 1 (66) | 1 (26) | 0 (12) | None | NA | NA | NA |
| Other triggers | 10 (66) | 5 (26) | 0 (12) | None | .63 | .15 | .10 |
| Recurrences | 30 (84) | 15 (42) | 9 (14) | 2 (6) | > .999 | .04 | .06 |
| Biochemical evaluation | |||||||
| Reduced C3 serum levels, 83 mg/dL or less* | 22 (71) | 16 (31) | 4 (12) | 3 (5) | .047 | .87 | .28 |
| Reduced C4 serum levels, 15 mg/dL or less* | 6 (69) | 1 (29) | 0 (12) | 0 (4) | .36 | .29 | .51 |
| Reduced CFH serum levels, 350 mg/L or less† | 1 (73) | 5 (34) | 0 (12) | 0 (5) | .005 | .68 | .16 |
| Reduced IF serum levels, below 70%‡ | 0 (57) | 0 (23) | 0 (10) | 0 (3) | NA | NA | NA |
IF mutation group has not been included in statistical analysis. The numbers of patients for whom data were available are reported in parentheses. Significant P values are in italics.
P1 indicates no mutation versus CFH mutation; P2, no mutation versus MCP mutation; P3, CFH mutation versus MCP mutation; and NA, not applicable.
C3 and C4 levels were measured by kinetic nephelometry.
CFH serum levels were measured by radial immunodiffusion assay.
IF serum levels were measured by ELISA.
Clinical findings
Data on characteristics, disease treatment, and outcome of all patients (including the new and the previously published cases from our registry) with mutations in MCP (MCP mut), CFH (CFH mut), and IF (IF mut) and with no mutation (non mut) are reported in Tables 5, 6, 7, 8, 9. Patients from family no. 024 and no. 130 and patient S199 192 have been excluded, since they carry both MCP and CFH and MCP and IF mutations, respectively.
Treatment and outcome of the first episode of non-Stx–HUS
Treatment or outcome . | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P1 . | P2 . | P3 . |
|---|---|---|---|---|---|---|---|
| Treatment of the first episode | |||||||
| No treatment | 11 (74) | 2 (34) | 4 (14) | 2 (6) | .18 | .21 | .03 |
| Plasma alone or in combination with drugs | 63 (74) | 32 (34) | 10 (14) | 4 (6) | .18 | .21 | .03 |
| Plasma, infusion or exchange | 34 (63) | 21 (32) | 7 (10) | 3 (4) | .28 | .34 | .80 |
| Plasma and drugs acting on the coagulation cascade | 6 (63) | 0 (32) | 2 (10) | 0 (4) | .07 | .32 | .009 |
| Plasma and drugs acting on the immune system | 14 (63) | 11 (32) | 1 (10) | 0 (4) | .20 | .37 | .14 |
| Plasma and both categories of drugs | 9 (63) | 0 (32) | 0 (10) | 1 (4) | .02 | .20 | NA |
| Supportive treatment | 59 (76) | 32 (39) | 7 (14) | 3 (6) | .58 | .03 | .02 |
| Outcome of the first episode | |||||||
| Complete remission | 28 (81) | 7 (40) | 12 (14) | 3 (6) | .05 | <.001 | <.001 |
| Partial remission | 30 (81) | 12 (40) | 1 (14) | 1 (6) | .44 | .028 | .085 |
| Dialysis | 19 (81) | 9 (40) | 1 (14) | 2 (6) | .91 | .17 | .20 |
| Death | 4 (81) | 12 (40) | 0 (14) | 0 (6) | <.001 | .39 | .02 |
Treatment or outcome . | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P1 . | P2 . | P3 . |
|---|---|---|---|---|---|---|---|
| Treatment of the first episode | |||||||
| No treatment | 11 (74) | 2 (34) | 4 (14) | 2 (6) | .18 | .21 | .03 |
| Plasma alone or in combination with drugs | 63 (74) | 32 (34) | 10 (14) | 4 (6) | .18 | .21 | .03 |
| Plasma, infusion or exchange | 34 (63) | 21 (32) | 7 (10) | 3 (4) | .28 | .34 | .80 |
| Plasma and drugs acting on the coagulation cascade | 6 (63) | 0 (32) | 2 (10) | 0 (4) | .07 | .32 | .009 |
| Plasma and drugs acting on the immune system | 14 (63) | 11 (32) | 1 (10) | 0 (4) | .20 | .37 | .14 |
| Plasma and both categories of drugs | 9 (63) | 0 (32) | 0 (10) | 1 (4) | .02 | .20 | NA |
| Supportive treatment | 59 (76) | 32 (39) | 7 (14) | 3 (6) | .58 | .03 | .02 |
| Outcome of the first episode | |||||||
| Complete remission | 28 (81) | 7 (40) | 12 (14) | 3 (6) | .05 | <.001 | <.001 |
| Partial remission | 30 (81) | 12 (40) | 1 (14) | 1 (6) | .44 | .028 | .085 |
| Dialysis | 19 (81) | 9 (40) | 1 (14) | 2 (6) | .91 | .17 | .20 |
| Death | 4 (81) | 12 (40) | 0 (14) | 0 (6) | <.001 | .39 | .02 |
IF mutation group has not been included in statistical analysis. The numbers of patients for whom data were available are reported in parentheses. Supportive treatment includes dialysis, blood transfusions, or concentrated red blood cell infusion. Plasma treatment includes infusion of 10 to 20 mL/kg/day and/or exchange of 1 to 2 plasma vol/day (or 30-40 mL/kg/day) for a total of 2 to 36 treatments in 2 days to 6 weeks. Complete remission is defined as normalization of both hematologic parameters and of renal function. Partial remission is defined as normalization of hematologic parameters with renal sequelae.
P1 indicates no mutation versus CFH mutation; P2, no mutation versus MCP mutation; P3, CFH mutation versus MCP mutation; NA, not applicable. Significant P values are in italics.
Long-term outcome of non-Stx–HUS patients
. | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P1 . | P2 . | P3 . |
|---|---|---|---|---|---|---|---|
| Patients with remission | 36 (84) | 9 (40) | 12 (14) | 2 (6) | .03 | .007 | <.001 |
| Patients with complete remission | 22 (36) | 3 (9) | 4 (12) | 2 (2) | .13 | .09 | >.999 |
| Patients with complete remission, after recurrences | 14 (36) | 6 (9) | 8 (12) | 0 (2) | .13 | .09 | >.999 |
| Patients with no remission | 48 (84) | 31 (40) | 2 (14) | 4 (6) | .03 | .003 | <.001 |
| Chronic renal insufficiency | 5 (48) | 3 (31) | 0 (2) | 0 (4) | .91 | .63 | .64 |
| ESRF | 32 (48) | 13 (31) | 2 (2) | 4 (4) | .03 | .32 | .29 |
| Death | 11 (48) | 15 (31) | 0 (2) | 0 (4) | .019 | .44 | .18 |
. | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P1 . | P2 . | P3 . |
|---|---|---|---|---|---|---|---|
| Patients with remission | 36 (84) | 9 (40) | 12 (14) | 2 (6) | .03 | .007 | <.001 |
| Patients with complete remission | 22 (36) | 3 (9) | 4 (12) | 2 (2) | .13 | .09 | >.999 |
| Patients with complete remission, after recurrences | 14 (36) | 6 (9) | 8 (12) | 0 (2) | .13 | .09 | >.999 |
| Patients with no remission | 48 (84) | 31 (40) | 2 (14) | 4 (6) | .03 | .003 | <.001 |
| Chronic renal insufficiency | 5 (48) | 3 (31) | 0 (2) | 0 (4) | .91 | .63 | .64 |
| ESRF | 32 (48) | 13 (31) | 2 (2) | 4 (4) | .03 | .32 | .29 |
| Death | 11 (48) | 15 (31) | 0 (2) | 0 (4) | .019 | .44 | .18 |
IF mutation group has not been included in statistical analysis. The numbers of patients for whom data were available are reported in parentheses.
P1 indicates no mutation versus CFH mutation; P2, no mutation versus MCP mutation; and P3, CFH mutation versus MCP mutation.
Treatment with plasma in non-Stx–HUS patients with CFH and MCP mutations
Episode . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P . |
|---|---|---|---|---|
| Treated episodes | 57 (61) | 23 (35) | 6 (8) | < .001 |
| Complete or partial remission | 38 (57) | 21 (23) | 3 (6) | .023 |
| No remission | 19 (57) | 2 (23) | 3 (6) | .023 |
| Nontreated episodes | 4 (61) | 12 (35) | 2 (8) | < .001 |
| Complete or partial remission | 4 (4) | 12 (12) | 1 (2) | .001 |
| No remission | 0 (4) | 0 (12) | 1 (2) | .001 |
Episode . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . | P . |
|---|---|---|---|---|
| Treated episodes | 57 (61) | 23 (35) | 6 (8) | < .001 |
| Complete or partial remission | 38 (57) | 21 (23) | 3 (6) | .023 |
| No remission | 19 (57) | 2 (23) | 3 (6) | .023 |
| Nontreated episodes | 4 (61) | 12 (35) | 2 (8) | < .001 |
| Complete or partial remission | 4 (4) | 12 (12) | 1 (2) | .001 |
| No remission | 0 (4) | 0 (12) | 1 (2) | .001 |
IF mutation group has not been included in statistical analysis. The numbers of patients for whom data were available are reported in parentheses. Plasma treatment includes infusion of 10 to 20 mL/kg/day and/or exchange of 1 to 2 plasma vol/day (or 30-40 mL/kg/day) for a total of 2 to 36 treatments in 2 days to 6 weeks.
P indicates CFH mutation versus MCP mutation. Significant P values are in italics.
Outcome of kidney transplantations in patients with non-Stx–HUS
. | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . |
|---|---|---|---|---|
| Kidney transplant recipients | 14 | 6 | 2 | 1 |
| Transplanted kidneys | 17 | 6 | 2 | 2 |
| Kidney outcome | ||||
| Good renal function at 1 y | 10 | 1 | 2 | 0 |
| Disease recurrence on the graft | 3 | 5 | 0 | 2 |
| Acute rejection | 4 | 0 | 0 | 0 |
. | No mutation, no. . | CFH mutation, no. . | MCP mutation, no. . | IF mutation, no. . |
|---|---|---|---|---|
| Kidney transplant recipients | 14 | 6 | 2 | 1 |
| Transplanted kidneys | 17 | 6 | 2 | 2 |
| Kidney outcome | ||||
| Good renal function at 1 y | 10 | 1 | 2 | 0 |
| Disease recurrence on the graft | 3 | 5 | 0 | 2 |
| Acute rejection | 4 | 0 | 0 | 0 |
P values are as follows: P1 indicates non mut versus CFH mut; P2, non mut versus MCP mut; P3, CFH mut versus MCP mut. Due to the small number of patients with IF mutations, they were analyzed descriptively without statistical comparisons.
The disease manifested during childhood in most patients (Table 5); no difference in disease onset among groups was observed. Putative triggering conditions were recognized in the majority of patients, infection being the most frequently associated condition.
One third of patients in non mut, CFH mut, and IF mut groups and two thirds in the MCP mut group had one or more disease recurrences. Lower than normal C3 serum levels were recorded in 30.9%, 51.6%, and 33.3% of non mut, CFH mut, and MCP mut patients, respectively and in 3 of 5 IF mut patients. C4 levels were normal in most patients.
During the first episode (Table 6), plasma infusion or exchange alone or in combination with drugs acting on the immune system and/or on the coagulation cascade was administered to 85% of non mut, 94% of CFH mut, 71% of MCP mut, and 67% of IF mut patients (P3 = .03). Eleven of 74 patients with no mutation, 2 of 34 with CFH, 4 of 14 with MCP (P3 = .03), and 2 of 6 with IF mutations received no therapy other than supportive treatment (dialysis, blood transfusion, or concentrated red blood cell infusion).
The first episode (Table 6) in MCP mut patients had a better prognosis than non mut and CFH mut groups: a complete remission was obtained in 34.5%, 17.5%, and 85.7% of non mut, CFH mut, and MCP mut patients, respectively (P1 = .05, P2 < .001, P3 < .001). In addition, only one MCP mut patient developed ESRF and none died during the acute episode, whereas the outcome was ESRF in 22% and death in 30% of CFH mut patients (P1 < .001, P3 = .02).
Even considering the long-term outcome (Table 7), MCP mut patients had the best prognosis. In fact, 86% of MCP mut patients retained normal renal function and had no residual hematologic abnormalities even after repeated disease recurrences compared with 22.5% of CFH mut, 42.8% of non mut (P1 = .03, P2 = .007, P3 < .001), and 33.3% of IF mut patients. Figure 6 reports a Kaplan-Meier cumulative survival curve indicating the fraction of the patients free of chronic renal insufficiency or chronic dialysis or death in MCP mut and CFH mut groups. One (MCP mut) versus 31 (CFH mut) events were observed in the 2 groups. The difference between the cumulative survival curves was statistically significant (P < .001).
The percentage of episodes treated with plasma infusion or plasmapheresis (Table 8) is significantly lower in MCP mut patients (65%) compared with CFH mut patients (93%; P < .001), confirming the data at onset (Table 7). In the MCP-mutated group, complete or partial remission (defined as hematologic normalization with renal sequelae) was achieved in 91% of plasma-treated episodes. Interestingly, 100% remissions were obtained in the nontreated episodes. In CFH mut patients, remission was obtained in 67% of plasma-treated episodes but also in all 4 episodes that were not treated. Plasma induced remission in 3 of 6 treated episodes in IF mut group.
Two patients with MCP mutations, 6 with CFH mutations, 1 with IF mutation (2 grafts), and 14 with no mutations underwent kidney transplantation (Table 9). The 2 patients with MCP mutations have good graft function at 10 years and 10 months after surgery, respectively. Five grafts in patients with CFH mutations and the 2 in the IF-mutated patient were lost because of disease recurrences within the first year after transplantation, and only one graft was well functioning at that time. Of note, 2 additional patients with CFH mutations (data not shown) received a combined kidney and liver transplant.31,32 Ten grafts in nonmutated patients were well functioning after 1 year, whereas the others were lost because of either disease recurrences (n = 3) or acute rejection (n = 4).
Discussion
Several mutations in CFH12-17,23,24,33 and a few mutations in MCP19,20,26 and IF18,25,26 have been reported so far in patients with non-Stx–HUS. However, large mutational screenings for MCP and IF were still lacking. In addition, no study has addressed comparison of clinical phenotype and response to treatment between patients with MCP, CFH, or IF mutations, so far. Here we report the results of MCP, CFH, and IF genetic screening in 156 non-Stx–HUS patients. Clinical characteristics of patients carrying mutations in MCP, CFH, and IF and of patients without mutations in these genes have been compared.
We found 14 independent mutational events in MCP, 28 in CFH (present data and Caprioli et al12,13 ), and 5 in IF. The majority of mutations are heterozygous. MCP and IF mutations are less frequent than CFH mutations; in fact the percentages of patients carrying MCP and IF mutations are around one third and one seventh the percentage of CFH mutation carriers, respectively. The mutation frequencies reported here are very comparable to those previously described in other cohorts22 ; however, we cannot exclude the possibility that we missed a few mutations, since the sensitivity of SSCP (86%-100%28,34,35 ) and DHPLC (93%-100%28,35 ) may not be absolute.
Cumulative fraction of patients free of events, defined as the combination of the occurrence of chronic renal insufficiency or initiation of dialysis or death, whichever occurred first after the onset of HUS (Kaplan-Meier) in non-Stx–HUS patients with MCP and CFH mutations from our registry.
Cumulative fraction of patients free of events, defined as the combination of the occurrence of chronic renal insufficiency or initiation of dialysis or death, whichever occurred first after the onset of HUS (Kaplan-Meier) in non-Stx–HUS patients with MCP and CFH mutations from our registry.
Ninety-three percent of MCP mutational events cluster in the 4 extracellular SCRs with C3b binding and cofactor activity, thus confirming the importance of this region for complement regulation. Eight of 10 MCP mutations cause quantitative deficiency of MCP. Five mutations result in truncated proteins, lacking the transmembrane C-terminus. The loss of the MCP C-terminus affects the cell-surface expression of MCP through failure of insertion of the mutant protein into the plasma membrane, as demonstrated by FACS analysis of PBMCs from mutation carriers (present data, Noris et al,19 and Richards et al20 ). The C1Y and C65R mutations cause severely reduced cell-surface MCP expression on PBMCs. Both mutations abrogate 1 of the 4 cysteines of SCR1 and SCR2, respectively, with loss of 1 disulphide bridge in each SCR. This abnormality likely causes an altered folding leading to protein retention in the endoplasmic reticulum with slow processing to the mature form, as documented by reduced protein levels and by the prevalence of the precursor form of MCP on Western blot of transfected CHO cells. The 238-242del+ D243N+P244S change in SCR4 determines the loss of Ser238 and results in reduced cell-surface MCP expression and in its intracellular retention as a precursor form. Deletion of Ser238 has been previously shown20 to cause almost complete retention of the protein intracellularly. The mutation also results in the loss of Asn239, which is an important site of N-glycosylation involved in cofactor activity and cytoprotective capacity of MCP.36 The F208C recombinant mutant shows a normal pattern on Western blot, suggesting a normal intracellular processing; however, it has severely reduced capability to bind C3b compared with wild-type protein, which may affect MCP complement regulatory properties. The latter interpretation fits with published mutagenesis experiments showing that substitution of F208 decreases C3b binding and abrogates cofactor activity.29 A normal pattern on Western blot was also seen for the A304V recombinant. C3b and C4b binding were not severely compromised in the high-expressing system. Perhaps in normal cells, the A304V mutant may not efficiently migrate to the surface and/or insert into the lipid bilayer. The mutation was not found in any of the 200 healthy controls (present data and Richards et al20 ) excluding the fact that the A304V is a polymorphism.
Ninety-three percent of CFH mutational events are spread over the 5 exons that encode the most C-terminal part of CFH, the majority of them clustering in SCR20, thus confirming previous data37-39 on the importance of the CFH C-terminus for its complement regulatory activity. The very C-terminal domains (SCR19-20) in fact contain a C3b binding site and a polyanion binding site, which are determinant for CFH contact with host endothelial cells and for surface cofactor activity, since deletion of this portion of the molecule causes loss of the capability of CFH to degrade endothelial-bound C3b.40 If we consider all the CFH mutational events (present data, Caprioli et al,12,13 Neumann et al,14 Dragon-Durey et al,15 Perez-Caballero et al,16 Richards et al,17 Davin et al,23 Heinen et al24 ), 4 mutational hot spots can be identified in SCR20 (Figure 7), namely amino acids 1183, 1191, 1197, and 1210. Functional studies by 2 independent groups38,41,42 documented that those amino acids are involved in binding of the protein to surface-bound C3b to heparin and to endothelial cells.
Functional studies were not done on IF mutants; however, we can infer that they affect neither mRNA levels nor protein secretion as documented by normal IF serum levels in mutation carriers. Most likely these changes might impair the capability of IF to cleave the alpha-chains of C3b and C4b, since most of them cluster in the light-chain serine protease domain.30
Amino acid position along CFH of all mutational events in non-Stx–HUS patients including published and present data. The x-axis indicates amino acid position.
Amino acid position along CFH of all mutational events in non-Stx–HUS patients including published and present data. The x-axis indicates amino acid position.
MCP, CFH, and IF functions are closely interrelated, as C3b cleaving by IF is dependent on MCP and CFH cofactor activity. MCP is highly expressed on glomerular endothelial cell surface and plays a major role in regulating glomerular C3 activation.43 Human glomerular endothelial cells and kidney glomerular basement membrane are rich in polyanionic molecules for CFH binding.44 Once bound to such sites, CFH acts as an outer barrier against complement attack. Thus, genetic defects of MCP, CFH, and IF in the presence of stimuli that activate the complement system cause an impaired protection of endothelial surface. As a consequence, more C3b reaches the endothelial cell surface, which is followed by the formation of the membrane attack complex and recruitment of inflammatory cells, all events that cause damage of endothelial cells and platelet adhesion.
Here we confirm previous data showing the incomplete penetrance of the disease phenotype in both MCP and CFH mutation carriers.12-26 It is likely that MCP, CFH, and IF mutations confer a predisposition to develop HUS, rather than directly causing the disease, and that a second hit is required for the full-blown manifestations of the disease. This possibility is supported by the observation that one third of patients did not develop the disease until adulthood. Nevertheless conditions that trigger complement activation either directly (bacterial and viral infections) or indirectly by causing endothelial insult (drugs, certain systemic diseases) could precipitate an acute event on the predisposed genetic background. Indeed, the onset of the disease was associated with an infectious event in all MCP-mutated, 70% of CFH-mutated, and 60% of IF-mutated patients (Table 5).
These findings have potential clinical implications: the identification of mutation carriers within a family could allow the selection of subjects at risk who should be monitored, particularly when exposed to triggering events such as pregnancy for fertile women and infections for children.
Patients with MCP mutations required a less intensive treatment than the other groups. Moreover a complete remission was generally obtained both at the presenting episode and after recurrences so that 86% of patients remained long-term dialysis free. However, there are some exceptions: as an example, one patient (S019 085) manifested extensive microvascular thrombosis and refractory hypertension during the presenting episode that resolved only after bilateral nephrectomy. On the other hand, patients with CFH mutations often presented with serious episodes that required intensive treatment and supportive care and 70% of them died or developed ESRF following the first episode or progressed to ESRF as a consequence of relapses. However, complete remission was obtained in few cases. Thus, although the genotype-phenotype correlation was not always straight, data indicate that MCP mutations are associated with a better prognosis than CFH mutations.
Plasma exchange and plasma infusion is generally the first-line therapy in non-Stx–HUS but debate still exists on its efficacy in the treatment of acute episodes.45,46 Thus, we investigated whether the response to treatment with plasma varied as a function of the genetic background. Plasma infusion or plasma exchange was used to treat 66% of the acute episodes in patients with MCP mutations. Remission was achieved in 91% of plasma-treated episodes but also in 100% of the nontreated episodes, suggesting that plasma does not have a great impact on the outcome of HUS in this group of patients. These findings can be explained, reasoning that MCP is a membrane-bound protein and theoretically plasma infusion or exchange would not correct the defect.
In cases with CFH mutations, 93% of the episodes were treated with plasma and remission was obtained in 67% of them. Similar results were observed in patients with IF mutations. Theoretically one should expect a better response to plasma treatment in CFH- and IF-mutated patients, being CFH and IF circulating proteins. However the amount of plasma administered should be high to provide sufficient wild-type CFH or IF to correct the genetic deficiency.
Whether kidney transplantation is an appropriate treatment in patients with non-Stx–HUS who had progressed to ESRF is debatable. Actually, around 50% of the patients who underwent renal transplantation had a recurrence of the disease in the graft,47-49 and graft failure occurred in more than 90% of them. Here we evaluated if mutation screening could help define graft prognosis. Kidney graft outcome was favorable in 2 patients with MCP mutations who experienced no disease recurrence and maintain a well-functioning graft at 10 years and 10 months after transplantation, respectively. Similar results were reported17 in 3 additional patients. Since MCP is a transmembrane protein highly expressed in the kidney, transplanting a normal kidney conceivably corrects the defect in these patients. In the 6 patients with CFH mutations and the 1 with IF mutation who were given a renal transplantation, the outcome instead was bad. Indeed in 6 of them, their graft failed because of disease recurrence. These results are consistent with previous data in literature showing a recurrence rate of 30% to 100% and 100% in CFH13,14,49 and IF18,25 mutation carriers, respectively. Since CFH and IF are mostly synthesized by the liver, the kidney transplantation did not correct the genetic defects and persistent CFH or IF deficiency predisposed to disease recurrence in the transplanted kidney.
Fifty-six percent of patients in this study carry neither MCP nor CFH nor IF mutations. They likely represent a genetically heterogeneous group, with variable response to therapy and clinical outcome. Alterations in other not yet identified genes encoding for complement regulatory proteins could have a role as well. On the other hand, the recent discovery of anti-CFH antibodies in the plasma of 3 children with sporadic non-Stx–HUS indicates that this disease can also be associated with an acquired autoimmune CFH defect.50
In conclusion, these findings underscore the influence of different genetic abnormalities on disease presentation, response to therapy, and outcome in non-Stx–HUS. Identification of MCP, CFH, and IF mutations could potentially translate into an improvement in the management and therapy of patients and will hopefully provide the way to design tailored treatments.
Appendix
Members of the International Registry of Recurrent and Familial HUS/TTP include the following:
Coordinators
G. Remuzzi, MD, P. Ruggenenti, MD (Clinical Research Center for Rare Diseases, Aldo e Cele Daccò, Ranica, Bergamo, Italy and Division of Nephrology and Dialysis, Azienda Ospedaliera, Ospedali Riuniti di Bergamo, Italy); M. Noris, Chem Pharm D (Clinical Research Center for Rare Diseases, Aldo e Cele Daccò, Ranica, Bergamo, Italy).
Investigators
Italy. M. Garozzo, MD (Division of Nephrology and Dialysis, S. Marta e S. Venera Hospital, Acireale, Catania); M. Antonelli, MD, F. Casucci, MD, F. Cazzato, MD (Division of Nephrology, Miulli Hospital, Acquaviva delle Fonti, Bari); I. M. Ratsch, MD (Pediatric Clinic, G. Salesi Hospital, Ancona); G. Claudiani, MD (Division of Hematology, S. Liberatore Hospital, Atri, Teramo); W. De Simone, MD (Division of Nephrology and Dialysis, S. Giuseppe Moscati Hospital, Avellino); P. Dattolo, MD, F. Pizzarelli, MD (Division of Nephrology and Dialysis, S. M. Annunziata Hospital, Bagno a Ripoli, Florence); R. Bellantuono, MD, T. De Palo, MD (Division of Nephrology and Dialysis, Giovanni XXIII Pediatric Hospital, Bari); N. Lattanzi, MD (Centro Emodialisi, Bari); M. Schiavoni, MD (Assistenza Emofilici e Coagulopatici, Ospedale Policlinico Consorziale, Bari); T. Barbui, MD (Division of Hematology, Azienda Ospedaliera, Ospedali Riuniti di Bergamo); G. Torre, MD (Pediatric Department, Azienda Ospedaliera, Ospedali Riuniti di Bergamo); A. M. Acquarolo, MD (II Rianimazione Spedali Civili, Azienda Ospedaliera, Brescia); O. Carli, MD, G. Gregorini, MD (Division of Nephrology and Dialysis, Spedali Civili, Azienda Ospedaliera, Brescia); G. Rossi, MD (Division of Hematology, Spedali Civili, Azienda Ospedaliera, Brescia); A. Cao, MD (Istituto di Clinica e Biologia dell'Età Evolutiva, Cagliari); C. Setzu, MD (Pediatric Division, G. Brotzu Hospital, Cagliari); A. Bonadonna, MD (Division of Nephrology and Dialysis, Presidio Ospedaliero di Camposampiero, Padova); C. Cascone, MD, G. Delfino, MD (Division of Nephrology and Dialysis, S. Giacomo Hospital, Castelfranco Veneto, Treviso); S. Li Volti, MD (Pediatric Department, Policlinico Hospital, Catania); C. Castellino, MD (Division of Hematology, Azienda Ospedaliera S. Croce e Carle, Cuneo); L. Calacoci, MD (Division of Immunohematology, S. Giovanni di Dio Hospital, Florence); C. Grimaldi, MD (Division of Internal Medicine and Nephrology, S. Giovanni di Dio Hospital, Florence); I. Pela, MD (Division of Nephrology, A. Meyer Hospital, Firenze); M. Salvadori, MD (Division of Nephrology and Dialysis, Careggi Hospital, Florence); E. Capussela, MD (Division of Hematology, Ospedali Riuniti di Foggia, Foggia); D. A. Procaccini, MD (Division of Nephrology and Dialysis, Ospedali Riuniti di Foggia); G. C. Barbano, MD, A. Canepa, MD, M. L. Degl'Innocenti, MD, A. Trivelli, MD (Division of Nephrology, G. Gaslini Pediatric Institute, Genoa); I. Fontana, MD (Transplant Center, S. Martino Hospital, Genoa); S. D'Ardia, MD (Division of Immunohematology, Ivrea Hospital, Ivrea, Turin); C. Marseglia, MD (Service of Nephrology and Dialysis, Carlo Poma Hospital, Mantova); A. Bettinelli, MD (Pediatric Division, S. Leopoldo Mandic Hospital, Merate, Lecco); R. Chimenz, MD (Division of Pediatric Nephrology, G. Martino Hospital, Messina); G. Ardissino, MD, A. Edefonti, MD (Division of Pediatric Nephrology, Dialysis and Transplant, De Marchi Pediatric Clinic, Milan); A. Lattuada, BiolSciD, E. Rossi, MD (Division of Hematology, L. Sacco Hospital, Milan); V. Rossi, MD (Division of Hematology, Niguarda Cà Granda Hospital, Milan); V. Toschi, MD (Transfusional Center, San Carlo Borromeo Hospital, Milan); L. Gaiani, MD, M. Leonelli, MD (Division of Nephrology, Dialysis and Transplant, Policlinico Hospital, Modena); D. Belotti, BiolSciD, E. Pogliani, MD (Division of Hematology and Transfusional Center, S. Gerardo Hospital, Monza, Milan); G. Masera, MD (Pediatric Department, S. Gerardo Hospital, Monza, Milan); M. R. Iannuzzi, MD (Division of Nephrology, A. Cardarelli Hospital, Naples); G. B. Capasso, MD, S. Scognamiglio, MD (Chair of Nephrology, Second University of Naples); G. Montini, MD, L. Murer, MD (Pediatric Division, Policlinico Hospital, Padova); A. Indovina, MD, R. Marcenò, MD (Division of Hematology, V. Cervello Hospital, Palermo); L. Amico, MD (Division of Nephrology and Dialysis, V. Cervello Hospital, Palermo); E. Trabassi, MD (Division of Nephrology and Dialysis, San Massimo Hospital, Penne, Pescara); G. Agnelli, MD (Division of Internal Medicine, University of Perugia); R. Caprioli, MD (Division of Nephrology and Dialysis, S. Chiara Hospital, Pisa); E. Nesti, MD (Division of Nephrology and Dialysis, S. Miniato Hospital, S. Miniato, Pisa); G. Garozzo, MD (Transfusional Center, M. P. Arezzo Hospital, Ragusa); E. Bresin, MD, E. Daina, MD, S. Gamba, Research Nurse (Clinical Research Center for Rare Diseases, Aldo e Cele Daccò, Ranica, Bergamo); M. Santostefano, MD (Division of Nephrology and Dialysis, Santa Maria delle Croci Hospital, Ravenna); G. Enia, MD, P. Finocchiaro, MD, C. Zoccali, MD (Division of Nephrology and Dialysis, Bianchi, Melacrino, Morelli Hospital, Reggio Calabria); V. Trapani Lombardo, MD (Division of Hematology, Bianchi, Melacrino, Morelli Hospital, Reggio Calabria); A. Amendola, MD, L. Dessanti, MD, F. Mandelli, MD, G. Meloni, MD (Department of Cellular Biotechnology and Hematology, La Sapienza University, Rome); A. De Feo, MD, M. Ferrannini, MD (Rome American Hospital); L. De Petris, MD, S. Rinaldi, MD, G. F. Rizzoni, MD (Division of Nephrology and Dialysis, Bambino Gesù Pediatric Hospital, Rome); T. Cicchetti, MD, G. Putortì, MD (Division of Nephrology and Dialysis, N. Giannettasio Hospital, Rossano Calabro, Cosenza); R. Paolini, MD (Medical Division, Rovigo Hospital, Rovigo); A. Pinto, MD (Division of Nephrology and Dialysis, S. G. di Dio e Ruggi d'Aragona Hospital, Salerno); A. Del Giudice, MD (Division of Nephrology, Casa Sollievo delle Sofferenza Hospital, S. Giovanni Rotondo, Foggia); P. R. Scalzulli, MD (Division of Hematology, Casa Sollievo delle Sofferenza Hospital, S. Giovanni Rotondo, Foggia); M. Sanna, MD (Division of Medical Pathology, Sassari Hospital); A. Amore, MD, G. Conti, MD, R. Coppo, MD, L. Peruzzi, MD (Division of Nephrology and Dialysis, Regina Margherita Pediatric Hospital, Turin); A. Khaled, MD (Division of Nephrology, S. Chiara Hospital, Trento); M. Pennesi, MD (Division of Pediatric Nephrology, Burlo Garofalo Pediatric Institut, Trieste); O. Amatruda, MD (Division of Nephrology, Fondazione Macchi Hospital, Varese); L. Tavecchia, MD (Division of Hematology, Borgo Roma Hospital, Verona).
Abroad. J. Ferraris, MD (Division of Nephrology, Hospital Italiano de Buenos Aires, Argentina); M. G. Caletti, MD, M. Adragua, MD (Juan P. Garrahan Hospital de Pediatria, Buenos Aires, Argentina); R. Wens, MD (Clinique de Nephrologie-Dialyse, CHU Brugmann, Brussels, Belgium); G. Filler, MD, K. Blyth, RN (Children's Hospital of Eastern Ontario, Ottawa, Canada); T. Ring, MD (Department of Nephrology, Aaolborg Hospital, Aaolborg, Denmark); C. Bührer, MD (Department of Neonatology, Charité Campus Virchow-Klinikum, Berlin, Germany); D. Müller, MD (Department of Pediatric Nephrology, Charité, Berlin, Germany); B. Hoppe, MD (University Children's Hospital, Cologne, Germany); C. V. Schnakenburg, MD (Department of Pediatrics, University Children's Hospital, Freiburg, Germany); D. Landau, MD (Division of Pediatric Nephrology, Soroka Medical Center, Beer-Sheba, Israel); I. Krause, MD (Dialysis Unit, Schneider Children's Medical Center, Petach-Teqva, Israel); R. Rahamimov, MD (Transplantation Department, Beilinson Medical Center, Petach-Teqva, Israel); P. Ponce, MD (Hospital Garcia de Orta, Almada, Portugal); J. Barbot, MD, M. Antunes, MD (Division of Hematology, Maria Pia Hospital, Porto, Portugal); M. S. Faria, MD (Depatment of Pediatric Nephrology, Maria Pia Hospital, Porto, Portugal); A. N. Lategann, MD (AMPATH Laboratories, Arcadia, Republic of South Africa); S. Al-Saadoun, MD, A. Manlangit, MD (Pediatric Nephrology, Riyadh Armed Forces Hospital, Riyadh, Saudi Arabia); A. Bock, MD (Nephrology, Kantonsspital Aargau, Switzerland); T. J. Neuhaus, MD, A. Schenk, MD (Nephrology Unit, University Children's Hospital, Zurich, Switzerland); A. Sharma, MD (Royal Liverpool and Broadgreen University Hospitals, Liverpool, United Kingdom); G. B. Haycock, MD (Pediatric Renal Unit, Guy's Hospital, London, United Kingdom); A. Katz, MD (Pediatric Nephrology, Children's Hospital, Birmingham, AL); V. R. Dharnidharka, MD (University of Florida, Division of Pediatric Nephrology, Gainesville); J. C. Lane, MD, C. B. Langman, MD (Division of Kidney Diseases, Children's Memorial Hospital, Chicago, IL); V. Kimonis, MD (Department of Pediatrics, Southern Illinois University School of Medicine, Springfield); L. Milner, MD (Division of Nephrology, Floating Hospital for Children, Boston, MA); C. E. Kashtan (Pediatrics, University of Minnesota Medical School, Minneapolis); L. Najera, MD, J. Steinke, MD (Pediatric Nephrology, Fairview University Medical Center, Minneapolis, MN); D. Milliner, MD (Mayo Clinic, Rochester, MN); B. Warady, MD (Children's Mercy Hospital, Kansas City, MO); L. Wrenshall, MD (Nebraska Medical Center, Omaha); K. Lieberman, MD (Pediatric Nephrology, Hackensack University Medical Center, NJ); R. Wallerstein, MD (Genetics Department, Hackensack University Medical Center, NJ); J. Listman, MD (State University of New York Upstate Medical University, Syracuse); S. B. Conley, MD (Department of Nephrology, St Christopher's Hospital for Children, Philadelphia, PA); R. Raafat, MD (Pediatric Nephrology, University of Texas Health Science Center, San Antonio); J. Gitomer, MD (Department of Nephrology, Marshfield Clinic, WI).
Laboratory analysis
F. Gaspari, ChemD (Clinical Research Center for Rare Diseases, Aldo e Cele Daccò, Ranica, Bergamo, Italy); C. Ottomano, MD, A. Vernocchi, MD (Division of Laboratory Analysis, Azienda Ospedaliera, Ospedali Riuniti di Bergamo, Italy).
Biochemical studies
P. Bettinaglio, BiolSciD, S. Bucchioni, BiolSciD, J. Caprioli, BiolSciD, F. Castelletti, BiotechD, D. Cugini, BiolSciD, G. Pianetti, Chemist (Clinical Research Center for Rare Diseases, Aldo e Cele Daccò, Ranica, Bergamo, Italy); C. Capoferri, Chem Pharm D, M. Galbusera, BiolSciD, S. Gastoldi, Chemist, D. Macconi, BiolSciD (M. Negri Institute for Pharmacological Research, Bergamo, Italy); P. F. Zipfel, MD (Hans Knoell Institute for Natural Products Research, Jena, Germany).
Statistical analysis
A. Perna, StatSciD (Clinical Research Center for Rare Diseases, Aldo e Cele Daccò, Ranica, Italy).
Prepublished online as Blood First Edition Paper, April 18, 2006; DOI 10.1182/blood-2005-10-007252.
A complete list of the members of the International Registry of Recurrent and Familial HUS/TTP appears in “Appendix.”
Supported by grants from “Comitato 30 ore per la vita,” from Telethon project GGP02162, from Associazione Ricerca Trapianto (ART), from Istituto Superiore di Sanità, and from the Foundation for Children with Atypical HUS along with the Nando Peretti Foundation. S. Brioschi, F.P., and S. Bucchioni received a fellowship from Fondazione Aiuto Ricerca Malattie Rare (ARMR). F.C., L.C., and G.M. are recipients of fellowships from ART. G.P. is a recipient of a fellowship from Association “Amitié Sans Frontiers.” D.K. is funded by the National Kidney Research Foundation and the Peel Medical Research Trust. C.J.F. is funded by National Institutes of Health (NIH) grant T32 AR07279. M.K.L. and J.P.A. are funded by NIH grant R01 AI37618.
J.C. and M.N. contributed to the conception and design of the study, analysis, and interpretation of the data and preparation of the manuscript; J.C. was also responsible for DHPLC analysis of IF; S. Brioschi performed research on MCP ; F.C. performed research on CFH ; P.B. contributed to experimental work on MCP, analysis of data, and writing of the paper; E.B. collected and analyzed clinical data and coordinated biologic sample collection and managing; F.P. participated in biochemical and genetic studies on complement and MCP ; G.P., S. Bucchioni, and G.M. performed DNA sequencing; L.C. and G.P. performed MCP FACS analysis; S.G. collected and managed biologic samples; C.M. participated on MCP and IF screening; C.J.F. and M.K.L. prepared and analyzed the MCP cell lines; D.K. performed the analysis of the splice site mutation; J.P.A. directed the studies on the transfected MCP cell lines and the splice site mutation and reviewed the final report; and G.R. participated in discussion and interpretation of the data and reviewed the final report.
J.C. and M.N. contributed equally to the work presented in this paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are deeply indebted to Federica Casiraghi for her invaluable contribution in setting up the ELISA for IF and to Dr Dario Cattaneo for his precious help with DHPLC. We thank Dr Borislav Dimitrov for statistical advice and Monica Lena for the management of patients and biologic samples.

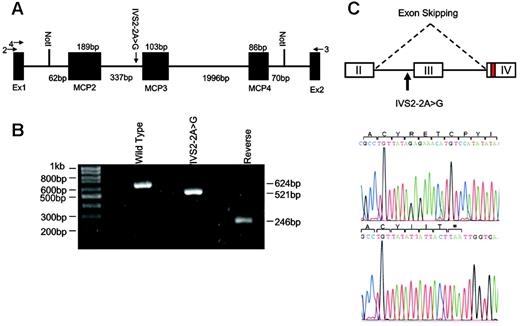
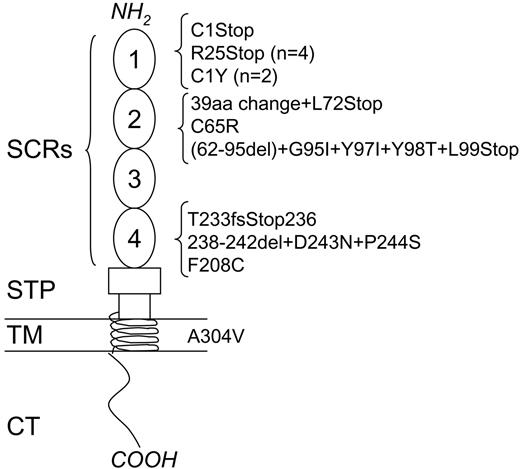
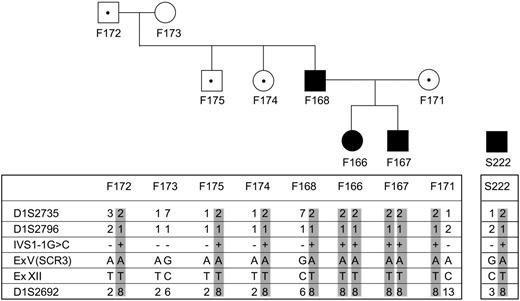
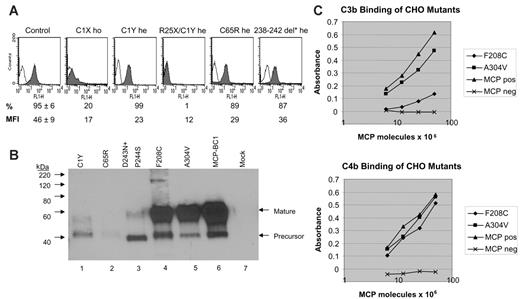
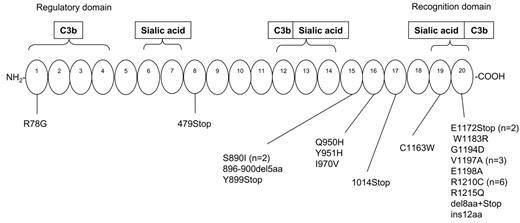
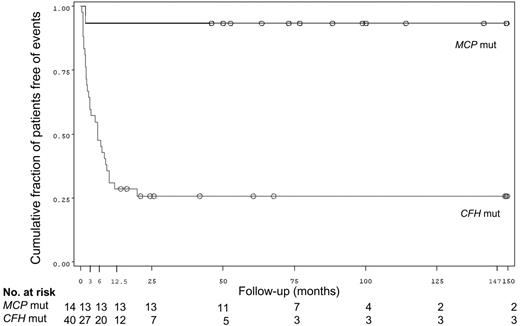
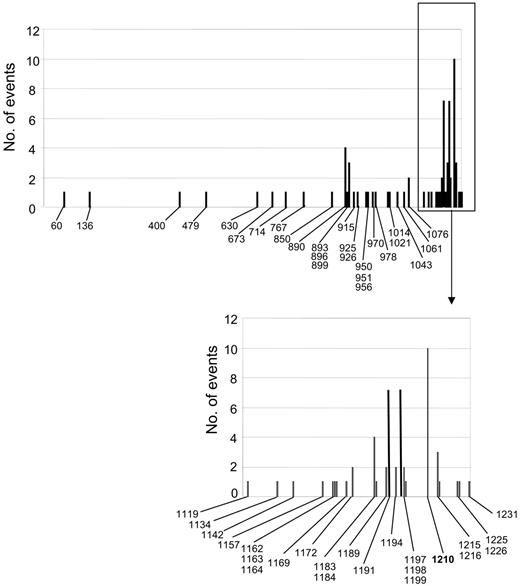
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal